Abstract
Background
Planar equilibrium radionuclide angiocardiography (ERNA) has been used as the gold standard for assessment of left ventricular (LV) function for over three decades. However, this imaging modality has recently gained less favor due to growing concerns about radiation exposure. We developed a novel approach that involves integrating short axis slices of gated bloodpool SPECT for quantification of LV function with improved signal-to-noise ratio and reduced radioactive dose while maintaining image quality and quantitative precision.
Methods
Twenty patients referred for ERNA underwent standard in vitro 99mTc-labeling of red blood cells (RBC), and were initially imaged following a low-dose (~ 8 mCi) injection using a dedicated cardiac SPECT camera, and then had planar imaging following a high-dose (~ 25 mCi) injection. Four different quantification methods were utilized to assess the LV function and were compared for quantitative precision and inter-observer reproducibility of the quantitative assessments.
Results
The Yale method resulted in the most consistent assessment of LV function compared with the gold standard high-dose ERNA method, along with excellent inter-observer reproducibility.
Conclusions
The new low-dose 99mTc-RBC imaging method provides precise quantification of LV function with a greater than 67% reduction in dose and may potentially improve assessment of regional function.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Planar equilibrium radionuclide angiocardiography (ERNA) has been used for the assessments of left ventricular (LV) volumes, ejection fraction (EF), and peak filling rate (PFR) for over three decades with high accuracy and precision in patients with known or suspected heart disease.1,2,3,4,5,6,7,-8 However, conventional planar ERNA has recently gained less favor due to the growing concerns of radiation exposure to patients and improvements in quantitative echocardiography. While new single-photon emission computerized tomography (SPECT) cameras equipped with solid-state detectors and pinhole collimators allows for low-dose imaging, these high-sensitivity SPECT cameras with improved resolution are not capable of acquiring traditional planar ERNA images, which was the established gold standard nuclear cardiac imaging method for assessment of the LV function. Recently, the LV volumes, EF, and PFR can be quantified from three-dimensional (3D) gated bloodpool SPECT with reasonable accuracy and precision based on the LV geometry, whereas the edges of LV cavity used for calculation of the LV volumes need to be predetermined via 3D image segmentation algorithms. The LV function quantification incorporated with the complex 3D segmentation may also suffer from suboptimal reproducibility due to the effects of image reconstruction parameters and variabilities of the LV edges derived from 3D gated bloodpool SPECT with limited image resolution.9,10 Alternatively, Wells et al.11 proposed a quasi-2D ERNA scheme by forwardly projecting 3D bloodpool SPECT to mimic the left anterior oblique (LAO) view of planar ERNA from which the end-diastolic LV volume (EDV) and EF can be calculated using Massardo’s method.12 While this LAO projection approach has a merit of mitigating the limitation of pinhole collimation, the selection of a “best” LAO view to visually separate the LV from the right ventricle (RV) is a challenging manual process which is very operator-dependent, causing another variation in ERNA quantification.
In the present study, we propose a novel semi-automatic approach to forwardly integrating (summing) the short axis (SA) slices, rather than the LAO view, from electrocardiographic (ECG)-gated 3D bloodpool SPECT images to generate quasi-2D planar ERNA and using a ERNA quantification method newly developed and previously validated in phantoms.13 The new method was compared with three other conventional quantification methods for assessments of EDV, end-systolic volume (ESV), LVEF, and PFR in patients undergone both high-dose 2D planar ERNA and low-dose 3D gated bloodpool SPECT.
Materials and Methods
Subjects and Red Blood Cell Labeling
Twenty patients (12 males, Age: 65 ± 8 year, Weight: 82 ± 21 kg, BMI: 28 ± 6 kg/m2) were randomly selected from the Cardiovascular Imaging Lab at the Yale New Haven Hospital, New Haven, CT for this study. Red blood cells (RBC) from these patients were labeled in vitro with 99mTc sodium pertechnetate (25 to 30 mCi) using the FDA-approved ULTRATAG® RBC kit (Mallinkcrodt Inc., St Louis, MO). The study protocol was adapted from the standard nuclear cardiac imaging protocols used in the Yale clinical cardiovascular imaging lab and was approved by Yale Institutional Review Board.
Three-Dimensional Bloodpool SPECT and Two-Dimensional Planar ERNA Imaging
Approximately 30 min post the RBC labeling process, patients were initially injected with approximately 8 mCi (low-dose) of 99mTc-labeled RBCs and were imaged for 6 min using a dedicated cardiac SPECT camera (Discovery NM 530c, GE Healthcare, Milwaukee, WI) equipped with 19 cadmium zinc telluride (CZT) detectors each being collimated with a tungsten pinhole collimator of 5 mm in diameter. All pinhole images were ECG-gated with 16 bins within the R–R intervals and were reconstructed using ten subsets and two iterations of the ordered subset expectation maximization algorithm with a matrix size of 70 × 70 and pixel size of 4 × 4 mm2. Immediately after the low-dose ECG-gated bloodpool SPECT imaging, patients were injected with an additional 17 mCi of remaining 99mTc-labeled RBCs to acquire high-dose (approximately 25 mCi) 2D planar ERNA using a different gamma camera equipped with conventional Sodium Iodide (NaI) detectors and low-energy/high-resolution parallel-hole collimators (MPS, GE Healthcare, Milwaukee, WI). The conventional 2D planar ERNA scans were ECG-gated with 24 bins within the cardiac cycle and were acquired for 10 min at the LAO position providing best RV and LV separation using a matrix size of 64 × 64 and pixel size of 4.8 × 4.8 mm2.
Generation of 2D Planar ERNA from 3D Gated Bloodpool SPECT
For each patient, two sets of quasi-2D planar ERNA were created from the ECG-gated bloodpool SPECT images. One set was generated by forwardly projecting the transaxial slices of ECG-gated bloodpool SPECT at the LAO view using commercial multigated software (Xeleris, GE Healthcare, Milwaukee, WI), while the other set was created from the SA slices of the ECG-gated bloodpool SPECT using the new approach illustrated in Figure 1. In brief, our algorithm for generating the quasi-2D ERNA for each ECG-gated bin includes five major steps as summarized below13:
Illustration of generating quasi-2D planar ERNA from the short axis slices of gated bloodpool SPECT images. The first (S1), last (SNS) slices and the total numbers of the short axis SPECT slices (NS= SNS − S1+1) used in the summing procedure were determined empirically via Gaussian fitting of the maximal counts (overlay pixels shown in black dots) being detected automatically from each of the short axis SPECT slices. ED, end-diastole; ES, end-systole; σ, standard deviation derived from the Gaussian fitting; LV, left ventricle; RV, right ventricle; Sm, slice number at the peak of the Gaussian curve
-
(a)
define the LV cavity regions automatically from the SA slices of gated bloodpool SPECT and detect the maximal counts in the LV regions slice-by-slice;
-
(b)
perform 1D Gaussian fitting to the sequence of the maximal counts determined above;
-
(c)
calculate the first apical (S1) and last basal (SNS) SA slices used for generating the quasi-2D ERNA based on statistics of the Gaussian curve generated in Step (b) as
where Sm represents the slice number at the peak of the Gaussian curve, σ is the standard deviation (SD) derived from the 1D Gaussian fitting, and α, β denote the over-relaxation parameters for optimization of the first and last SA slice selections, respectively;
-
(d)
calculate the total number of SA slices used in the generation of quasi-2D ERNA as
and
-
(e)
integrate (sum) the total numbers (NS) of gated bloodpool SA slices determined in Step (d) for all 16 ECG-gated bins to generate a sequence of quasi-2D ERNA for EDV, ESV, LVEF, and PFR calculations.
The over-relaxation parameters in Eqs. (1) and (2) above were adapted from our previous phantom study13 and were determined empirically as α = 1.5 and β = 1.0 for the present patient study.
Calculations of EDV, ESV, LVEF, and PFR from 2D Planar ERNA
The EDV was calculated from the conventional 2D (high-dose) ERNA and forwardly projected 2D (low-dose) ERNA using commercial software (Xeleris, GE Healthcare, Milwaukee, WI). Also, the quasi-2D ERNA integrated from the bloodpool SPECT SA slices was quantified using the new Yale method developed for the present study,13 in which the EDV were calculated as
where \( {\mathcal{M}} \) represents the pixel size (cm) of quasi-2D ERNA, \( {\mathcal{R}} \) is the ratio of the total counts to the maximal counts derived from the region of interest (ROI) of the LV in the quasi-2D ERNA, and NS is obtained from Eq. (3).12,13 The LV ROI above and background in the end-diastole (ED) and end-systole (ES) were determined semi-automatically using a region-growing algorithm adapted for this study13 and the LVEF was calculated based on the background-corrected counts derived from the ED and ES ROIs as
The ESV and PFR were calculated from the background-corrected count profiles with a 4-homonic smoothing process.14
Calculations of EDV, ESV, LVEF, and PFR from 3D Bloodpool SPECT
The EDV, ESV, LVEF, and PFR were also calculated from 3D gated bloodpool SPECT images using commercial quantitative bloodpool SPECT (QBS) software (Cedar Sinai, Los Angeles, CA). The 3D edges of LV cavity were determined directly from the 3D ECG-gated bloodpool SPECT images5,15,16,17,18,19,-20 and the EDV and ESV were calculated based on the LV edges determined by the QBS software mentioned above. The PFR was derived from the slopes of LV volume curves within the cardiac cycle and the LVEF was calculated by
Comparisons of EDV, ESV, LVEF, and PFR and Evaluation of Inter-observer Reproducibility
The EDV, ESV, LVEF, and PFR quantified from (a) low-dose (~ 8 mCi) forwardly projected LAO ERNA using GE-multigated method denoted by Method 2 or M2, (b) low-dose quasi-2D ERNA using the Yale method termed as Method 3 or M3,13 as well as from low-dose full 3D gated bloodpool SPECT using (d) Cedar Sinai-QBS denoted by Method 4 or M4,17 were compared to those quantified from high-dose (~ 25 mCi) LAO planar ERNA (gold standard for this study, termed as Method 1 or M1) using conventional GE-Xeleris software. The four different methods mentioned above are reiterated in Table 1. The EDV, ESV, LVEF, and PFR were further quantified by two different operators using the Yale method to assess inter-observer reproducibility of the quantitative data.
Statistical Analysis
Data were expressed as mean ± SD. Paired t test with Bonferroni corrections was used for the assessment of difference between two measures. P < 0.017 (3 pair comparisons) was considered statistically significant between two measures. Analyses of linear regression and estimation difference between two measures were utilized to assess agreements between the quantitative data. Bland–Altman plots21 were used to assess agreements of two quantitative measures resulted from two different quantification methods and to evaluate inter-observer reproducibility of the quantitative data processed by two different operators.
Results
Precisions of EDV, ESV, LVEF, and PFR Quantified via Four Different Methods
The EDV, ESV, LVEF, and PFR quantified from high-dose 2D planar ERNA (~ 25 mCi, gold standard for this study) and from low-dose (~ 8 mCi) forwardly projected LAO SPECT ERNA, quasi-2D SA SPECT ERNA, and full 3D SPECT ERNA using four different quantification methods are shown in Figure 2. As seen, the EDV, ESV, LVEF, and PFR quantified using Yale method (M3) are similar to those calculated from the high-dose 2D planar ERNA (P = ns). However, the EDVs and ESVs quantified using the other two methods (M2 and M4) are significantly smaller than those quantified with M1 (Figure 2A, B). The LVEFs quantified from the low-dose 3D SPECT ERNA using the QBS software (M4) are significantly higher than those quantified from the high-dose 2D planar ERNA (Figure 2C), whereas PFRs calculated using all four methods are similar (Figure 2D).
Evaluations of Linear Correlations and Differences in EDV, ESV, LVEF, and PFR Assessments
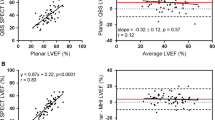
Figures 3, 4, 5, and 6 show the linear regressions and estimation differences in comparisons of the EDV, ESV, LVEF, and PFR quantified via the four methods (M1 to M4) in which the parameters (EDV, ESV, LVEF, and PFR) quantified using M2 to M4 were compared with those quantified using the gold standard M1. The correlations of EDVs calculated via M2 to M4 vs M1 are similar, the correlation coefficients (R2) ranged 0.56 to 0.68, as demonstrated in Figure 3A to C, whereas the Yale method (M3) resulted in the smallest mean estimation difference of EDV (Figure 3D). Similar patterns of the quantitative results are also observed in the comparisons of ESV and LVEF (Figures 4 and 5). Notice that the mean difference of LVEFs calculated via M4 vs that via M1 is markedly greater as compared to the other 2 methods (M2 to M3). In comparisons of the PFRs calculated via M2 to M4 vs M1, the correlations and mean estimation differences are similar as seen in Figure 6. Bland–Altman plots for the assessments of agreement between the Yale Method (M3) and the conventional gold standard method (M1) for the quantifications of EDV, ESV, LVEF, and PFR are demonstrated in Figure 7. As shown, the mean differences in EDV (1.2 mL), ESV (4.71 mL), LVEF (− 0.6%), and PFR (0.06 EDV/s) are all small between the two methods.
Evaluation of Inter-observer Reproducibility for EDV, ESV, LVEF, and PFR Quantified via Yale Method
Figures 8 and 9, respectively, show the linear regression and Bland–Altman plots for the analyses of inter-observer reproducibility for the EDV, ESV, LVEF, and PFR quantified by Operator 1 (O1) and Operator 2 (O2) using the Yale method (M3). The correlations of EDVs (Figure 8A), ESVs (Figure 8B), LVEFs (Figure 8C), and PFRs (Figure 8D) quantified by O1 and O2 are excellent (R2 > 0.9) and the linear regression lines are close to the line of identity as seen in Figure 8. The mean differences of EDV (− 2.75 mL), ESV (− 3.09 mL), LVEF (1.95%), and PFR (0.14 EDV/s) quantified by O1 and O2 using the Yale method (M3) are minimal as demonstrated in Figure 9.
Discussions
We have developed a new method for quantification of the LV function from low-dose short axis slices of ECG-gated bloodpool SPECT. With the new method, the radiation dose can be reduced considerably at least by 67% from ~ 25 mCi to ~ 8 mCi. The new method incorporated with the quasi-2D ERNA images being integrated from the short axis slices of 3D bloodpool SPECT outperforms the other conventional methods for quantification of the EDV, ESV, and LVEF.
Merit of Using the Short Axis Slices of Bloodpool SPECT to Generate Quasi-2D ERNA
In the current study, we utilized the SA rather than LAO view of ECG-gated bloodpool SPECT images to generate a sequence of quasi-2D ERNA for quantification of the LV function because in practicality the SA slices are usually well aligned along the long axis of the LV post SPECT reconstruction and slice reorientation processes. As such, the LV can be clearly separated from the RV and other extracardiac organs in the SA slices. We believe that the use of the short axis rather than transaxial slices of 3D gated bloodpool SPECT to generate the quasi-2D ERNA has a substantial merit of circumventing the selection uncertainty and variability for a “best” LAO view from the forwardly projected transaxial slices.11
Discrepancy in EDV, ESV, and LVEF Assessments Using 2D ERNA vs Full 3D gated Bloodpool SPECT
We noticed in the present study that the EDV and ESV calculated via the geometry-based quantification method (M4) from full 3D ECG-gated bloodpool SPECT were lower (Figure 2A, B) and LVEF was higher (Figure 2C) than those quantified from the conventional 2D ERNA (M1). Thus, the values of EDV, ESV, and LVEF may not be inter-changeable between the values calculated from the quasi-2D ERNA and full 3D gated bloodpool SPECT.10,22 Also, note that we used the high-dose 2D ERNA as the gold standard for those comparisons because the 2D ERNA technique has been used in clinical practice over 30 years and the quantitative analyses of the LV function parameters (LV volumes and LVEF) are highly reproducible and thus are well accepted in nuclear cardiac imaging community.
Clinical Implications of Precise EDV and LVEF Quantification from ERNA
While LVEF is the most common index of the LV function used in clinical practice,23 the index of LV volumes has drawn an increased attention in clinical trials.2 Previously study by Solomon et al. showed that patients with critical myocardial infarction had a marked increase in EDV and decrease in LVEF particularly for patients with heart failure.24 Although echocardiography remains a non-invasive imaging modality of choice for evaluation of the LV function due to its broad availability and relatively low cost in clinical use, the quantification of EDV and LVEF from 2D echocardiography may suffer from low reproducibility in the assessment of EDV and LVEF. While 3D real-time echocardiography has recently become a clinically feasible method for assessing the EDV and LVEF, this 3D imaging modality is not widely available.
In contrast, ERNA has been a well-established non-invasive technique over three decades for evaluations of the RV and LV functions with extremely reliable and reproducible quantification of EDV and LVEF.6,7,14,25 Unlike echocardiography which predominately relies on the geometrical changes in the LV to calculate the LV volumes, ERNA takes advantage of radionuclide count changes in the LV to estimate the LV volumes. In the new method introduced herein, we further adopted a semi-automated region-growing algorithm to determine the count-based region changes in the LV during the cardiac cycle rather than the operator-defined LV ROIs used in 2D or 3D echocardiography and other conventional radionuclide imaging methods. The less operator-dependent region-growing algorithm associated with the geometry-independent scenario used in our methods has resulted in extraordinary precision and reproducibility in the quantitative assessments of EDV, ESV, LVEF, and PFR. Ultimately, we believe that the excellent reliability of the new quantitative approach introduced herein may also enhance one’s confidence in the EDV assessment and further strengthen our understanding of the impact of LV remodeling changes in heart failure patients.23,26
Study Limitations
As aforementioned, we used high-dose 2D ERNA as a gold standard for the comparisons performed in the present study since this imaging technique has been well established and the highly reliable and reproducible quantitative LV volumes and LVEF have been well accepted for over three decades in the nuclear cardiac imaging community. For comparisons of quantitative accuracy of the LV volumes, however, a true gold standard, such as that derived from cardiac magnetic resonance imaging, may be required to reduce potential bias in the comparisons. Nevertheless, we do not expect much variation in the quantitative accuracy and precision of the LV volumes for the present as well as future patient studies since our methods for the quantification of LV volumes have been validated in our previous phantom study.13 Finally, we also recognize that the number of patients selected for the current study may be limited although the 20 patients selected had reasonably high image quality and a wide range of LV volumes and LVEFs adequately for a reliable comparison.
New Knowledge Gained
The present patient study demonstrates that the novel scheme of using the short axis slices of ECG-gated SPECT bloodpool images associated with the region-growing approach evidently improves the precision and reproducibility for the assessment of LV function. The new Yale ERNA quantification method can be used reliably to assess the LV volumes within the entire cardiac cycle, leading to exceedingly precise and reproducible quantification of the LV function.
Conclusions
The new method has a potential of providing higher quantitative precision and reproducibility for EDV, ESV, and LVEF assessments than other conventional methods. Quantification of the LV function from high-sensitivity solid-state SPECT with the new method allows for at least two-third of radioactive dose reduction while maintaining high reliability and reproducibility for quantitative assessments of the LV function.
Abbreviations
- CZT:
-
Cadmium zinc telluride
- EDV:
-
End-diastolic volume
- EF:
-
Ejection fraction
- ERNA:
-
Equilibrium radionuclide angiography
- ESV:
-
End-systolic volume
- PFR:
-
Peak filling rate
- QBS:
-
Quantitative bloodpool SPECT
- RBC:
-
Red blood cell
- SPECT:
-
Single-photon emission computerized tomography
- 99mTc:
-
Technetium-99m
References
Alexanderson E, Marquez MF. Evaluation of ventricular function using nuclear cardiology. Part I: Methods and study techniques. Arch Inst Cardiol Mex 2000;70:417-23.
Anand IS, Florea VG, Solomon SD, Konstam MA, Udelson JE. Noninvasive assessment of left ventricular remodeling: Concepts, techniques, and implications for clinical trials. J Card Fail 2002;8:S452-64.
Barrett BT, McGraw PV, Murray LA, Murgatroyd P. Anterior chamber depth measurement in clinical practice. Optom Vis Sci 1996;73:482-6.
Cohen MV, Yang XM, Neumann T, Heusch G, Downey JM. Favorable remodeling enhances recovery of regional myocardial function in the weeks after infarction in ischemically preconditioned hearts. Circulation 2000;102:579-83.
De Bondt P, Nichols K, Vandenberghe S, Segers P, De Winter O, Van de Wiele C, et al. Validation of gated blood-pool SPECT cardiac measurements tested using a biventricular dynamic physical phantom. J Nucl Med 2003;44:967-72.
Wackers FJ. Equilibrium gated radionuclide angiocardiography: Its invention, rise, and decline and comeback? J Nucl Cardiol 2016;23:362-5.
Wackers FJT. Has LVEF changed beyond chance? Limits of agreement of radiotracer-derived LVEF. J Nucl Cardiol 2015;22:1244-6.
Mitra D, Basu S. Equilibrium radionuclide angiocardiography: Its usefulness in current practice and potential future applications. World J Radiol 2012;4:421-30.
Jensen MM, Schmidt U, Huang C, Zerahn B. Gated tomographic radionuclide angiography using cadmium-zinc-telluride detector gamma camera; comparison to traditional gamma cameras. J Nucl Cardiol 2014;21:384-96.
Wright GA, Thackray S, Howey S, Cleland JG. Left ventricular ejection fraction and volumes from gated blood-pool SPECT: comparison with planar gated blood-pool imaging and assessment of repeatability in patients with heart failure. J Nucl Med 2003;44:494-8.
Wells RG, Marvin B, Kovalski G, Ruddy TD. Planar radionuclide angiography with a dedicated cardiac SPECT camera. J Nucl Cardiol 2013;20:358-66.
Massardo T, Gal RA, Grenier RP, Schmidt DH, Port SC. Left ventricular volume calculation using a count-based ratio method applied to multigated radionucide angiography. J Nucl Med 1990;31:450-6.
Hashemi-Zonouz T, Wu J, Sandoval V, Allahverdi SH, Fazzone-Chettiar R, Liu Y-H. A new approach to quantification of end-diastolic volume and ejection fraction from SPECT equilibrium radionuclide angiocardiography: Methodology and phantom validation. J Med Biol Eng 2019;39:393-402.
Lee F, Fetterman R, Zaret B, Wackers F. Rapid radionuclide-derived systolic and diastolic cardiac function using cycle-dependent background correction and Fourier analysis. Proc Comput Cardiol 1985;8:443-6.
Ficaro EP, Lee BC, Kritzman JN, Corbett JR. Corridor4DM: The Michigan method for quantitative nuclear cardiology. J Nucl Cardiol 2007;14:455-65.
Garcia EV, Faber TL, Cooke CD, Folks RD, Chen J, Santana C. The increasing role of quantification in clinical nuclear cardiology: The Emory approach. J Nucl Cardiol 2007;14:420-32.
Germano G, Kavanagh PB, Slomka PJ, Van Kriekinge SD, Pollard G, Berman DS. Quantitation in gated perfusion SPECT imaging: The Cedars-Sinai approach. J Nucl Cardiol 2007;14:433-54.
Van Kriekinge SD, Berman DS, Germano G. Automatic quantification of left ventricular ejection fraction from gated blood pool SPECT. J Nucl Cardiol 1999;6:498-506.
Vanhove C, Franken PR. Left ventricular ejection fraction and volumes from gated blood pool tomography: Comparison between two automatic algorithms that work in three-dimensional space. J Nucl Cardiol 2001;8:466-71.
Vanhove C, Walgraeve N, De Geeter F, Franken PR. Gated myocardial perfusion tomography versus gated blood pool tomography for the calculation of left ventricular volumes and ejection fraction. Eur J Nucl Med Mol Imaging 2002;29:735-41.
Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;1:307-10.
Wood PW, Choy JB, Nanda NC, Becher H. Left ventricular ejection fraction and volumes: it depends on the imaging method. Echocardiography 2014;31:87-100.
Konstam MA, Kramer DG, Patel AR, Maron MS, Udelson JE. Left ventricular remodeling in heart failure: current concepts in clinical significance and assessment. JACC Cardiovasc Imaging 2011;4:98-108.
Solomon SD, Glynn RJ, Greaves S, Ajani U, Rouleau JL, Menapace F, et al. Recovery of ventricular function after myocardial infarction in the reperfusion era: The healing and early afterload reducing therapy study. Ann Intern Med 2001;134:451-8.
Jovin IS, Ebisu K, Liu YH, Finta LA, Oprea AD, Brandt CA, et al. Left ventricular ejection fraction and left ventricular end-diastolic volume in patients with diastolic dysfunction. Congest Heart Fail 2013;19:130-4.
Konstam MA, Udelson JE, Anand IS, Cohn JN. Ventricular remodeling in heart failure: A credible surrogate endpoint. J Card Fail 2003;9:350-3.
Acknowledgements
The authors would like to thank Drs. Maria J. Salas, Raymond Russell, Taraneh Hashemi-Zonouz, and Sule Hur Allahverdi for their guidance and technical assistance in this patient study.
Disclosures
This study was supported in part by a Research Grant (16-00248, LIU) from the Connecticut Bioscience Innovation Fund (Connecticut Innovations, Rocky Hill, CT). The other co-authors of this paper have nothing to disclose pertaining to the present study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors of this article have provided a PowerPointfile, available for download at SpringerLink, which summarizes the contents of the paper and is free for re-use at meetings and presentations. Search for the article DOI on SpringerLink.com.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, YH., Fazzone-Chettiar, R., Sandoval, V. et al. New approach for quantification of left ventricular function from low-dose gated bloodpool SPECT: Validation and comparison with conventional methods in patients. J. Nucl. Cardiol. 28, 939–950 (2021). https://doi.org/10.1007/s12350-019-01823-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12350-019-01823-8