Abstract
Background
Pancreatic cancer presents extremely poor prognosis due to the difficulty of early diagnosis, low resection rate, and high rates of recurrence and metastasis. Immune checkpoint blockades have been widely used in many cancer types but showed limited efficacy in pancreatic cancer. The current study aimed to evaluate the landscape of tumor microenvironment (TME) of pancreatic cancer and identify the potential markers of prognosis and immunotherapy efficacy which might contribute to improve the targeted therapy strategy and efficacy in pancreatic cancer in the context of predictive, preventive, and personalized medicine (PPPM).
Methods
In the current study, a total of 382 pancreatic samples from the datasets of Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) were selected. LM22 gene signature matrix was applied to quantify the fraction of immune cells based on “CIBERSORT” algorithm. Weighted Gene Co-expression Network Analysis (WGCNA) and Molecular Complex Detection (MCODE) algorithm was applied to confirm the hub-network of immune-resistance phenotype. A nomogram model based on COX and Logistic regression was constructed to evaluate the prognostic value and the predictive value of hub-gene in immune-response. The role of transmembrane protein 92 (TMEM92) in regulating cell proliferation was evaluated by MTS assay. Western blot and Real-time PCR were applied to assess the biological effects of PD-L1 inhibition by TMEM92. Moreover, the effect of TMEM92 in immunotherapy was evaluated with PBMC co-culture and by MTS assay.
Results
Two tumor-infiltrating immune cell (TIIC) phenotypes were identified and a weighted gene co-expression network was constructed to confirm the 167 gene signatures correlated with immune-resistance TIIC subtype. TMEM92 was further identified as a core gene of 167 gene signature network based on MCODE algorithm. High TMEM92 expression was significantly correlated with unfavorable prognosis, characterizing by immune resistance. A nomogram model and external validation confirmed that TMEM92 was an independent prognostic factor in pancreatic cancer. An elevated tumor mutation burden (TMB), mostly is consistent with commonly mutations of KRAS and TP53, was found in the high TMEM92 group. The predictive role of TMEM92 in immunotherapeutic response was also confirmed by IMvigor210 datasets. In addition, the specific biological roles of TMEM92 in cancer was explored in vitro. The results showed that abnormal overexpression of TMEM92 was significantly associated with the poor survival rate of pancreatic cancer. Moreover, we demonstrated that TMEM92 inhibit tumour immune responses of the anti-PD-1 antibody with PBMC co-culture.
Conclusion
The current study explored for the first time the immune-resistance phenotype of pancreatic cancer and identified TMEM92 as an innovative marker in predicting clinical outcomes and immunotherapeutic efficacy. These findings not only help to recognize high-risk and immune-resistance population which could be supplied targeted prevention, but also provide personalized medical services by intervening TMEM92 function to improve the prognosis of pancreatic cancer. In addition, the biological role of TMEM92 might reveal the potential molecular mechanisms of pancreatic cancer and lead to a novel sight for development of a PPPM approach for pancreatic cancer management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Challenges of pancreatic cancer management
Pancreatic cancer is one of lethal malignancies with a 5-year overall survival rate of approximately 9% [1]. The incidence is rising at a rate of 0.5 to 1.0% per year, and pancreatic cancer is projected to become the second-leading cause of cancer death by 2030 [2]. Pancreatic cancer presents extremely poor prognosis which due to difficulty of early diagnosis, low resection rate, and high rates of recurrence and metastasis. Most patients were diagnosed at an advanced stage and few of them could benefit from gemcitabine-based combination therapy which only slightly improved the 5-year survival rate. Thus, it is urgent to explore novel valuable markers for targeted treatment and prognosis monitoring of pancreatic cancer.
The role of tumor microenvironment in context of predictive, preventive, and personalized medicine
Since the concept of predictive, preventive, and personalized medicine (PPPM) has been proposed in malignant tumours, the prevention, drug resistance monitoring and genomic-guided therapy have made great improvement in cancers treatment Cancer screening method and therapeutic target prediction based on genomic sequencing provided reliable evidence on early diagnosis and personalized medical treatment [3]. Human epidermal growth factor receptor 2 (Her-2) and Kirsten rat sarcoma viral oncogene homologue gene (KRAS) have been widely used for predictive diagnosis and treatment response in gastric and colorectal cancer. Unlike other gastrointestinal tumor targeted therapies for pancreatic cancer are limited due to the lack of effective targets. In recent years, immunotherapy against tumor by activating the immune system has achieved great success. The programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) pathway is one of the most studied immune checkpoint pathways, which can inactivate the immune response in the tumor microenvironment [4]. Immune checkpoint inhibitors (ICIs) such as anti-PD1/PD-L1 antibodies have shown clinical effects in the treatment of several malignant tumors [5,6,7,8,9]; however, ICIs are generally ineffective in pancreatic cancer [10]. The main reason for this phenomenon was that there are many interstitium in pancreatic cancer which accounts for more than 90% of the total tumor volume [11] and the effector T cells are poorly infiltrated in the tumor microenvironment (TME) of pancreatic cancer [12]. On the other hand, PD-L1 expressed on the surface of malignant tumor cells can directly inhibit the antitumor activity of CD8 + T cells [13]. Therefore, the exploration of the TME and the method of regulating the expression level of PD-L1 in pancreatic cancer is extremely urgent. In addition, the classification and immune phenotype annotation of pancreatic cancer based on TME have rarely investigated, which is critical for screening potential populations and developing preventive, personalized immunotherapy strategies and might improve the clinical application value of PPPM in pancreatic cancer.
Transmembrane (TMEM) family is a type of protein which spans the entire width of lipid bilayers [14]. Recently, increasing studies have focused on the role of TMEMs in cancer [15,16,17]. In hepatocellular carcinoma, TMEM106c is associated with malignant features and poor prognosis, while TMEM205 is correlated with immune cell infiltrates [18, 19]. TMEM92 can act as an adaptor for E3 ubiquitin ligase to support the degradation of β-catenin and E-cadherin. MiR-23a and miR-24 overexpression leads to gene silencing of TMEM92 and results in mesothelial cell integration of pancreatic cancer [20]. So far, whether TMEM92 is involved in the immunosuppressive mechanism of pancreatic cancer is unclear.
Working hypothesis in the framework of personalization of medical services in pancreatic cancer
We hypothesize that the TME-based immune phenotype annotation is highly correlated with clinical pathological parameters of pancreatic cancer and immune-related gene signature is propitious to advantage group identification, risk prediction and personalized management of pancreatic cancer. We expect that the immune characteristic signature could provide targeted prevention and personalized medical services to improve the survival outcome and to reverse the immune-resistance characters of pancreatic cancer. Collectively, the results of current study will be guiding the personalized treatment of pancreatic cancer in the context of PPPM.
Methods
Data collection of pancreatic cancer
The data of GSE62452, GSE85916, and GSE57495 were downloaded from the Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/) and TCGA-PAAD were derived from UCSC browser (https://xenabrowser.net/), respectively. The FPKM value of TCGA-PAAD datasets was converted into TPMs which is consistent with other microarrays from GEO datasets (GSE62452, GSE85916, GSE57495) as a method of data normalization. The “ComBat” algorithm was conducted to decrease the probability of batch effects from non-biological technical biases between the different datasets. A total of 382 pancreatic cancer patients with both survival parameters and gene expression profile were selected for further analysis. The data of somatic mutation corresponding to the TCGA-PAAD datasets were downloaded from TCGA portal (https://portal.gdc.cancer.gov). “Maftool” package in R was used to determine driver genes in TCGA-PAAD and the somatic variations in driver genes were estimated for different expression of hub-gene. IMvigor210 dataset was obtained from http://research-pub.gene.com/IMvigor210CoreBiologies, and 298 urothelial cancer patients with both gene expression data and clinical parameters were chosen to further analysis.
Clustering of tumor-infiltrating immune cells
LM22 gene signature matrix was applied to quantify the fraction of immune cells in pancreatic cancer base on “CIBERSORT” algorithm in R project. Stromal and immune content in pancreatic cancer sample were measured by ESTIMATE algorithm. Samples with distinct TIIC patterns were clustered by hierarchical agglomerative clustering. The package of “Consensu Cluster Plus” in R was used to implement the unsupervised clustering method based on Ward’s linkage and Euclidean [21] and the procedure was repeated 1000 times to improve the stability of classification.
Construction of co-expression network and protein–protein network.
Weighted gene co-expression network analysis (WGCNA) algorithm was conducted to construct the co-expression network as described in previous study [22]. Pearson’s test was applied to evaluate the correlation between module epigengenes and immune activities in pancreatic cancer. Protein–protein interaction network of immune-related module was conducted by Cytoscape (v3.8.2). The gene module which negatively correlated with TIIC patterns was selected and defined as immune suppressive gene signature in pancreatic cancer. Furthermore, the hub-gene was identified from gene modules based on topological features and molecular complex detection algorithm (MCODE).
Enrichment analysis
The Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways participating in the potential biological function of immune suppressive gene signature were enriched by “Cluster Profiler” package with a criteria FDR < 0.05. Gene-set enrichment analysis (GSEA) was used to further investigated the hug-gene functions in pancreatic cancer. The P values of GSEA were based on 10,000 permutations and followed by adjusted for multiple testing to control the FDR.
Statistical analyses
Wilcoxon rank-sum test and unpaired Student t tests were applied to compare two groups with or without normally distributed variables respectively. For more than two groups’ comparison, Kruskal–Wallis tests was performed [23]. Survival curves were constructed by the Kaplan–Meier (KM) method and compared with the log-rank test. X-tile software was applied to establish the optimal cutoff value [24]. The correlation coefficient was calculated by Spearman analysis. The correlation between TIIC score subtypes and somatic mutation was evaluated by chi-square test. A nomogram model was constructed by rms package according to the multivariate COX regression model and Logistic regression model. The predicting accuracy is evaluated by C-index. Calibration curve was act as the internal validation of nomogram. All statistical analyses were performed by R project (https://www.r-project.org/) or SPSS software (version 26.0). Two-sided P < 0.05 was treated as statistically significant.
Cell validation
Western blot
Western blotting was performed as our previously described [25]. The primary antibodies used were as follows: anti-TMEM92 (HPA063009, MedChemExpress Corporation, NJ, USA) and anti-PD-L1 (Cell Signalling Technology, Beverly, MA, USA), GAPDH (Proteintech, China). The result was analyzed by NIH Image J software.
Cell lines and cell viability
The human pancreatic adenocarcinoma cell lines Capan-2,SW1990 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and maintained in RPMI 1640 medium that contained 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 mg/ml) under 5% CO2 at 37 ℃. Cell proliferation was measured by the MTS assay (Promega, USA).
Transient transfection
TMEM92 overexpression plasmid and the empty vector control were designed and provided by Obio Technology Corp., Ltd. (Shanghai, China). In brief, Capan-2, SW1990 cells with a cell density of 1.0 × 105 cells/well were cultured in a six-well plate on the day before transfection. The cells were transfected with 1.5 μg TMEM92 overexpression plasmid or the empty vector control respectively according to the manufacturer’s instructions on the following day. Overexpression efficiency of TMEM92 was evaluated by quantitative reverse transcription real-time polymerase chain reaction (qRT-PCR) and Western blotting.
RNA extraction and qRT-PCR
Total RNA extracted as described previously [26]. The conditions for real-time PCR included initial activation were 30 s at 95 °C, followed by 45 cycles at 95 °C for 5 s, and 58 °C for 25 s (Applied Biosystems. 7500 Real-Time PCR Systems, Thermo fisher, IL, USA). Transcripts of 18 s was used as the internal control. The PCR primers used were as follows:
-
TMEM92 forward, 5′-GCAGCCAAATGTGGTCTCATCC-3′;
-
TMEM92 reverse, 5′-GCAAAAGACGGACAGGATGACC-3′;
-
PD-L1 forward: 5′-TTTCAATGTGACCAGCAC-3′;
-
PD-L1 reverse: 5′-GGCATAATAAGATGGCTC-3′;
-
18S forward: 5′-CCCGGGGAGGTAGTGACGAAAAAT-3′;
-
18S reverse: 5′-CGCCCGCCCGCTCCCAAGAT-3′.
PBMC
Peripheral blood mononuclear cell (PBMC) samples from healthy donors were obtained from the First Hospital of China Medical University (CMU). All procedures were approved by the Research Ethics Committee of the First Hospital of CMU, and written informed consent was obtained from all health donors. Isolation of these PBMCs were isolated using a Ficoll density-based centrifugation [27]. The PBMC were used for co-cultures with transfected TMEM92 cell lines and control group.
Immune checkpoint blocking antibodies—in vitro
The PD-1 (nivolumab) blocking Abs were kindly provided by BMS in vitro experiments. Concentrations of the blocking Abs (10 g/mL) were based on those described in the literature [28].
Results
Landscape of tumor-infiltrating immune cells in pancreatic cancer
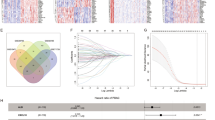
A total of 382 pancreatic cancer samples from the combined-cohort (TCGA-PAAD, GSE57495, GSE62452, GSE85916) with both clinical parameters and expression profile was collected (Table S1). The CIBERSORT was conducted to evaluate the activity of immune cells in tumor tissue (Table S2), and the immune scores and tumor purity was calculated by employing the ESTIMATE algorithm. Unsupervised hierarchical clustering of total samples with matched tumor-infiltrating immune cell (TIIC) profiles was performed using the “Consesus Cluster Plus” package in R project and classified the pancreatic cancer patients was classified into two independent TIIC subtypes (Fig. 1A, Fig. S1A-D ) and the landscape of immune cell interaction was visualized by correlation coefficient heatmap (Fig. 1B). The survival outcomes were significant difference in two TIIC subtypes (log-rank test, p < 0.001; Fig. 1C). To further investigate the biological difference among these subtypes which leading to distinct prognostic outcomes, we evaluated the composition of immune cells of tumor microenvironment in TIIC clusters. In the TIIC cluster A which associated with better overall survival, CD8 T cells, naive B cells, activated memory CD4 T cells, Monocytes, M1 macrophages and Resting mast cells were highly infiltrated. In contrast, in TIIC cluster B which associated with poor overall survival, resting memory CD4 T cells, regulatory T cells, active NK cells, M0 and M2 macrophages and neutrophils exhibited high density. In addition, TIIC cluster A showed significant higher immune and stromal score (Kruskal–Wallis test: P < 0.001, P < 0.01, respectively) (Fig. 1D). Thus, TIIC cluster A was considered as the type of immune-sensitive cluster whereas TIIC cluster B was considered as the immune-resistance subtype.
The landscape of tumor-infiltrating immune cells in pancreatic cancer. A Unsupervised hierarchical clustering of tumor-infiltrating immune cells for total 382 pancreatic patients. B The correlation coefficient heatmap of immune cell interactions. C Overall survival of immune TIIC Clusters by Kaplan–Meier analysis (log-rank test p < 0.001). D Comparison of the fraction of immune cell infiltration in different TIIC clusters (Kruskal–Wallis test). *p < 0.05; **p < 0.01; ***p < 0.001
Identification of gene subtypes and functional annotation
In order to investigate the underlying biological features of different immune subtypes, WGCNA was conducted to identify the transcriptome changes among distinct TIIC subtypes. The co-expression network was close to scale-free distribution by choosing the threshold value 8 (Fig. 2A). A total of 12 modules were identified which shown that the green module was most positively relevant to TIIC cluster B and unfavorable prognosis (Fig. 2B, Fig. S1E–G). To confirm the gene signature correlated with immune-resistance TIIC cluster B, 167 epigengenes included in the green module were further investigated (Table S3). The unsupervised hierarchical clustering of 167 epigengenes was performed and classified the total population into two gene clusters which termed as gene cluster A and B. As presented in the heatmap, the gene cluster A was highly associated with TIIC cluster A. In contrast, TIIC cluster B were associated with gene cluster B (Fig. 2C). The prognostic value of TIIC gene clusters was assessed by survival analysis. We noticed that the favorable survival outcome was found in gene cluster A when compared to gene cluster B (log-rank test, p < 0.001; Fig. 2D) which is consistent with the immune cluster profile. Moreover, CD8 T cells, naive B cells, monocytes and resting mast cells were increasingly infiltrated in gene cluster A. The gene cluster B exhibited higher infiltration of regulatory T cells, M0 and M2 macrophages which act as hazard factors in predicting tumor prognosis and immune response (Fig. 2E) [29, 30]. These results demonstrated that 167 gene signatures which upregulated in gene cluster B might accurately predict TIIC subtypes and immune-resistance activities. Furthermore, GO and KEGG enrichment analysis of 167 gene signatures was performed by Cluster Profiler packages in R project as shown in Fig. 2F. PI3K-AKT signaling pathway, focal adhesion, and ECM-receptor interaction were significantly enriched. To identify the vital genes for immune-resistance phenotype of pancreatic cancer, a protein–protein network (PPI) of 167 epigengenes was constructed. The hub-gene network which consist of 42 nodes and 720 edges was determined by using three topological parameters (degree, betweenness, and closeness) and MCODE algorithm (Fig. 2G) (Table S4). As the seed gene of 42 hub-genes network, TMEM92 was selected to further investigation.
Identification of gene subtypes and functional annotation. A Identification of the best soft-thresholding power value. B Module-trait associations were evaluated by correlations between module eigengene and clinical traits. (C) Unsupervised hierarchical clustering of 167 epigengenes from green module to separated patients into two groups: gene clusters A and B. D Overall survival of different gene clusters by Kaplan–Meier analysis (log-rank test p < 0.001). E Comparison of the proportion of immune cell infiltration in different gene clusters (Kruskal–Wallis test). *p < 0.05; **p < 0.01; ***p < 0.001. F GO and KEGG enrichment analysis of 167 gene signatures. G Hub-network was selected from PPI network base on topological features and MCODE which consist of 42 nodes and 720 edges
Transcriptome traits and clinical characteristics of TMEM92
Firstly, the association between gene cluster and TMEM92 expression level were represented in Fig. 3A. In order to evaluate the immune activity and tolerance characterizing of TMEM92, we analyzed the expression of selected immune-related gene signatures in the 382 pancreatic cancer samples. CD274, HAVCR2, CTLA4, PDCD1, LAG3, and IDO1 were considered as immune-checkpoint-related signatures; while GZMB, CD8A, CXCL9, CXCL10, PRF1, TNF, TBX2, IFNG, and GZMA were considered as immune-activity-relevant signatures [31, 32].The result indicated that the majority of immune-related genes were significantly downregulated in high TMEM92 expression group when compared to low expression group (Fig. 3B). Secondly, the prognostic value of TMEM92 was evaluated by Kaplan–Meier plotter. In total population, the patients with high TMEM92 expression exhibited unfavorable survival outcomes than those with low expression (log-rank test, p < 0.001, Fig. 3C). To confirm the predictive value of TMEM92, a nomogram model of overall survival based on multivariate analysis was performed (Fig. 3D). The C-index of overall survival prediction was 0.62 (95%CI, 0.56–0.68). The survival probability at 1 year and 3 years were represented by the calibration curve which demonstrated the consistency between nomogram prediction and objective observation (Fig. S1H-I). In addition, we further investigated the effect of TMEM92in patients with different treatment response in TCGA cohort. Interestingly, patients with CR/PR response and low TMEM92 expression had the best OS. In contrast, the worst prognosis was found in patients with high TMEM92 expression and poor response to primary treatment (log-rank test, p = 0.001, Fig. 3E).The prognostic value of TMEM92 was further validated by each independent dataset (TCGA cohort: log-rank test, p = 0.002; GSE57495:log-rank test, p = 0.037; GSE62452:log-rank test, p = 0.01; GSE85916: log-rank test, p = 0.308, Fig. 3F). Besides, GSEA analysis revealed that the MTOR, Neurotrophin, Adipocytokine and T cell receptor signaling pathways were highly enriched in low TMEM92 expression group, and base Excision repair, cell cycle, pentose phosphate, and P53 signaling pathways were enriched in high TMEM92 group (Fig. 3G). These data indicated that TMEM92 closely related with immune-resistance phenotype and unfavorable survival in pancreatic cancer.
Transcriptome traits and clinical characteristics of TIIC score. A Alluvial diagram of gene clusters in groups with different TMEM92 expression level and survival status. B The expression level of immune-activation-relevant genes (GZMB, CD8A, CXCL9, CXCL10, PRF1, TNF, TBX2, IFNG, and GZMA) and immune-checkpoint-relevant genes (CD274, HAVCR2, CTLA4, PDCD1, LAG3, and IDO1) in different TMEM92 expression groups. C Overall survival of different TMEM92 expression in total cohort by Kaplan–Meier method (log-rank test p < 0.001). D The probability of survival at 1, 3, and 5 years was determined by correspondence between the total points and survival axis. E Kaplan–Meier analysis for patients in the TCGA-PAAD cohort stratified by objective response (OR) and TMEM92 expression (log-rank test p = 0.001). F Overall survival of different TMEM92 expression in TCGA cohort (log-rank test, p = 0.002); GSE57495 (log-rank test, p = 0.037); GSE62452 (log-rank test, p = 0.01); GSE85916 (log-rank test, p = 0.308). G Enrichment plots showing the MTOR, neurotrophin, adipocytokine, and T cell receptor signaling pathways were highly enriched in low TMEM92 expression group, and base excision repair, cell cycle, pentose phosphate, and P53 signaling pathways were enriched in high TMEM92 group
The correlation of TMB and TMEM92
Tumor mutation burden (TMB) was confirmed as the predictor of immunotherapeutic efficacy in many cancer types and used for screening benefit population of immunotherapy [31, 33, 34]. We explored the correlation of TMEM92 and TMB in TCGA cohort to clarify the genetic changes in different expression groups. The results indicated that patients with high TMEM92 expression presented higher TMB than patients with low TMEM92 expression (Wilcoxon test p = 4.2e − 3; Fig. 4A) and TMEM92 was positively correlated with the TMB (Spearman correlation: R = 0.31, p = 1.6e − 4; Fig. 4B). Then, we evaluated the prognostic impact of TMB and TMEM92. Stratified survival analysis demonstrated that patients hold high TMEM92 expression levels and high TMB levels had the worst clinical outcomes than other subgroups (log-rank test, p = 0.002; Fig. 4C). We next investigated the distributions of somatic alterations between the high and low TMEM92 expression groups. The top 20 genes which have the highest alteration frequency were selected by maftools approach [35]. Among these driver genes, the alteration frequency of KRAS and TP53 were significantly higher in the high TMEM92 group (chi-square test: p < 0.001, p < 0.05, respectively). Previous researches have demonstrated that the mutant of KRAS, TP53, CDKN2A, and SMAD4 were the most commonly mutated genes detected in pancreatic cancer patients [36] which consists with our findings in both high and low TMEM92 expression groups (Fig. 4D, E). Besides, KRAS and TP53 mutations constitute the main part of all sample mutations in TCGA cohort, and a low prevalence for targetable mutations was found in most pancreatic cancers which might be a critical reason for less efficacy of immunotherapy [37]. Therefore, these data demonstrated that the TMEM92 might treated as an independent predictive factor in therapeutic response than TMB in pancreatic cancer.
The correlation of TMB and TMEM92. A The difference of TMB between different TMEM92 expression groups. Wilcoxon test, p = 4.2e − 3. B The correlation of TMEM92 and mutation load in TCGA-PAAD dataset. Spearman R = 0.31, p = 1.6e − 4. C Kaplan–Meier analysis for patients of TCGA-PAAD cohort stratified by TMB and TMEM92 expression (log-rank test p = 0.002). D The oncoPrint was constructed based on low TMEM92 expression level. E The oncoPrint was constructed based on high TMEM92 expression level
The predictive value of TMEM92 in immunotherapy
In order to confirm the value of the TMEM92 in predicting the response of immunotherapy, the patients in IMvigor210 cohort was assigned to high and low TMEM92 expression groups. In the IMvigor210 cohort, patients with high TMEM92 level exhibited significantly worse OS than those with low expression level (log-rank test, p < 0.001; Fig. 5A). Notably, the patients with objective response in immunotherapy were mostly found in the lowTMEM92 expression group (chi-square test, p = 0.0017; Fig. 5B) which also presented higher objective response rate compare to high expression group (two-sided Fisher exact test, p = 0.002; Fig. 5C). In addition, multivariate logistic regression analysis was performed and a nomogram of TMEM92 in predicting immune-resistance response was constructed as shown in Fig. 5D. The C-index of the nomogram was 0.62 which represented the model was well calibrated (Fig. 5E). Furthermore, the area under the ROC curve (AUC) was 0.63 (95% confidence interval CI: 0.55–0.70, p = 0.002, Fig. 5F). Taken together, the results demonstrated that the TMEM92 act as an immune-resistance factors in predicting immunotherapeutic response.
The predictive value of TMEM92 in immunotherapy. A Survival analysis of patients with different TMEM92 expression in the IMvigor210 cohort (log-rank test p < 0.001). B The TMEM92 expression level in groups with different clinical response status to immunotherapy in the IMvigor210 cohort. Wilcoxon test, p = 0.0017. C Rate of clinical response (CR/PR, SD/PD) to immunotherapy in different TMEM92 groups in the IMvigor210 cohort. D Logistic nomogram of TMEM92 in predicting immune-resistance response. E The calibration plots for the nomogram in the IMvigor210 cohort. F Receiver operating characteristic curve for TMEM92 expression level and immune-resistance response
TMEM92 promoted tumor growth in pancreatic cancer and increased the efficacy of immune checkpoint blockade therapy
The relationship between the expression level of TMEM92 in pancreatic cancer was further evaluated. To understand the biological functions of TMEM92 in pancreatic cancer cells, TMEM92 protein expression was measured in four pancreatic cancer cell lines (Fig. 6A). SW1990 and Capan-2 cells were chosen for the gain of function experiments due to the lowest expression levels of TMEM92 in these pancreatic cancer cell lines. TMEM92 was overexpressed in SW1990 and Capan-2 cells lines using TMEM92 overexpression plasmid and overexpression efficiency was tested by qRT-PCR and Western blotting assays (Fig. 6B, C).To investigate whether TMEM92 could promote pancreatic cancer proliferation. MTS assays showed that TMEM92 increase the growth capability of pancreatic cancer cells (Fig. 6D). As TMEM92 is critical for immune-resistance phenotype in pancreatic cancer, we detected the relationship between TMEM92 and PD-L1. We found that the increasing expression of TMEM92 can down-regulated PD-L1 both at pre- and post- transcriptional levels in pancreatic cancer cells (Fig. 6E, F). For the efficacy of immune checkpoint blockade therapy of TMEM92, OE-NC, or OE-TMEM92 SW1990 cells were placed in co-cultures with PBMCs from healthy donors after 72 h of treatment with PD-1 blocking Abs. We found that TMEM92 decreased the efficacy of immune checkpoint blockade therapy (Fig. 6G).
TMEM92 promoted tumor growth in pancreatic cancer and decreased the efficacy of immune checkpoint blockade therapy. A Protein expression ofTMEM92 in four pancreatic cancer cell lines by Western blotting. B, C SW1990 and Capan-2 cells were infected with TMEM92 overexpression plasmid or the empty vector control. TMEM92 expression levels were detected by qRT-PCR (B) and Western blotting analyses (C). D SW1990 and Capan-2 cells were transfected with pcDNA3.1 vector or TMEM92 plasmid. MTS assay was performed 24, 48, 72, 96 h after transfection. E, F SW1990 and Capan-2 cells infected with pcDNA3.1 or TMEM92 plasmids were harvested after 48 h for RT-PCR analysis (E) and Western blotting analysis (F). G Cell viability of PBMC from healthy donors co-culture with SW1990 infected with pcDNA3.1 or TMEM92 plasmids. TMEM92 overexppression groups were compared with pcDNA3.1 transfection group. Each bar represents the mean ± SD of three independent experiments. Statistical analyses were performed with one-way ANOVA followed by Tukey’s multiple comparison’s tests. Compared groups were shown in the figures. **p < 0.01
Discussion
PPPM concept in pancreatic cancer
Seeking predictive, preventive, and personalized medical strategies was the aim of modern cancer management. With the progress of cancer-screening and prevention technologies, the diagnosis and cure rates of cancer have been greatly improved. For advanced cancer, how to monitor the resistance to therapy and perform genotype-guided treatment were the primary goal of PPPM [3, 38]. Pancreatic cancer usually accompanied with poor prognosis due to the lack of predictive markers of early diagnosis and the limitation of response in standard treatment. With the development of immunotherapy, immune checkpoint inhibitors such as blockades targeting PD-1/PD-L1 or CTLA-4 have improved the therapeutic effects in various malignant tumor. However, ICIs exhibit no efficacy in pancreatic cancer which are regarded as immune-resistant tumors [39]. The reason for the immunosuppression character of pancreatic tumors is attributed mostly to the complexity of TME [40, 41]. Many research have reported that most of the enhanced immune cells in the TME of pancreatic cancer are immunosuppressive related cells, such as myelogenous suppressor cells (MDSC), M2 macrophages (M2), and regulatory T cells (Treg) [42, 43]. However, a small set of pancreatic patients still obtained clinical response from ICI treatment which suggest that individual difference exist throughout the diagnosis and treatment of pancreatic cancer. Thus, it is urgent to explore a novel predictive marker based on characterizing TME. By conducting the landscape of TME and classify the total population into immune-related subtypes, it is essential for clarifying the potential reason for immune-resistance characters of pancreatic cancer and providing basis for personalized treatment. These are consistent with the purpose of PPPM in pancreatic cancer.
Roles of tumor microenvironment and TMEM92 in pancreatic cancer
In this study, we analyzed the tumor-infiltrating immune cells in a combined cohort of 382 pancreatic samples and separated them into two independent immune clusters. The TIIC cluster with favorable OS was highly infiltrated by CD8 T cells, naive B cells, activated memory CD4 T cells, follicular helper T cells, and M1 macrophages. In contrast, the TIIC cluster B with unfavorable OS was highly infiltrated with regulatory T cells, M0 and M2 macrophages which is consistent with previous research [44]. However, except for infiltrating of immunosuppressive related cells, even patients with activated T cells also showed resistant to immunotherapy, indicting the necessity to reveal the immune-resistance phenotype in pancreatic cancer and the underlying mechanisms. Thus, we further focused on the molecular characterization of pancreatic cancer TME in regulating immune activity by combining the TIIC profiles and gene expression profiles to recognize specific subgroup of immune-resistance phenotype. Based on WGCNA and TIIC clusters, we identified 167 gene-signature which related to immune-resistance characters and unfavorable prognosis.
In order to seek the critical symbol of immune-resistance phenotype in pancreatic cancer, we constructed a PPI network of 167 gene-signatures. TMEM92, a member of TMEM family, was identified as the seed gene for further investigation. Many TMEMs are abnormally expressed in cancer. Some are related to the malignant behavior of tumors, while others are correlated with poor prognosis, which could be used as prognostic indicators. Recent research indicated that TMEM229a inhibits the progression of non-small cell lung cancer by blocking the ERK signaling pathway [45]. TMEM180 is associated with poor survival in stage III colorectal cancer [46]. In addition, TMEM92 could promote cell growth, invasion, and migration in breast cancer cells. Furthermore, depletion of TMEM92 could downregulated N-cadherin, Vimentin, and Snail levels which resulting in epithelial-mesenchymal transition (EMT) inactivated [47]. In the current study, survival analysis and a nomogram model revealed that TMEM92 is an independent prognostic factor of pancreatic cancer. External validation by an independent cohort further confirmed the prognostic value of TMEM92. In the meantime, genes involved in immune-checkpoint response and immune activity were significantly downregulated in the high TMEM92 expression group. We also found that TMEM92 is positively associated with TMB and neoantigen load in TCGA dataset. KRAS and TP53 were the most frequent mutant genes detected in the high TMEM92 group, in line with the previous research, e.g., KRAS mutated in almost 95% of pancreatic ductal adenocarcinoma (pancreatic cancer) and exhibited unfavorable prognosis [48]. Although TMB is correlated with immunotherapeutic response in many cancer types, whereas in pancreatic cancer, TMB was mostly contributed by KRAS and TP53 with the absence of neoantigen load [37]. There are no therapeutics which target driver mutations in pancreatic cancer that occur at > 20% prevalence [37]. Therefore, TMEM92 might act as a predictor of immune response independent from TMB in pancreatic cancer. We further analyzed the predictive value of TMEM92 for immunotherapy in IMvigor210 cohort which underwent anti-PD-1/PD-L1 agent treatment. Unfavorable OS and immunotherapeutic non-responders were both found in high TMEM92 expression group which indicated that patients with high TMEM92 might be resistant to checkpoint blockade therapy. However, the application of the IMvigor210 cohort which consists of 298 urothelial cancer samples as an external validation has to be intenerated with caution due to the limitations of the lack of datasets related to the response to immunotherapy in pancreatic cancer. Consequently, the role of TMEM92 in predicting immunotherapeutic efficacy should be further investigated in a pancreatic cancer cohort.
The exploration of biological function of TMEM92 in pancreatic cancer showed that silencing TMEM92 could notably suppress the viability capacities of pancreatic cancer cells. Considering there is no evidence of TMEMs in regulating tumor immune activities, we further investigated the potential mechanism of TMEM92 for its immune-resistance role in pancreatic cancer. The interaction between PD-L1 on malignant cells and PD-1 on T cells is the critical theoretical basis of applying ICIs; our results demonstrated that PD-L1 were selectively down-regulated after TMEM92 was overexpressed in SW1990 and Capan-2 cells. Meanwhile, TMEM92 decreased the tumor inhibition effect of the anti-PD-1 antibody in vitro with PBMC co-culture. These results suggested that TMEM92 may play a key role in regulating the antitumor efficiency of immune checkpoint-based therapy. However, one might realize that, the prognostic and predictive values of TMEM92 were confirmed only by online datasets and cell models, it is essential to be further validated by large-scale of prospective clinical researches of pancreatic cancer. Furthermore, the mechanism of TMEM92 in regulating immune activities should be further investigated in pancreatic cancer cells and tissue samples and to construct the inhibitors of TMEM92 to investigate the potential application value in targeted prevention in pancreatic cancer patients.
The expected impact of current study on PPPM of pancreatic cancer
How advanced patients could benefit from immunotherapy has been paid a lot of attention in the fields of pancreatic cancer treatment. Under the traditional reactive cancer treatment model, the exploration of novel immunotherapeutic strategies has not achieved the expected clinical benefits. Besides, in the absence of effective markers, application of immunotherapy may not only be ineffective, but also increase the side effects and economic burden on patients [38]. Therefore, the current study aims to screen a potential biomarker to predict the immune response and stratify high-risk groups in pancreatic cancer treatment, which will increase the efficiency of immunotherapy and reduce the cost of treatment. These findings improve the strategies of targeted prevention and personalized treatment in pancreatic cancer and promote the paradigm shift from reactive medical services to PPPM.
Conclusions and expert recommendations
Several researches demonstrated that the lack of immunogenicity of pancreatic cancer are leading to poor response for immunotherapy [49]. TME consists of malignant cells and tumor-infiltrating immune cells (TIICs) which have been confirmed to reflect the efficacy of immunotherapy. However, there has neither evidence to clarify the TME infiltration patterns in pancreatic cancer, nor the application value of TME in the context of PPPM. Thus, investigation of TME and potential mechanisms could benefit for reversing immune-resistance character and enhance immune-based treatment effects in pancreatic cancer. The current study investigated the TIIC landscape of pancreatic cancer and presented a comprehensive scene of immunomodulation response. The assessment of infiltrating patterns of TME supply novel perceptions on correlation between tumor and immune response. We suggest that the classification according to TIICs could be applied as predictive markers for immune response in pancreatic cancer patients. In addition, TME-based immune-related gene signature which is correlated with poor prognosis and resistance for immunotherapy was identified in this study.
By recognizing immune-resistance phenotype, we for the first time identified TMEM92 as the candidate marker in pancreatic cancer immunotherapeutic response. In addition, this current study revealed that overexpressed TMEM92 increases the survival rates in pancreatic cancer cells. Moreover, TMEM92 decreased tumor immune responses of the anti-PD-1 antibody in vitro with PBMC co-culture. These data confirmed that TMEM92 could be a new prognostic biomarker of pancreatic cancer and a novel predictive target for immune checkpoint blockade and personalized treatment. We recommend to promote the application of TIIC based stratification in combination with pathological parameters in pancreatic cancer and convert into clinical practice according to the concept of PPPM. For the further application of TMEM92 gene signature in the context of PPPM in pancreatic cancer management, we recommend the following:
Predictive diagnostics: Population stratification based on TME has been proved to be effective in many cancer types [50, 51]. For pancreatic cancer, the current study identified a gene-signature (TMEM92) according to TIICs classification which was correlated with immune-activities and unfavorable overall survival. It is a candidate marker in predicting patient outcomes and recognizing immune-resistance subpopulation which can be applied for predictive diagnosis of pancreatic cancer.
Targeted prevention: TMEM92 abnormal expression enhanced the ability of proliferation and reduced the response rate of immunotherapy in pancreatic cancer cells. The inhibition of TMEM92 might be the novel target for targeted therapy and improving immunotherapeutic efficacy. In contrast, low expression of TMEM92 might act as a basis of application of immunotherapy in pancreatic cancer. Thus, TMEM92 could help us to recognize high-risk groups in order to apply targeted prevention and further provide targeted therapeutic strategies by intervene TMEM92 function.
Personalization of medical care: The expression of TMEM92 reflects the immune and prognostic characters of pancreatic cancer patients. By utilizing the next-generation sequencing (NGS) with multi-omics strategies, patients are stratified into characteristic subgroups based on TMEM92 status which might improve the application of personalized management in pancreatic cancer treatment from the perspectives of PPPM.
Data availability
The datasets generated and/or analyzed during the current study are available in the TCGA, GEO, and IMvigor210 repository: TCGA (https://portal.gdc.cancer.gov); GEO (GSE57495: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57495; GSE62452:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62452; GSE85916: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85916); IMvigor210 (http://research-pub.gene.com/IMvigor210CoreBiologies).
Code availability
All software applications used are included in this article.
Abbreviations
- TME:
-
Tumor microenvironment
- TIIC:
-
Tumor-infiltrating immune cells
- WGCNA:
-
Weighted gene co-expression network analysis
- MCODE:
-
Molecular complex detection
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PD-1:
-
Programmed cell death protein 1
- PD-L1:
-
Programmed cell death ligand 1
- TMEM92:
-
Transmembrane protein 92
- TMB:
-
Tumor mutation burden
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics. CA Cancer J Clin. 2021;71(1):7–33.
Park W, Chawla A, O’Reilly EM. Pancreatic cancer: a review. JAMA. 2021;326(9):851–62.
Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, et al. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6(1):9.
Nagaraju GP, Malla RR, Basha R, Motofei IG. Contemporary clinical trials in pancreatic cancer immunotherapy targeting PD-1 and PD-L1. Semin Cancer Biol. 2021; S1044–579X(21)00270–4.
Uhlitz F, Zamarin D. Rejuvenating dysfunctional T cells in ovarian cancer: CD28 is the license to kill. Cancer Cell. 2021;S1535–6108(21):00562–6.
Zhang B, Vogelzang A, Miyajima M, Sugiura Y, Wu Y, Chamoto K, et al. B cell-derived GABA elicits IL-10+ macrophages to limit anti-tumour immunity. Nature. 2021;599(7885):471–6.
Au L, Hatipoglu E, Robert de Massy M, Litchfield K, Beattie G, Rowan A, et al. TRACERx Renal Consortium. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell. 2021;39(11):1497–518.
Nguyen NT, Huang K, Zeng H, Jing J, Wang R, Fang S, et al. Nano-optogenetic engineering of CAR T cells for precision immunotherapy with enhanced safety. Nat Nanotechnol. 2021;16(12):1424–34.
Scheiner B, Pomej K, Kirstein MM, Hucke F, Finkelmeier F, Waidmann O, et al. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy - development and validation of the CRAFITY score. J Hepatol. 2021;76(2):353–63.
Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–5.
Mazur PK, Siveke JT. Genetically engineered mouse models of pancreatic cancer: unravelling tumour biology and progressing translational oncology. Gut. 2012;61(10):1488–500.
Stromnes IM, Hulbert A, Pierce RH, Greenberg PD, Hingorani SR. T-cell localization, activation, and clonal expansion in human pancreatic ductal adenocarcinoma. Cancer Immunol Res. 2017;5(11):978–91.
Jiang ZB, Huang JM, Xie YJ, Zhang YZ, Chang C, Lai HL, et al. Evodiamine suppresses non-small cell lung cancer by elevating CD8+ T cells and downregulating the MUC1-C/PD-L1 axis. J Exp Clin Cancer Res. 2020;39(1):249.
Schmit K, Michiels C. TMEM proteins in cancer: a review. Front Pharmacol. 2018;9:1345.
Koteluk O, Bielicka A, Lemańska Ż, Jóźwiak K, Klawiter W, Mackiewicz A, et al. The landscape of transmembrane protein family members in head and neck cancers: their biological role and diagnostic utility. Cancers (Basel). 2021;13(19):4737.
Carroll J, He J, Ding S, Fearnley IM, Walker JE. TMEM70 and TMEM242 help to assemble the rotor ring of human ATP synthase and interact with assembly factors for complex I. Proc Natl Acad Sci U S A. 2021;118(13):e2100558118.
Luo S, Wang X, Bai M, Jiang W, Zhang Z, Chen Y, et al. The conserved autoimmune-disease risk gene TMEM39A regulates lysosome dynamics. Proc Natl Acad Sci U S A. 2021;118(6):e2011379118.
Duan J, Qian Y, Fu X, Chen M, Liu K, Liu H, et al. TMEM106C contributes to the malignant characteristics and poor prognosis of hepatocellular carcinoma. Aging (Albany NY). 2021;13(4):5585–606.
Rao J, Wu X, Zhou X, Deng R, Ma Y. TMEM205 is an independent prognostic factor and is associated with immune cell infiltrates in hepatocellular carcinoma. Front Genet. 2020;11:575776.
Listing H, Mardin WA, Wohlfromm S, Mees ST, Haier J. MiR-23a/-24-induced gene silencing results in mesothelial cell integration of pancreatic cancer. Br J Cancer. 2015;112(1):131–9.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Zhang S, Zang D, Cheng Y, Li Z, Yang B, Guo T. Identification of key gene and pathways for the prediction of peritoneal metastasis of gastric cancer by co-expression analysis. J Cancer. 2020;11(10):3041–51.
Hazra A, Gogtay N. Biostatistics series module 3: comparing groups: numerical variables. Indian J Dermatol. 2016;61(3):251–60.
Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–9.
Xu L, Zhang Y, Liu J, Qu J, Hu X, Zhang F, et al. TRAIL-activated EGFR by Cbl-b-regulated EGFR redistribution in lipid rafts antagonises TRAIL-induced apoptosis in gastric cancer cells. Eur J Cancer. 2012;48(17):3288–99.
Zang D, Zhang C, Li C, Fan Y, Li Z, Hou K, et al. LPPR4 promotes peritoneal metastasis via Sp1/integrin α/FAK signaling in gastric cancer. Am J Cancer Res. 2020;10(3):1026–44.
Marcq E, Audenaerde JRV, Waele J, Jacobs J, Loenhout JV, Cavents G, et al. Building a bridge between chemotherapy and immunotherapy in malignant pleural mesothelioma: investigating the effect of chemotherapy on immune checkpoint expression. Int J Mol Sci. 2019;20(17):4182.
Marcq E, Van Audenaerde JRM, De Waele J, Merlin C, Pauwels P, van Meerbeeck JP, et al. The search for an interesting partner to combine with PD-L1 blockade in mesothelioma: focus on Tim-3 and LAG-3. Cancers. 2021;13(2):282.
Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. CELL RES. 2017;27(1):109–18.
Jairath NK, Farha MW, Jairath R, Harms PW, Tsoi LC, Tejasvi T. Prognostic value of intratumoral lymphocyte-to-monocyte ratio and M0 macrophage enrichment in tumor immune microenvironment of melanoma. Melanoma Manag. 2020; 7(4):Mmt51.
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44.
Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–40.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY). 2015;348(6230):124–8.
Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56.
Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022.
Dreyer SB, Chang DK, Bailey P, Biankin AV. Pancreatic cancer genomes: implications for clinical management and therapeutic development. Clin Cancer Res. 2017;23(7):1638–46.
Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation - EPMA position paper 2016. EPMA J. 2016;7(1):23.
Foley K, Kim V, Jaffee E, Zheng L. Current progress in immunotherapy for pancreatic cancer. Cancer Lett. 2016;381(1):244–51.
Pico de Coaña Y, Choudhury A, Kiessling R. Checkpoint blockade for cancer therapy: revitalizing a suppressed immune system. Trends Mol Med. 2015;21(8):482–91.
Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and prevention of pancreatic cancer. Trends Cancer. 2018;4(6):418–28.
Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 2013;108(4):914–23.
Mahajan UM, Langhoff E, Goni E, Costello E, Greenhalf W, Halloran C, et al. Immune cell and stromal signature associated with progression-free survival of patients with resected pancreatic ductal adenocarcinoma. Gastroenterology. 2018;155(5):1625–39.
Stromnes IM, Hulbert A, Pierce RH, Greenberg PD, Hingorani SR. T-cell localization, activation, and clonal expansion in human pancreatic ductal adenocarcinoma. Cancer Immunol Res. 2017;5(11):978–91.
Zhang X, He Y, Jiang Y, Bao Y, Chen Q, Xie D, et al. TMEM229A suppresses non-small cell lung cancer progression via inactivating the ERK pathway. Oncol Rep. 2021;46(2):176.
Shiraishi T, Ikeda K, Tsukada Y, Nishizawa Y, Sasaki T, Ito M, et al. High expression of TMEM180, a novel tumour marker, is associated with poor survival in stage III colorectal cancer. BMC Cancer. 2021;21(1):302.
Lin MZ, Teng LL, Sun XL, Zhang LP, Chen F, Yu LJ. Transmembrane protein 92 performs a tumor-promoting function in breast carcinoma by contributing to the cell growth, invasion, migration and epithelial-mesenchymal transition. Tissue Cell. 2020;67:101415.
Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17(3):153–68.
Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49(1):178–93.
Zhang X, Shi M, Chen T, Zhang B. Characterization of the immune cell infiltration landscape in head and neck squamous cell carcinoma to aid immunotherapy. Mol Ther Nucleic Acids. 2020;22:298–309.
Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi M, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7(5):737–50.
Funding
This work was supported by Science and Technology Plan Project of Liaoning Province (2021-BS-102); National Natural Science Foundation of China (81801661).
Author information
Authors and Affiliations
Contributions
Guan Wang and Simeng Zhang designed the experiments. Guan Wang, Simeng Zhang, and Qiaoyun Chu wrote the manuscript. Mengzhu Lv and Ce Li performed the experiments. Simeng Zhang and Xing Wan performed the bioinformatics analysis. All authors contributed to the study design and data interpretation and have reviewed the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Data retrieved from the GEO and TCGA controlled-access database was collected using tumors from patients who provided informed consent based on guidelines laid out by the GEO and TCGA Ethics, Law and Policy Group. This study was approved by the Human Ethics Review Committee of China Medical University, and all procedures were conducted in accordance with ethical principles (protocol #: 2015PS63K).
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13167_2022_287_MOESM1_ESM.jpg
Supplementary file1 (JPG 3538 KB) Supplementary Figure 1. (A-D) Consensus matrixes of 382 pancreatic cancer samples for each k (k = 2–5) in classifying TIIC clusters, displaying the clustering stability using 1000 iterations of hierarchical clustering. (E) 2 pairs of gene modules were merged according to their similarity base on the threshold (Red line). (F) The genedendrogram was constructed by hierarchical clustering base on dissTOM of genes. (G) The correlation of epigengenes in green module with TIIC cluster trait. (H-I) Probability of survival at 1 (H) and 3 (I) years in the total cohort.
Rights and permissions
About this article
Cite this article
Zhang, S., Wan, X., Lv, M. et al. TMEM92 acts as an immune-resistance and prognostic marker in pancreatic cancer from the perspective of predictive, preventive, and personalized medicine. EPMA Journal 13, 519–534 (2022). https://doi.org/10.1007/s13167-022-00287-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-022-00287-0