Abstract
In this study, bacterial community compositions in seven different estuarine sediments of Poyang Lake were analyzed using 16S rRNA gene-targeted metagenomic approach. Remarkable differences in the bacterial diversity were observed in these different estuarine sediments. Le, Chang and Rao river samples exhibited the higher bacterial diversity; the Fu river sample showed the less diversity. Bacterial richness and diversity were positively regulated by sediment inorganic phosphorus, and nitrite nitrogen, total phosphorus and inorganic phosphorus were found to be important drivers for bacterial community compositions. Proteobacteria, Acidobacteria, Firmicutes, Chloroflexi, Bacteroidetes, Planctomycetes, Gemmatimonadetes, Actinobacteria, Nitrospirae, and Verrucomicrobia were the major components of sediment bacterial communities. Among them, Proteobacteria was the most dominant phylum, followed by Acidobacteria and Firmicutes. Our study gives a comprehensive insight into the structure of bacterial community of the different estuarine sediments of Poyang Lake, indicating that the environmental factors played a key role in influencing the bacterial community composition in the freshwater ecosystem.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microorganisms are crucial components in buried sediments and may account for more than 30 % of the biomass of the earth (Whitman et al. 1998). In freshwater lakes and other aquatic ecosystem sediments, microorganisms are important for decomposing, transforming organic matter (OM) and remineralizing nutrients, suggesting their crucial role in the earth’s biogeochemical cycles (Zhao et al. 2012; Jurgens et al. 2000). Consequently, investigating the composition of the bacterial community is vital to understand the metabolic processes in freshwater and other aquatic ecosystems (Spring et al. 2000).
Previous studies indicated that environmental physicochemical factors, such as nutrient availability, sedimentation, salinity, OM, pH and temperature, could influence the microbial structures in aquatic ecosystems and were considered the major driving forces of microbial communities (Stepanauskas et al. 2003; Wu et al. 2008; Tijdens et al. 2008). Former studies showed that there are diverse, abundant, frequently exchanged and highly active microorganisms in sediment environments, and the environment presents varied environmental gradients (Huang et al. 2014). Therefore, sediments may provide a suitable natural environment for surveying the adaptations and shifts of microbial communities (Stepanauskas et al. 2003; Wu et al. 2008).
Currently, the excess use of nitrogen fertilizer, municipal wastewater and industrial effluent discharge has led to the eutrophication of aquatic ecosystems (McLaughlin et al. 2015; Haukka et al. 2006). In China, 50 % of the freshwater lakes investigated were eutrophic (Jin et al. 2005).
Poyang Lake is the largest fresh water lake in China, and it is located in the northern part of Jiangxi province, at the southern bank of the Yangtze River. The Gan (GJ), Fu (FH), Xin (XJ), Rao (RH) and Xiu (UH) rivers flow into Poyang Lake, and it undergoes a great change in water level over the course of a year. The Le (LH) and Chang (CJ) rivers flow into the Rao River. The lake is the natural habitat of migratory birds, and it also offers important spawning grounds and food for many fishes (Wu et al. 2012). However, in recent years, the area of the lake fluctuates dramatically between the wet and dry seasons, the seasonal decline begins earlier and lasts longer (Zhang et al. 2011; Min and Zhan 2012), the size of the lake has been decreasing overall, the eutrophication degree is more and more serious because of industrialization and urbanization, and the water quality has deteriorated (Wu et al. 2011; Deng et al. 2011; Zhen et al. 2011).
Numerous studies have been conducted to investigate the water quality and the biodiversity of the fishes, birds, plants, and bacterioplankton communities of Poyang Lake (Wu et al. 2012; Liang et al. 2015; Yao et al. 2015; Fang et al. 2006; Guo et al. 2008). Most of these studies did not investigate the bacterial diversity and communities in the different estuarine sediments of Poyang Lake.
In this study, we aimed to determine the total bacterial diversities and community compositions in the different estuarine sediments of Poyang Lake by using 16S rRNA gene-targeted metagenomic approach. Furthermore, we analyzed the environmental physicochemical properties to investigate which factors have a significant impact on bacterial diversity, richness and community structures in those sediment samples.
Materials and methods
Ethics statement
The Jiangxi provincial water resources bureau concerned with protection of the Poyang Lake and Rivers. And it issued the permission for our sample collection. Our samples just contain many microorganisms, did not involve endangered or protected species.
Site description, sample collection
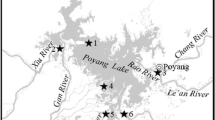
Sediment samples were collected from seven typical estuaries of Poyang Lake (Fig. 1). The longitude and latitude of these estuaries are shown in Table 1. The up to 5 cm sediments were collected in these different estuaries in October 2014. At the same site, five samples were collected and mixed together for homogenization. All samples were divided into two parts: one for immediate chemical analysis and another stored at −80 °C for DNA extraction.
Chemical analysis
Air-dried sediment/soil samples that had been sieved using a 2.0-mm sieve were used for analysis. At each station, in situ measurements of sediment temperature were recorded with a portable electronic thermometer. Sediment pH was determined by a multi-parameter pH meter (Sartorius PB-10, Germany) with a soil to water ratio of 1:10. Sediment total nitrogen (TN) was determined by the semi-micro Kjeldahl method (Fawcett 1954). Sediment inorganic nitrogen, including Ammonia–Nitrogen (NH4 +-N), Nitrite-Nitrogen (NO2 −-N) and Nitrate-Nitrogen (NO3 −-N), were extracted in a 1 mol L−1 KCl solution. The concentrations of ammonia and nitrite in the soil extract were measured using colorimetric methods (Schütz and Nuñer 2007), and nitrate was measured using the UV spectrometric method (Ding et al. 2014). Total phosphorus (TP) was analyzed by the ammonium molybdate method (Ebina et al. 1983), and inorganic phosphorus (IP) was measured according to Aspila et al. (1976). Chemical analyses were performed in triplicate.
DNA extraction from sediment samples
DNA extraction from 0.5 g of each sediment sample was performed by using a MoBio UltraClean™Soil DNA isolation kit (SanDiego, CA, USA) following the manufacturer’s instructions. Finally, the DNA was eluted with TE buffer. The amount and purity of DNA were determined by using a NanoDrop® Spectrophotometer ND-1000 (Thermo Fisher Scientific, USA) based on the absorbency of A260 and the ratio of A260/A280, respectively. The extracted total microbial DNA was stored at −80 °C prior to analysis.
Amplicon generation
The variable V4 region of the bacterial 16S rRNA gene was amplified with the general 16S rRNA gene primers 515F and 806R containing the specific barcode sequence. The forward primer (515F) was 5′-GTTTCGGTGCCAGCMGCCGCGGTAA-3′, where the sequence of the barcode is shown in italics. The reverse primer (806R) was 5′-GTGAAAGGACTACHVGGGTWTCTAAT -3′, where the sequence of the barcode is shown in italics. All PCR reactions were carried out in 30 μLs with 15μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of forward and reverse primers and approximately 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 60 s. Finally, extension occurred for 10 min at 72 °C.
PCR product quantification, qualification and purification
We mixed the same volume of 1X loading buffer (containing SYB green) with PCR products and ran electrophoresis on a 2 % agarose gel for detection. PCR products were mixed in equidensity ratios. Then, mixture PCR products were purified with a GeneJET Gel Extraction Kit (Thermo Scientific).
Library preparation and sequencing
Sequencing libraries were generated using the NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina MiSeq platform.
Data analysis
Paired-end reads from the original DNA fragments were merged using FLASH (Magoč and Salzberg 2011), a very fast and accurate analysis tool that was designed to merge paired-end reads when at least some of the reads overlap the read generated from the opposite end of the same DNA fragment. Paired-end reads were assigned to each sample according to the unique barcodes.
Sequences analyses were performed by the UPARSE software package (Uparse v7.0.1001, http://drive5.com/uparse/) (Edgar 2013) using the UPARSE-OTU and UPARSE-OTUref algorithms. In-house Perl scripts were used to analyze alpha (within samples) and beta (among samples) diversity. Sequences with ≥97 % similarity were assigned to the same OTUs. We picked representative sequences for each OTU and used the RDP classifier (Version 2.2, http://sourceforge.net/projects/rdp-classifier/) (Wang et al. 2007) to annotate taxonomic information for each representative sequence. To compute Alpha Diversity, we rarified the OTU table and calculated three metrics: Chao1 (estimates the species abundance), Observed Species (estimates the number of unique OTUs found in each sample), and the Shannon index. Rarefaction curves were generated based on these three metrics.
A graphical representation of the relative abundance of bacterial diversity from phylum to species can be visualized using a Krona chart (Ondov et al. 2011).
Statistical analysis
A Kendall’s correlation between environment factors and bacterial richness, diversity and composition were conducted using SPSS 16.0 software.
Nucleotide sequence accession numbers
The nucleotide sequences of bacterial 16S ribosome DNA gene fragments have been deposited at the NCBI Sequence Read Archive under accession numbers SRR2837836 (LS sample), SRR2854158 (CS sample), SRR2854159 (RS sample), SRR2854634 (XS sample), SRR2886900 (FS sample), SRR2886926 (GS sample), and SRR2886938 (US sample).
Results
Sediment chemical properties
Physicochemical characteristics of sediment samples collected from seven sites varied differently depending on the parameters measured (Table 1). The temperature of the different sites ranged from 17.4 to 20.7 °C, and the pH values ranged from 6.09 to 6.76. Among them, temperature and pH values of Le river were lower than that of other sites (Table 1). The average concentrations of the TN in these estuarine sediments ranged from 0.71 to 1.31 g kg−1, but the differences were not significant different. The concentration of NH4 +-N ranged from 40.31 to 79.39 mg kg−1, of which Le river sample showed the highest one. However, the concentration of NO2 −-N and NO3 −-N were extremely low, they ranged from 0.05 to 0.29 mg kg−1 and 0.53 to 4.31 mg kg−1, respectively (Table 1). TP and IP concentrations were additional evaluation indexes of the sediment. In our study, the concentrations of TP and IP ranged from 0.23 to 0.41 g/kg and 0.17 to 0.34 g/kg, respectively (Table 1).
Diversities of bacterial communities
To determine the bacterial community of these different estuarine sediments of Poyang Lake, metagenomic technology was adopted targeting the V4 region of the bacterial 16S rDNA gene with the Illumina Miseq platform. A total of 301,415 effective tags were obtained from the seven sediment samples, with an average length of 291 nucleotides. Each of the 7 libraries contained between 24,751 and 66,133 tags. The efficient percentage ranged from 71.80 to 75.47 % (Table 2). And Q20 and Q30 values were above 99.55 and 98.19 %, respectively. All further analyses were performed on these effective tags. At a >97 % sequence identity threshold, 2261, 2020, 1990, 2413, 1499, 1829 and 1594 OTUs were identified in LS (sediment of Le river), CS (sediment of Chang river), RS (sediment of Rao river), XS (sediment of Xin river), FS (sediment of Fu river), GS (sediment of Gan river) and US (sediment of Xiu river), respectively (Table 2), of which the XS sample possessed the highest OTUs, followed by the LS sample. OTUs were determined to calculate the richness, diversity and rarefaction curves of the microbial communities. The rarefaction curves showed a similar pattern for all the samples, and suggested that the bacterial community was well represented since they became gentle while the number of sequences analyzed increased (Fig. 2).
Richness indexes of Chao1, evaluated at 97 % similarity, showed a similar comparative trend in predicting number of OTUs. Sample from LS had the highest richness (Chao1 = 2557.32), while sample from FS had the lowest one (Chao1 = 1964.46). The highest sediment bacterial diversity (Shannon index = 9.06) was found at LS sample, while the lowest one was at FS (Shannon index = 7.17) (Table 2). A similar tendency was observed for Simpson index, we found that LS and CS samples showed higher bacterial diversity (Simpson index = 0.993, 0.994), and FS sample was the lowest one (Simpson index = 0.960).
Bacterial community structure
In this study, the use of the 16S rRNA gene analyses resulted in the identification of 10 different phyla in all samples (the proportion ranged from 94.11 % to 98.19 %), including Proteobacteria, Acidobacteria, Firmicutes, Chloroflexi, Bacteroidetes, Planctomycetes, Gemmatimonadetes, Actinobacteria, Nitrospirae, and Verrucomicrobia. The majority of bacterial sequences in all samples belonged to these phyla Proteobacteria, Acidobacteria and Firmicutes, which represented 38.28 to 69.81 %, 2.83 to 21.09 %, and 7.68 to 18.20 % of each total sequences, respectively (Fig. 3).
At the class level, the bacterial taxa were distributed in Betaproteobacteria, Alphaproteobacteria, Acidobacteria, Gammaproteobacteria, Clostridia, Deltaproteobacteria, Anaerolineae, Bacilli, Sphingobacteria and Planctomycetia (the proportion ranged from 68.70 to 90.49 %). Among them, Betaproteobacteria (15.65–40.66 %) was the most dominant bacterial class in all samples except XS sample (Fig. 4). Acidobacteria (17.33 %) proved to be predominant in XS library, followed by Betaproteobacteria (13.90 %).
At the genus level (the lowest level assigned), Janthinobacterium, Pseudomonas, Acinetobacter, Brevundimonas, Clostridium, Geobacter, Kaistobacter, Exiguobacterium, Rhodoplanes, and Polaromonas were the top 10 dominant genera in all samples. Janthinobacterium exhibited more proportional representation in GS (23.81 %), US (24.85 %) and FS (16.19 %) samples, Pseudomonas was more abundant in GS (9.50 %) and FS (8.16 %) samples, Acinetobacter showed more abundant in XS (7.93 %) sample, Brevundimonas showed more abundant in FS sample (6.47 %), and Clostridium exhibited more proportional representation in GS sample (5.07 %) (Fig. 5).
Influential factors on bacterial communities
Kendall’s correlation analysis indicated that the bacterial OTU number and Chao1 estimator were positively correlated to the levels of sediment inorganic phosphorus (P < 0.05). However, bacterial Shannon diversity illustrated no significant correlation with the determined parameters (P > 0.05) (Table 3).
Sediment nitrite nitrogen was negatively correlated with relative abundance of Proteobacteria (P < 0.05). Chloroflexi showed a significant positive correlation with both sediment total phosphorus and inorganic phosphorus (P < 0.05). Nitrospirae was positively affected by sediment total nitrogen, and Verrucomicrobia showed positive correlation with sediment nitrite nitrogen, total phosphorus and inorganic phosphorus (P < 0.05) (Table 3).
Discussion
Bacterial community diversity and composition in the Poyang Lake has been well documented (Wu et al. 2012). However, the bacterial communities in different estuarine sediments of Poyang Lake were overlooked for a long time, and which bacteria take part in the important nutrient cyclings in these sediments were still unknown. This is the first comprehensive study on the overall composition of bacterial community in these estuarine sediments of Poyang Lake.
Chemical properties of these seven different estuarine sediments of Poyang Lake
Poyang Lake, the largest freshwater lake in China, is the most important mainstream connected with the Yangtze River. It plays a crucial role in the maintenance of the aquatic biota of the Yangtze Basin (Wang et al. 2007), and it is located in a globally important ecological area (Liang et al. 2015). The Gan, Fu, Xin, Rao and Xiu rivers flow into Poyang Lake. The water regime of Poyang Lake is controlled by the five tributaries (Shankman et al. 2006). Determining the environmental physicochemical properties and bacterial community compositions of these different estuaries of Poyang Lake may be helpful in assessing, understanding and maintaining the water quality of Poyang Lake.
From our study, we found that the Le River showed the highest total nitrogen and ammonia nitrogen concentration. The high total nitrogen and ammonia nitrogen content of this river may be caused by the number of fisheries in Poyang County. As we know, protein-rich wastes from aquaculture systems could increase total nitrogen, ammonia nitrogen and total organic carbon (Mook et al. 2012). Additionally, we found that the total phosphorus and inorganic phosphorus content of the Xin and Le rivers were higher than the other estuaries. This may be due to the mining of the phosphate rock mountain in Shangrao City, which is located upstream of the Xin river.
Bacterial diversities in estuarine sediments
In this study, the bacterial diversities in estuarine sediments of Poyang Lake were obtained using Illumina Miseq sequencing. Our analysis revealed a high biodiversity in these estuarine ecosystems, reflecting the rich chemical features of these environments (Qu et al. 2008). Among them, LS and XS showed higher bacterial richness and diversity, this maybe due to the higher concentrations of nitrogen and phosphorus in these two samples. Beside that, Kendall’s correlation analysis also indicated that bacterial diversity and richness were found to be likely positively affected by sediment inorganic phosphorus. Previous studies have indicated that in freshwater lacustrine sediment ecosystems, greater amounts of nutrients are thought to support richer and more diverse bacterial communities through increased niche partitioning (Dykhuizen 1998). And, phosphorus, as a major nutrient for aquatic ecology, has been recognized as the most critical nutrient limiting lake productivity, it has significant impacted on the bacteria community structure in lake sediment (Doricha et al. 1984; Jin et al. 2005; Song et al. 2012). Our results suggested that abundant nitrogen and phosphorus might cause high bacterial richness and diversity. Other chemical factors, such as total organic carbon, metal ion, etc., might also regulate the distribution of sediment bacteria in these estuaries. This still need further research.
Bacterial community structure in estuarine sediments
As we know, “Global Dispersal” is the major conflicting hypothesis in microbial biodiversity studies (Martiny et al. 2006). It suggests that microorganisms are ubiquitous and have few barriers to gene flow resulting in similar microbial communities across different spatial scales and habitats. Since this study showed the similar bacterial communities in seven different estuarine sediments that have been described in other soil/sediment environmental, this finding further supports the hypothesis. Previous studies indicated that the soil/sediment bacterial communities were normally comprised of the 9 major bacterial phyla: Proteobacteria, Actinobacteria, Acidobacteria, Chloroflexi, Bacteroidetes, Firmicutes, Planctomycetes, Verrucomicrobia and Gemmatimonadetes (Janssen 2006). In our study, the bacterial communities of all estuarine sediment samples were also found to belong to those 9 bacterial phyla, and among them, bacterial communities in all samples were predominated by Proteobacteria, a metabolically diverse group of Gram-negative bacteria frequently found in freshwater sediment (Zhang et al. 2015; Fierer et al. 2007). Acidobacteria and Firmicutes were the secondary and tertiary phyla in these estuarine sediment samples. Acidobacteria and Firmicutes are ubiquitous in freshwater lake sediment and are known to degrade a variety of organic compounds (Tamaki et al. 2005; Kirchman 2002). So, their dominance of the sediment of this eutrophic estuary is not surprising. Our findings may indicate that members of the predominant taxa are well adapted to the surficial layer of freshwater sediments regardless of geographical location, and the predominance of Proteobacteria, Acidobacteria and Firmicutes in all samples suggested that they may be actively involved in the functioning and processes of estuarine sediment of Poyang Lake.
At the genus level, Acinetobacter was richer in XS sample. Acinetobacter was reported as one of commonly detected phosphorus-accumulating microorganisms in sediment (Martins et al. 2011), which may explain their concentration in phosphorus enriched sediment sample of Xin river. Besides, we also found that Janthinobacterium was abundant in GS, US and FS samples, previous studies have showed that Janthinobacterium may be included in anaerobic metabolism linked to denitrification cycling (McTaggart et al. 2014). The presence of Janthinobacterium species may be valuable for the denitrification cycling in GS, US, and FS samples.
Influential factors on bacterial communities
Generally, many studies have reported that environmental factors, such as total nitrogen, phosphorus and other nutrient elements, have impacts on bacterial colonization in sediment samples (Stepanauskas et al. 2003; Wu et al. 2008; Tijdens et al. 2008; Angeloni et al. 2006; Zeng et al. 2009). When the nutrient elements increased, the biomass of phytoplankton, algae and bacteria also increased, as there is a mutualistic relationship among these organisms (Hietala et al. 2004; Wetzel 1983). Similar results were found in our study. Nitrite nitrogen, total phosphorus and inorganic phosphorus were found to be important drivers for the distribution of sediment Proteobacteria, Chloroflexi, Nitrospirae and Verrucomicrobia communities in estuarine sediments of Poyang Lake. The results of the present study support the idea that environmental changes could be evaluated using the composition variation of bacterial communities. Although other environmental variables were not statistically significant, an influence of synergistic reaction of these nutrients on the distribution of bacterial community structures still cannot be excluded. More information is needed in this area.
In conclusion, as the first study of the bacterial diversity and community of the different estuarine sediments of Poyang Lake, our study demonstrated that the bacterial diversities varied among the different samples. LS and XS samples exhibited the higher bacterial richness and diversity, and the FS sample showed the least diversity, which was in accordance with the environmental nutrient content. Bacterial diversity was affected by sediment inorganic phosphorus. Nitrite nitrogen, total phosphorus and inorganic phosphorus were also found to be important drivers for bacterial community compositions. Proteobacteria predominated in all sediment bacterial communities, followed by Acidobacteria and Firmicutes, they might be involved in various biogeochemical processes in sediment. Our study points to the interactions between bacterial community composition and sediment environmental physicochemical factors, which could shed light on the functional roles of microorganisms involved in the earth’s biogeochemical cycles in the freshwater lake ecosystem.
References
Angeloni NL, Jankowski KJ, Tuchman NC et al (2006) Effects of an invasive cattail species (Typha x glauca) on sediment nitrogen and microbial community composition in a freshwater wetland. FEMS Microbiol Lett 263:86–92
Aspila KI, Agemain H, Chau A (1976) A semi-automated method for the determination of inorganic, organic and total phosphorus in sediments. Analyst 101:187–197
Deng X, Zhao Y, Wu F et al (2011) Analysis of the trade-off between economic growth and the reduction of nitrogen and phosphorus emissions in the Poyang Lake Watershed, China. Ecol Model 222:330–336
Ding K, Wen X, Chen L et al (2014) Abundance and distribution of ammonia-oxidizing archaea in Tibetan and Yunnan plateau agricultural soils of China. Front Env Sci En 8:693–702
Doricha RA, Nelsona DW, Sommers LE (1984) Availability of phosphorus to algae from eroded soil fractions. Agr Ecosyst Environ 11:253–264
Dykhuizen DE (1998) Santa Rosalia revisited: why are there so many species of bacteria? Antonie van Leeuwenhoek 73:25–33
Ebina J, Tsutsui T, Shirai T (1983) Simultaneous determination of total nitrogen and total phosphorus in water using peroxodisulfate oxidation. Water Res 17:1721–1726
Edgar Robert C (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Fang JY, Wang ZH, Zhao SQ et al (2006) Biodiversity changes in the lakes of the Central Yangtze. Front Ecol Environ 4:369–377
Fawcett JK (1954) The semi-micro kjeldahl method for the determination of nitrogen. J Med Lab Technol 01:1–22
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Guo H, Hu Q, Jiang T (2008) Annual and seasonal streamflow responses to climate and land-cover changes in the Poyang Lake basin, China. J Hydrol 355:106–122
Haukka K, Kolmonen E, Hyder R et al (2006) Effect of nutrient loading on bacterioplankton community composition in Lake Mesocosms. Microb Ecol 51:137–146
Hietala J, Vakkilainen K, Kairesalo T (2004) Community resistance and change to nutrient enrichment and fish manipulation in a vegetated lake littoral. Freshwat Biol 49:1525–1537
Huang R, Zhao DY, Jiang CL et al (2014) Heterogeneity of bacterial community compositions in surface sediments of three lake zones in lake Taihu. Proc Natl Acad Sci, India, Sect B Biol Sci 85:465–474
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Jin XC, Xu QJ, Huang CZ (2005a) Current status and future tendency of lake eutrophication in China. Sci China C Life Sci 48:948–954
Jin X, Wang S, Pang Y et al (2005b) The adsorption of phosphate on different trophic lake sediments. Colloid Surface A 254:241–248
Jurgens G, Glöckner FO, Amann R et al (2000) Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol Ecol 34:45–56
Kirchman DL (2002) The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100
Liang Y, Xiao HY, Liu XZ et al (2015) Spatial and temporal water quality characteristics of Poyang Lake Migratory Bird Sanctuary in China. Chin J Geochem 34:38–46
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Martins G, Terada A, Ribeiro DC et al (2011) Structure and activity of lacustrine sediment bacteria involved in nutrient and iron cycles. FEMS Microbiol Ecol 77:666–679
Martiny JB, Bohannan BJ, Brown JH et al (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112
McLaughlin RW, Wang SB, Zhou W et al (2015) A Comparison of the bacterial diversity of two shallow freshwater lakes in China. Proc Natl Acad Sci, India, Sect B Biol Sci 85:137–146
McTaggart TL, Shapiro N, Woyke T et al (2014) Draft genome of Janthinobacterium sp. RA13 isolated from lake Washington sediment. Genome Announc 3(1):e01588-14
Min Q, Zhan L (2012) Characteristics of low-water changes in Lake Poyang during 1952-2011. J Lake Sci 24:675–678
Mook WT, Chakrabarti MH, Aroua MK et al (2012) Removal of total ammonia nitrogen (TAN), nitrate and total organic carbon (TOC) from aquaculture wastewater using electrochemical technology: a review. Desalination 285:1–13
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385
Qu JH, Yuan HL, Wang ET et al (2008) Bacterial diversity in sediments of the eutrophic Guanting Reservoir, China, estimated by analyses of 16S rDNA sequence. Biodivers Conserv 17:1667–1683
Schütz JH, Nuñer APDO (2007) Growth and survival of dorado Salminus brasiliensis (Pisces, Characidae) post-larvae cultivated with different types of food and photoperiods. Braz Arch Biol Technol 50:435–444
Shankman D, Keim BD, Song J (2006) Flood frequency in China’s Poyang Lake region: trends and teleconnections. Int J Climatol 26:1255–1266
Song H, Li Z, Du B et al (2012) Bacterial communities in sediments of the shallow Lake Dongping in China. J Appl Microbiol 112:79–89
Spring S, Schulze R, Overmann J et al (2000) Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: molecular and cultivation studies. FEMS Microbiol Rev 24:573–590
Stepanauskas R, Moran MA, Bergamaschi BA (2003) Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat Microb Ecol 31:85–98
Tamaki H, Sekiguchi Y, Hanada S et al (2005) Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl Environ Microbiol 71:2162–2169
Tijdens M, Hoogveld HL, Kamst-van Agterveld MP et al (2008) Population dynamics and diversity of viruses, bacteria and phytoplankton in a shallow eutrophic lake. Microb Ecol 56:29–42
Wang HZ, Xu QQ, Cui YD et al (2007) Macrozoobenthic community of Poyang Lake, the largest freshwater lake of China, in the Yangtze floodplain. Limnology 8:65–71
Wetzel RG (1983) Limnology. Saunders College, New York
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95:6578–6583
Wu L, Kellogg L, Devol AH et al (2008) Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Appl Environ Microbiol 74:4516–4529
Wu L, Li M, Guo Y et al (2011) Influence of three gorges project on water quality of Poyang Lake. Procedia Environ Sci 10:1496–1501
Wu L, Ge G, Zhu GF et al (2012) Diversity and composition of the bacterial community of Poyang Lake (China) as determined by 16S rRNA gene sequence analysis. World J Microbiol Biotechnol 28:233–244
Yao X, Wang SR, Ni ZK et al (2015) The response of water quality variation in Poyang Lake (Jiangxi, People’s Republic of China) to hydrological changes using historical data and DOM fluorescence. Environ Sci Pollut Res 22:3032–3042
Zeng J, Yang L, Li J et al (2009) Vertical distribution of bacterial community structure in the sediments of two eutrophic lakes revealed by denaturing gradient gel electrophoresis (DGGE) and multivariate analysis techniques. World J Microbiol Biotechnol 25:225–233
Zhang Q, Sun P, Chen X et al (2011) Hydrological extremes in the Poyang Lake basin, China: changing properties, causes and impacts. Hydrol Process 25:3121–3130
Zhang JX, Yang YY, Zhao L et al (2015) Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl Microbiol Biotechnol 99:3291–3302
Zhao DY, Huang R, Zeng J et al (2012) Diversity analysis of bacterial community compositions in sediments of urban lakes by terminal restriction fragment length polymorphism (T-RFLP). World J Microbiol Biotechnol 28:3159–3170
Zhen L, Li F, Huang H et al (2011) Households’ willingness to reduce pollution threats in the Poyang Lake region, southern China. J Geochem Explor 110:15–22
Acknowledgments
This research was supported by the special funds of the national academy alliance for academy construction (2012-1) and the sci-tech support plan of Jiangxi province (2008BB26100).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sheng, P., Yu, Y., Zhang, G. et al. Bacterial diversity and distribution in seven different estuarine sediments of Poyang Lake, China. Environ Earth Sci 75, 479 (2016). https://doi.org/10.1007/s12665-016-5346-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12665-016-5346-6