Abstract
Zinc (Zn) is a vital micronutrient from the perspective of biofortification and biotic stress endurance in pigeonpea. The ZIP transporters with domain (Pfam: PF02535) regulate uptake and transport of metal ions, including Zn, in consonance with plant metal homeostasis. Genome-wide analysis in pigeonpea identified 19 non-redundant members of ZIP family (CcZIP) that were analyzed for gene structure, conserved motifs and homology besides other structural and biochemical parameters. Intra-specific as well as the inter-specific phylogenetic relationships of these 19 CcZIPs were elucidated by comparison with ZIP proteins of Arabidopsis thaliana, Medicago truncatula, Phaseolus vulgaris and Glycine max. In addition to gene structure, the cis-regulatory elements (CREs) in the promoter region were also identified. It revealed several stress responsive CREs that might be regulatory for differential expression of CcZIP proteins. Expression analysis showed that both CcZIP3 and CcZIP15, having zinc deficiency responsive element, up-regulated in the reproductive leaf tissues and down-regulated in matured green pods of the pod borer resistant genotypes with higher zinc content. Alternately, the expression of CcZIP6 and CcZIP13 was higher in matured green pods than reproductive leaves of the resistant genotypes. These findings on differential expression indicate the possible role of these CcZIPs on the mobilization of Zn from leaves to pods, phloem loading and unloading, and higher accumulation of seed zinc in pod borer resistant genotypes used in this study. Further functional characterization of CcZIP genes could shed light on their role in bio-fortification and genetic improvement to inhibit the pod borer herbivory in pigeonpea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pigeonpea [Cajanus cajan (L.) Millsp.; Family: Fabaceae] is a grain legume crop, and this crop has been growing predominantly in India with an annual production of 3.3 million tons which is more than 70% of global production (FAOSTAT 2017). The yield of pigeonpea is compromised because of the losses incurred due to several biotic and abiotic stresses, and among the biotic stresses pod borer complex (a group of pod borers) causes huge loss in the net production (Grover and Pental 2003; Sharma et al. 2010). An earlier study has reported that pigeonpea cultivars with high zinc and iron content could confer tolerance to pod borer infestation and was also a deterrent to pod borer herbivory (Kaur et al. 2014). The dehulled split grain of dried matured seeds and tender green pods are consumed along with cereal foods as a source of dietary proteins in many parts of the world, including India. Nearly 48% of the human population, those dependent upon vegetarian diets, suffer from zinc deficiency. Most of them showed diverse symptoms, including low immunity, retarded brain development, abnormal child growth, and delayed cognitive development (Hambidge 2000; Clemens 2014). Thus, bio-fortification of zinc in the edible parts, including seeds of pigeonpea is quite important not only to combat the malnourishment of zinc (Zn) micronutrient but also to increase the plant immunity against biotic stresses.
Plant growth and development requires an optimal amount of Zn supply from the soil to drive several metabolic processes, such as photosynthesis, cellular respiration, growth and differentiation etc., as this micronutrient serves both structural and catalytic roles either by acting as a structural component of several proteins and enzymes or catalyzing cellular functions by binding as cations and transporting across the membranes (Sinclair and Krämer 2012). Zn is the important structural component of zinc finger proteins, and these transcription factors were upregulated in response to both abiotic and biotic stresses in several plant systems, including Arabidopsis thaliana, wheat and potato, and in turn, their expressions make them stress-tolerant (Schweizer et al. 2013; Lawrence et al. 2014; Zang et al. 2016; Wang et al. 2017). Moreover, Zn as a cofactor also acts as a protective agent against the oxidation of several cell components, including membrane proteins, metalloproteins and lipids, and has defensive properties against several biotic and abiotic stresses in different crop plants (Cabot et al. 2019). One such metalloprotein and the anti-oxidative enzyme ‘superoxide dismutase’ contain Zn as a cofactor. It catalyzes the dismutation of superoxide radicals to molecular oxygen and H2O2 to induce the plant primary immune response which further regulates several intra- and inter-cellular signaling pathways required for plant defense system under zinc stress conditions (Cabot et al. 2019). Even after insect and pathogen attack, the plants with high and low zinc content respond differently by altering alcohol dehydrogenase and carbonic anhydrase activities, and overexpression of metallothionein and proteins for reactive oxygen species (ROS) detoxification (Wongpia and Lomthaisong 2010; Shivashankar et al. 2015; Zang et al. 2016; Ćalić et al. 2017). Zinc content in edible plant parts also inhibits gut amylase of the pod borer (Helicoverpa armigera) in pigenopea. The content of Zn was also reported to be higher in the seeds and pods of moderately resistant cultivars than those of susceptible cultivars in pigeonpea (Kaur et al. 2014).
The plants obtain Zn from the soil as divalent cations and transport to the seeds through a cascade of intercellular symplast to symplast movement via various apoplastic spaces, and it involves solubilization of Zn in the soil, Zn uptake into roots, xylem loading in roots and unloading in leaves, phloem loading and phloem unloading in the developing seeds (Olsen and Palmgren 2014; Lira-Morales et al. 2019). Zinc homeostasis in both strategy-I and -II model plants, A. thaliana and Oryza sativa, had been already documented (Ishimaru et al. 2011; Bashir et al. 2012; Sinclair and Krämer 2012; Olsen and Palmgren 2014; Ricachenevsky et al. 2015; Kawakami and Bhullar 2018; Pita-Barbosa et al. 2019; Lira-Morales et al. 2019). Although the molecular mechanisms and genes involved during solubilization and uptake of zinc, xylem loading in roots and phloem loading for transport and distribution into plant organs and subcellular compartments have been unfolded in model plants, the genes responsive to post phloem loading and unloading of zinc to developing seeds in dicots have not been elucidated till date (Ricachenevsky et al. 2015; Kawakami and Bhullar 2018; Pita-Barbosa et al. 2019). As with other metal ions, the acquisition and cellular movement of zinc require various chelators and membrane-bound transporters to import and export across the biological membranes, including tonoplast and chloroplast (Bashir et al. 2016; Vigani and Hanikenne 2018). Several such membrane proteins capable of uptake and transport of metal ions in consonance with plant metal homeostasis have been characterized in A. thaliana and O. sativa (Ricachenevsky et al. 2015; Kawakami and Bhullar 2018; Lira-Morales et al. 2019), which include members of the zinc-regulated transporter (ZRT) and iron-regulated transporter (IRT) like protein (ZIP), cation diffusion facilitator (CDF), plant cadmium resistance (PCR), vacuolar iron transporter (VIT), and heavy metal ATPase (HMA) gene families. Lira-Morales et al. (2019) also proposed a ZIP protein regulation pathway in A. thaliana under different conditions with emphasis on the basic region of leucine zipper (bZIP) transcription factors, the response of bZIP to Zn availability and occurrence of zinc-deficiency-related cis-elements (ZDRE).
Among all the members, ZIP gene family are the first heavy metal transporters reported in plants, and their expression is mainly regulated by the Zn content in the plant parts and soil (Grotz et al. 1998; Ricachenevsky et al. 2015). The plant ZIP proteins have 6–9 transmembrane domains, with eight being the predominant ones, histidine-rich variable loop between III and IV transmembrane domains, and their carboxyand amino terminal ends are mainly at the outer surface of the plasma membrane (Guerinot 2000). The genes with ZIP domain have been identified and characterized from several plants, including A. thaliana, O. sativa, Medicago truncatula, Phaseolus vulgaris, Zea mays, Setaria italica and Poncirus trifoliata (Grotz et al. 1998; Chen et al. 2008; Stephens et al. 2011; Astudillo et al. 2013; Li et al. 2013; Jain et al. 2013; Mondal et al. 2014; Alagarasan et al. 2017; Fu et al. 2017). The possible role of these ZIP proteins have been elucidated in uptake and transport of Zn cations to the cytoplasm and their translocation to different plant parts (Sinclair and Krämer 2012; Kawakami and Bhullar 2018). Such functions of ZIP genes are also affirmed through gene expression analysis and yeast complementation studies (Krishna et al. 2020). Even the analysis of ZIP gene family in different species also demonstrated their importance in Zn uptake, transport and accumulation under zinc-deficient conditions (Assunção et al. 2010; Stephens et al. 2011; Jain et al. 2013; Astudillo-Reyes et al. 2015; Lilay et al. 2020). One member of the ZIP family (PvZIP12) was overexpressed in the P. vulgaris genotypes with higher zinc content (Astudillo-Reyes et al. 2015). In M. truncatula, three genes (MtZIP1, MtZIP5 and MtZIP7) regulating the zinc transport and homeostasis have also been identified (Stephens et al. 2011). Jain et al. (2013) also hypothesized the transcriptional regulation of AtZIP4, AtZIP9 and AtZIP12 by Zn in soil and their role in zinc homeostasis in A. thaliana.
As reported earlier, the decoded genome sequence is quite essential for functional genomics research including in silico characterization of various gene families in different plant systems. The draft genome sequence of the C. cajan also provides a valuable resource for in silico analysis of the gene families in this species. Although several gene families have already been characterized in C. cajan (Malviya et al. 2015; Singh et al. 2019), the genome-wide analysis of the ZIP gene family is not yet reported in pigeonpea. Therefore, we describe here the in silico identification of non-redundant members of genes containing ZIP domain; characterization of their biochemical properties, genomic organization and motif analysis; elucidation of intra- and inter-specific phylogenetic relationship; and expression analysis of ZIP genes in the genotypes with contrasting host response to pod borer and seed zinc content.
Materials and methods
In silico identification and characterization of CcZIP genes
The complete sequence assembly, coding sequence and protein sequence of pigeonpea were downloaded from the public database (http://gigadb.org/dataset/100028), and a total of 48,680 predicted genes in the C. cajan Gene Model V5.0 along with their location on the genome (chromosomes and scaffolds) were loaded to a local MySQL database for easy retrieval during the present study. The Hidden Markov Model (HMM) profile of the ZIP domain (PF02535) from the Pfam database was used as a query for the identification of ZIP proteins in C. cajan and their corresponding genes (CcZIP) were also obtained from Gene Model V5.0 using the HMMER 3.3 programme with E-value ≤ 10–3 (El-Gebali et al. 2018). Further, the identified CcZIP proteins were also searched against the conserved domain database (CDD) and ensured that each CcZIP must have the ZIP domain PF02535 as described earlier (Marchler-Bauer et al. 2015). Various biochemical parameters, such as length of the protein sequence, molecular weight, isoelectric point (pI) and grand average of hydropathicity (GRAVY) values of these CcZIP proteins were determined using protein analysis module in Bio-Python (Chapman and Chang 2000). All 19 predicted CcZIP proteins were aligned using the multiple sequence comparison by log-expectation (MUSCLE) programme in MEGA-X software to exclude overlapping CcZIP genes, if any (Edgar 2004; Kumar et al. 2018). Multiple sequence alignment was further carried out using MCOFFE to explore the relationship between the CcZIP paralogs (Moretti et al. 2007), and subsequently, TMCOFFE was used to align the transmembrane domains with discovering the regions present in the helix, outside and inside of the plasma-membrane (Floden et al. 2016). The topology and distribution of transmembrane helices (TMHs), pore-lining helices (PLHs) and signal peptides were determined by using MEMPACK of PSIPRED protein analysis workbench (Nugent et al. 2011), and the findings were also affirmed by reanalyzing the occurrence of TMHs and signal peptides using TMHMM and SignalP 4.1 server, respectively (Krogh et al. 2001; Petersen et al. 2011). The post-translational modification signature sequences of the CcZIP proteins were also determined using ScanProsite (de Castro et al. 2006). Subcellular localization of each CcZIP gene were also predicted using ProtComp v9.0 programme. The nomenclature of CcZIP genes (CcZIP1 to CcZIP19) was given as per their occurrence on pigeonpea chromosome (CcLG01 to CcLG11) followed by sequence scaffolds (Scaffold-000020 to Scaffold-137616) in ascending order following the genome information (Varshney et al. 2012).
Determination of CcZIP gene structure
The predicted coding sequences (CDS) of CcZIP genes were submitted to the gene structure display server, GSDS 2.0, along with the phylogenetic tree of the CcZIP paralogs to determine the genomic organization of exon and intron (Hu et al. 2014). Further, the promoter sequences from −1000 to +1 bp of each of the CcZIP genes were extracted from the pigeonpea sequence assembly, and were analyzed for cis-regulatory elements (CREs) and functional motifs in the promoter using the Softberry Nsite-PL programme along with RegSite PL database of plant regulatory elements (Shahmuradov and Solovyev 2015). The position of significant stress-responsive CREs were graphically represented using GSDS 2.0.
Analysis of conserved motifs in CcZIP proteins
The conserved motifs present in 19 CcZIP proteins were identified using the MEME 4.11.2 Suite server considering the parameters, viz. occurrence of motifs with any number of repetitions, the length of motifs should be between 6 and 60 amino acids, only motifs with E-value ≤ 10−20 and a maximum of 10 motifs per sequence (Bailey et al. 2009). The occurrence of individual motifs in CcZIP sequences was also obtained using MAST module of MEME 4.11.2 Suite. The functional annotations of these motifs were carried out using the InterProScan (Quevillon et al. 2005), and the sequence logos of conserved motifs across 19 CcZIPs were also generated using WebLogo (Crooks et al. 2004).
Phylogenetic analysis of CcZIP proteins
Non-redundant ZIP proteins with domain PF02535 of three legumes viz. M. truncatula, G. max and P. vulgaris were retrieved from the Phytozyme database (www.phytozome.net; Supplementary Table 1). Subsequently, the conserved sequences of these retrieved proteins along with the A. thaliana ZIP proteins and 19 CcZIPs were aligned using the MUSCLE programme in MEGA X software with default parameters (Kumar et al. 2018). Based on their conserved sequence alignment, the rooted phylogenetic tree was also constructed following maximum likelihood method using MEGA-X software (Kumar et al. 2018). The interspecific evolutionary relationships of 19 CcZIPs with the ZIP family of A. thaliana and three legumes mentioned above were also delineated. In contrast, the intra-specific phylogenetic relationships among 19 CcZIP genes were established on the basis of the ZIP domain only, keeping the rest of the parameters unchanged.
In silico expression patterns for CcZIP
In sillico tissue-specific expression patterns of CcZIPs, their transcript abundance data (expressed as log2 transformed FPKM) were retrieved from the C. cajan gene expression atlas (CcGEA; Pazhamala et al. 2017). The expression profile of all CcZIP transcripts, except CcZIP10 (due to the unavailability of its expression data in CcGEA), in thirty different tissues of C. cajan were visualized as the heat map.
Differential expression analysis of CcZIP genes by qRT PCR
The expression levels of CcZIP transcripts were determined by quantitative real-time polymerase chain reaction (qRT-PCR) analysis among the genotypes with contrasting host response to pod borer and seed zinc content, viz. C. cajan cv. ICPL87 (susceptible control), C. cajan acc. ICP28, C. cajan acc. ICP-26, C. scarabaeoides acc. ICPW90 and C. scarabaeoides acc. ICPW94. The matured green pods and reproductive apical leaves (during the first flush of flowering) were sampled using liquid nitrogen from the plants grown in the greenhouse with identical soil and environmental conditions. Total RNA was extracted from 100 mg of matured green pod and reproductive leaf tissues using G-Sure RNA extraction kit (GCC Biotech, India) following manufacturer’s instructions. The quality and yield of RNA (DNAase-treated) were determined by Nanodrop 2000 spectrophotometer (Thermo Scientific, USA), and was validated by agarose gel (1.4% v/v) electrophoresis in 3-N-morpholino-propane sulfonic acid (MOPS) buffer. The purified RNA (1 μg) was used for the synthesis of c-DNA using 1 μl of Verso reverse transcriptase enzyme (200 U l−1), dNTP mix (2.5 mM each dNTP) and 250 ng oligo(dT) primer following the instructions of Verso cDNA synthesis kit (Thermo Scientific, USA). The integrity and quality of cDNA were affirmed by tubulin (TUB6, Gene ID: B9R897) gene amplification using master mix (Qiagen India Pvt. Ltd, India) and primers (F: 5′GCCCTGACAACTTCGTCTTC3′ and R: 5′GCAGTTTTCAGCCTCTTTGC3′ (Sinha et al. 2015). The resulting cDNA samples were diluted in nuclease-free water (1:10), and 2 μl of the diluted cDNA sample was used for quantitative real-time assay using Quanti-Fast SYBR green PCR master mix (Qiagen India Pvt. Ltd, India) and the primers described in the Supplementary Table 2 to determine the relative expression of CcZIP genes among the genotypes with contrasting host response to H. armigera along with susceptible control. The quantitative RT-PCR analysis was performed in 96-well plate using CFX connect Real-Time PCR detection system (Biorad, USA) with a total reaction volume of 10 μl (2 μl of cDNA, 5 μl of SYBR Green PCR master mix, 0.5 μl of each forward and reverse primer, and 2 μl of ddH2O). Thermal cycling conditions involved a pre-incubation at 95 °C for 7 min followed by 35 cycles of 3-step amplification at 95 °C for 10 s, 55 °C for 20 s and 72 °C for 20 s. The expression of CcZIP genes in the samples was normalized with that of TUB6 as an endogenous control (Sinha et al. 2015). The qRT PCR analysis was performed with three technical and three biological replicates, and the specificity of qRT PCR assay was confirmed by melting curve analysis. The relative expression of CcZIP transcripts in each sample was calculated by measuring ΔCT value for each of the CcZIP genes with respect to the endogenous control ‘TUB6’ and was represented as 2−ΔCT value (Livak and Schmittgen 2001).
Estimation of zinc content
Matured green pods and reproductive apical leaves from the five genotypes were harvested in triplicate and air-dried. The dried samples were ground to a fine powder using bio-homogenizer (Pelican Equipment Ltd., Chennai, India), and 0.5 g of the powder of each sample was digested in an infra digestion system (KES12IL, Pelican Equipment, Chennai, India) using triple acid mix (9 nitric acid: 2 sulphuric acid: 1 perchloric acid) till the colourless solution was obtained. The colourless solution was made up to 100 ml using deionized water and filtered through Whatman filter paper (No. 40). The absorbance of the solution was measured using Atomic Absorption Spectrophotometer (Varian AA240, Varian, Palo Alto, CA, USA), and the content of zinc was estimated using the standard curve method (Kundu et al. 2017).
Results and discussion
The micronutrient zinc either acts as the cofactor or as a structural component in functional subunits of several proteins and enzymes essential for the growth and development of plants (Sinclair and Krämer 2012). In addition, zinc protects plant cells from oxidative stress-mediated by scavenging ROS and plays a significant role in plant signalling as an intracellular secondary messenger (Yamasaki et al. 2007; Cabot et al. 2019). The acquisition, uptake, distribution and accumulation of Zn in dicot plants involves several members of heavy metal transporter gene families. Thus characterization of these gene families is essential to understand the homeostasis as well as biofortification of zinc in plant parts. This study reports the identification and characterization of 19 members of ZIP family transporters (CcZIPs) in C. cajan, and their possible role on seed Zn content.
In silico identification of CcZIP genes and their characterization
The genome-wide search using the ZIP domain (PF02535) as query against the 48,680 C. cajan protein sequences (Gene Model V5.0) identified 19 non-redundant CcZIP genes containing the ZIP domain with an E-value 10–3 or less (Table 1). These were numbered as CcZIP1 to CcZIP19 based on their location in the chromosomes, followed by sequence scaffolds in ascending order. Among the CcZIP genes, six were localized in four chromosomes of pigeonpea (CcLG-01, -03, -09 and -10), and the rest 13 CcZIP genes were identified in the sequence scaffolds (Table 1). The occurrence of CcZIP genes is attributed to the non-inclusion of several sequence scaffolds to pigeonpea chromosomes (CcLG01 to -11) due to coverage of 72.6% of genomic sequence in the first draft genome sequence of pigeonpea (Varshney et al. 2012).
The full-length CDS of CcZIP genes containing open reading frames (ORF) were ranged between 252 to 598 amino acids, and the molecular weight of CcZIP proteins also varied from 26.5 kDa (CcZIP11) to 62.1 kDa (CcZIP8). Among them, size of only three CcZIP proteins (CcZIP2, CcZIP4 and CcZIP8) appeared to be quite large beyond the molecular weight of 50 KDa (Table 1). The predicted length of 19 CcZIP genes was found to be varied between 795 bp (CcZIP11) and 15,312 bp (CcZIP6), and their ORF length varied between 759 bp (CcZIP11) and 1797 bp (CcZIP8). All 19 CcZIP genes were predicted to be functional in pigeonpea because they all have an initiation codon and culminate with a stop codon. The theoretical pI values of CcZIP proteins were estimated to be within the range of 5.82 (CcZIP12) to 9.39 (CcZIP07), and these acidic or basic properties of CcZIP proteins might influence the differential response of pigeonpea to the abiotic and biotic stresses encountered during growth and development (Allagulova et al. 2003). The amino acid composition analysis revealed that all the CcZIP proteins have a positive GRAVY value (0.239–0.821) except CcZIP4 (− 0.082; Table 1) which could be attributed to the presence of hydrophobic amino acids in the membrane-spanning ZIP proteins as reported in several plant species (Grotz et al. 1998; Astudillo et al. 2013; Mondal et al. 2014; Krishna et al. 2020).
The PROSITE analysis of these CcZIP proteins predicted several peptide sequences concerned with post-translational modifications and other specific features, such as N-glycosylation site, phosphorylation site, and N-myristoylation, amidation and ATP synthase-A signature sequences (Table 2). In this study, N-myristoylation signature sequence was predominantly present in all CcZIP proteins (six in CcZIP16 to 24 in CcZIP8) as the significant post-translational modification sequence. In addition, multiple phosphorylation sites have been documented in CcZIP proteins similar to the ZIP genes in several legume plants and model species (Krishna et al. 2020; Tingholm et al. 2020). These signature sequences and post-translation modification sites might act as substrates for several kinases, including casein kinase II, protein kinase-C, cAMP- kinase and cGMP- kinase. The predominance of N-myristoylation sequence in CcZIP proteins could be attributed to their role in conformational stability and modulation of different functions in many cellular pathways, especially during signal transduction, inter-cellular export and membrane transport of zinc and other heavy metals in pigeonpea as evident in different plants (Zaun et al. 2012; Mondal et al. 2014).
Analysis of gene structure, promoter sequences and motifs of CcZIPs
Similar to other gene families, the structural diversity among the members of the ZIP gene family depends on the number of introns and exons and their length. In terms of the exon–intron arrangement, the members CcZIPs were highly diverse, and the number of introns varied from zero in CcZIP11 to a maximum of 11 introns in CcZIP5 (Supplementary Fig. 1). Moreover, this variation in gene sequence was also well reflected in the multiple sequence alignment plots among the members CcZIP proteins (Fig. 1), whose length was varying between 252 and 598 AA with a minimum similarity of 15.82% between CcZIP4 and CcZIP8 to a maximum 94.65% identity between CcZIP13 to CcZIP18 protein (Supplementary Table 3). The structural diversity of CcZIP proteins were also well corroborated with the earlier report on the diversity of ZIP proteins in A. thaliana, P. vulgaris, O. sativa, Z. mays and P. trifoliata (Grotz et al. 1998; Chen et al. 2008; Astudillo et al. 2013; Mondal et al. 2014; Fu et al. 2017; Krishna et al. 2020). Such structural diversity of ZIP genes across species could be attributed to their multifunctional role, particularly in cellular movement and transport of several cations (Krishna et al. 2020). In addition, the diversity of ZIP proteins suggests different responses during adaptation to biotic and abiotic stresses encountered during the growth and development of different plant species (Cabot et al. 2019).
The MEMPACK of PSIPRED, TMHMM server and SignalP 4.1 server delineated the number of transmembrane helices (TMHs), pore line helices (PLHs) and signal peptides present in the CcZIP proteins (Table 3; Fig. 2a). The number of TMHs present in the maximum number of CcZIP proteins ranged from 6 to 9 as expected (Guerinot 2000; Li et al. 2013) baring three viz. CcZIP2 (13), CcZIP8 (13) and CcZIP10 (5). These increased or decreased number of TMHs in ZIP proteins could be attributed to either sequence duplication or deletion followed by genomic reorganization (D’Ovidio et al. 2004). Similar variation in the number of TMHs had also been reported in maize (Mondal et al. 2014). As expected, most of the TMHs of CcZIP protein embedded varying numbers (2–5) of PLHs, which might facilitate the sensing and movement of ions across the membranes in response to different stress (Maksaev et al. 2018). Moreover, 11 CcZIPs (CcZIP2, CcZIP4, CcZIP6, CcZIP8, CcZIP9, CcZIP10, CcZIP14, CcZIP15, CcZIP16, CcZIP17 and CcZIP19) possessed signal peptide sequence (Table 3), which suggest the involvement of these CcZIP proteins in the cellular movement of heavy metal ions including Zn2+ as reported in several plant species (Krishna et al. 2020). ProtCompv9.0 analysis showed that most of the CcZIP proteins are located in the plasma membrane, except for CcZIP5 and CcZIP7 proteins, which were predicted to be in endoplasmic reticulum (Table 3). Similar to the present findings, the ZIP transporters were also located in different cell organelles in different species, and this is mostly attributed to their role in zinc homeostasis in plants concerning low and high zinc soils as well as a stress response (Tiong et al. 2015; Bashir et al. 2016; Cabot et al. 2019).
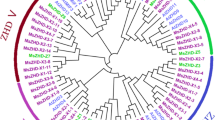
Intraspecific relationship and structure of CcZIP proteins and functional motifs of 19 CcZIPs. (a) A phylogenetic tree with 100 bootstrap replications along with schematic protein sequence showing transmembrane helices, pore lining helices and signal peptide sequences, (b) Conserved motif composition of each CcZIP protein. Motifs 1–10 are displayed as differently colored boxes with the corresponding sequence information for each motif
The cis-regulatory elements (CRE) are important signature sequences involved in transcriptional regulation of genes and remain very important during plant growth and development during several biotic and abiotic stresses in their habitat by modulating gene expression. Analysis of the putative promoter sequence of CcZIP genes, 1,000 bp upstream of the transcription initiation site using Nsite-PL detected an array of CREs regulating plant development, plant hormonal response, biotic and abiotic stress induction, and secondary metabolism in addition to basal gene expression (Supplementary Table 4; Supplementary Fig. 2). The promoter of CcZIP16 gene contains the maximum number (43) of CREs, whereas CcZIP7 has only eight CREs. Some of these predicted CREs are stress-responsive, such as metal regulatory element (MRE), light regulatory element (LREs), ABA-responsive elements (ABRE), zinc deficiency-related elements (ZDRE), low-temperature responsive elements (LTRE) and TGACG-motif. In addition, some of the hormone signalling stress responsive CREs, such as methyl jasmonate responsive element (MejA-RE), ethylene responsive element (ERE) and auxin responsive element (ARE) (Supplementary Table 4; Fig. 3). The promoter sequences of two members of pigeonpea ZIP family, CcZIP3 and CcZIP15, also have ZDRE cis-element as reported earlier in the members of ZIP family of A. thaliana and Z. mays, and these ZDREs might regulate the transcription of ZIP gene(s) under zinc deficiency stress (Assunção et al. 2010; Jain et al. 2013; Mondal et al. 2014). The putative CREs for biotic and abiotic stress induction were predicted at multiple sites in the promoter regions of CcZIP genes reported in soybean. These CREs might have been involved in pathogen infection, herbivory attacks and abiotic stress response (Wang et al. 2015). In addition to stress-responsive CREs, several cis-elements associated with cellular development, plant metabolism, cell cycle regulation and hormonal development were also predicted. The presence of these CREs is indicative of their possible involvement in regulating CcZIP gene expression during cellular growth and development in response to different biotic and abiotic environments (Cabot et al. 2019). The motif analysis using MEME suite also identified 10 conserved motifs, containing 29–60 amino acid residues, among the CcZIP proteins and the prime function of these motifs were annotated to be zinc transporter, iron transporter and a component of TMH (Fig. 2b; Supplementary Table 5). Similar conserved motifs for cation transport are also reported in ZIP proteins of A. thaliana, P. vulgaris, O. sativa and Z. mays (Chen et al. 2008; Astudillo et al. 2013; Mondal et al. 2014; Krishna et al. 2020). The sequence logo of these conserved motifs generated by WebLogo is presented (Supplementary Fig. 3). As described above, most of the CcZIPs were found to have 6–9 TMHs with an average of 7.49, and these predicted TMHs have C-terminal and N-terminal ends present inside and outside the surface of the plasma membrane, respectively. Another important feature of the ZIP proteins is a variable hydrophobic loop located between TMH-III and -IV (Guerinot 2000), and this feature was also quite evident among the 19 members CcZIP proteins (Fig. 2). This variable hydrophobic loop is characterized by conserved histidine residues (H-x-H-x-H) and is predicted to be the cytoplasmic metal ion binding site (Eng et al. 1998; Guerinot 2000). Similar to AtZIP7, AtZIP8 and AtZIP11 in A. thaliana, MtZIP1 and MtZIP7 in M. truncatula, and PtZIP2 in P. trifoliata, CcZIP11 and CcZIP12 lack this His-rich region between TM-III and TM-IV, and in lieu of this His residues were found in their TM-III as an alternative metal-binding site as reported in A. thaliana and P. trifoliata (Eng et al. 1998; Fu et al. 2017). Four CcZIP proteins (CcZIP2, CcZIP5, CcZIP7 and CcZIP8) did not have His rich region (H-x-H-x-H), and instead polar amino acid residues were present adjacent to either TM-III or TM-IV domains as reported in Z. mays, P. vulgaris and Solanum tuberosum (Astudillo et al. 2013; Mondal et al. 2014; Li et al. 2020). It was also reported that either deletion or mutation in H-x-H-x-H region of TjZNT1 (a ZIP transporter) did not affect Zn2+ and Cd2+ transport activity, rather it increased the specificity for Zn2+ in Thlapsi japonicum (Nishida et al. 2008). Moreover, Kawachi et al. (2008) showed that yeast cell containing mutant AtMTP1, a vacuolar Zn2+/H+ antiporter of A. thaliana lacking 32 residues in the histidine rich loop, became hyper-resistant to Zn2+ and resistant to Co2+. Contrasting to these reports, three ZIP genes in P. vulgaris (PvZIP6, PvZIP7 and PvZIP18) lacking these His rich region (H-x-H-x-H) remained non-functional (Astudillo et al. 2013). CcZIP4 has more significant number of histidine residues in the variable loop between TM-III and TM-IV, and it implies multiple metal ion binding abilities as reported in the case of StZIP12 of S. tuberosum (Li et al. 2020). Kawachi et al. (2008) also proposed that the histidine-rich loop of AtMTP1 functions as buffering pocket of Zn2+ and acts as a sensor to determine zinc level at the cytoplasmic surface. These structural variations of CcZIP proteins are also well corroborated with the earlier studies on ZIP proteins in several dicots and monocot species (Chen et al. 2008; Astudillo et al. 2013; Mondal et al. 2014; Fu et al. 2017; Krishna et al. 2020; Li et al. 2020).
Phylogenetic analysis of CcZIP proteins
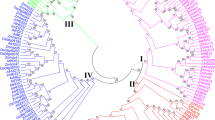
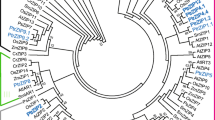
The intraspecific genetic relationship among 19 CcZIP proteins of pigeonpea depicted in a phylogenetic tree showed the clustering of these CcZIP proteins into six major groups, mostly engendered during evolution to the subgroups identified by motif analysis (Fig. 2a). Similar kind of intraspecific diversity of ZIP genes were also made in P. vulgaris, Z. mays and S. tuberosum (Astudillo et al. 2013; Mondal et al. 2014; Li et al. 2020). The interspecific evolutionary relationships of ZIP family members of C. cajan along with three other legume species (G. max, P. vulgaris and M. truncatula) and model dicot A. thaliana was deduced, and consequently a phylogenetic tree was drawn (Fig. 4). This phylogenetic tree grouped 101 genes containing ZIP domain (PF02535) into three major clusters (A, B and C), and major cluster-A has eight sub-clusters (A1–A8) which mostly corroborated with the presence of conserved motifs barring few cases (Fig. 4). In this study, we considered ten conserved motifs (MEME-1 to 10) generated by MEME suite and among them, seven (MEME-1 to -5, MEME-9 and -10) were annotated to be the core component of zinc transporter protein (Supplementary Table 5) because of their involvement in the binding and transport of Zn cations as reported in other plants (Krishna et al. 2020). The 19 CcZIP genes fell under two (A and B) out of three clusters in the interspecific phylogenetic tree (Fig. 4). The sub-cluster-A1 consisted of 14 genes containing ZIP domains, including three CcZIP members (CcZIP4, CcZIP5 and CcZIP7), all of which possessed MEME-2 to -7 motif in common. Similarly, cluster-B and sub-cluster-A2, -A3, -A4, -A5, -A7 and -A8 contained three to seven common motifs in different permutations and combinations in corroboration with their grouping under phylogenetic trees, respectively. A similar kind of clustering of zinc transporters was also reported in both dicots and monocot plants (Krishna et al. 2020). In some clusters few ZIP genes possessed different motif(s) in addition to common motifs, and this kind of heterogeneous motif distribution has also been noticed during phylogenetic classification of different gene families in several dicots and monocots, including P. vulgaris and Z. mays (Astudillo et al. 2013; Mondal et al. 2014; Krishna et al. 2020).
Expression patterns of CcZIP genes in pigeonpea
In silico expression patterns for 18 CcZIP genes (except CcZIP10) were determined using the C. cajan gene expression atlas (CcGEA; (Pazhamala et al. 2017), and tissue-specific differential expression was observed for all the CcZIP genes analyzed (Fig. 5). Among all, the CcZIP9 and CcZIP16 meagerly expressed in all tissue except reproductive buds and nodules, whereas the CcZIP1, CcZIP2, CcZIP3, CcZIP4, CcZIP5, CcZIP8, CcZIP11 and CcZIP14 showed moderate to high-level expression in both the tissues of C. cajan cv. Asha. The rest of the eight CcZIP genes showed quite heterogeneous and tissue-specific expression. In silico expression analysis of CcZIP proteins under a controlled environment revealed that CcZIP3, CcZIP5, CcZIP6 and CcZIP8 genes consistently showed higher expression (> 30 Log2 transformed FPKM) among four kind of roots, viz. seedling roots (SR), vegetative roots (VR), reproductive roots (RR) and senescence roots (SsR). Whereas CcZIP19 showed higher expression in all three kinds of roots except SsR, CcZIP15 showed comparatively higher expression in both SR and RR, and CcZIP14 showed higher expression in VR and RR. CcZIP4 and CcZIP13 showed higher expression in VR and RR, respectively. In both vegetative and reproductive leaves CcZIP6, CcZIP8 and CcZIP14 showed higher expression, whereas in vegetative leaves, CcZIP4 and CcZIP19, and in reproductive leaves CcZIP3, CcZIP5, CcZIP13, CcZIP15 and CcZIP17 showed higher expression, respectively. Similarly, in both vegetative and reproductive shoot apical meristems (SAM) CcZIP19 is expressed at par, whereas in vegetative SAM CcZIP4 and CcZIP14, and in reproductive SAM CcZIP11 showed higher expression, respectively. Likewise, in both reproductive immature and mature reproductive pods CcZIP5, CcZIP8, CcZIP12 and CcZIP14 are expressed at par, whereas in mature pods, CcZIP3 and CcZIP18 showed higher expression. On comparison of expression of CcZIP genes among different kinds of roots, leaves, SAMs and pods, it was found that CcZIP13 consistently showed higher expression both in reproductive roots and leaves, and CcZIP18 showed higher expression in reproductive matured pods (Supplementary Table 6). The above findings indicate the putative role of CcZIP13 in the acquisition of zinc from the soil and its mobilization and distribution at the onset of the reproductive phase (Jain et al. 2013) and the role of CcZIP18 on phloem unloading in the pods (Olsen and Palmgren 2014). The in silico differential expression of CcZIPs were also well correlated with in vivo expression data of reproductive leaves and matured green pods using qRT-PCR analysis. This varied expression of CcZIPs in both tissues among different genotypes is likely to be associated with the regulation of transcription factors under abiotic environment, stress and stages of development such as bZIP19 and bZIP23 in A. thaliana under Zn deficit condition (Assuncao et al. 2010; Lira-Morales et al. 2019).
In silico analysis showing abundance of 18 CcZIP transcripts in 30 tissues of pigeonpea on the basis of data obtained from gene expression atlas (CcGEA; Pazhmala et al. 2017)
As erstwhile discussed, zinc is the structural and regulatory component of several inter-related metabolic pathways, and it plays an essential role even in embryo and endosperm development (Vallee and Falchuk 1993). The Zinc transporters proteins, particularly P1-B-ATPase pumps, are essential for the export and accumulation of zinc inside the seeds of A. thaliana (Olsen et al. 2016). Even in rice and maize, several ZIP genes regulating cellular movement and phloem unloading of zinc either in the endosperm or in the kernel had been identified (Krishna et al. 2020). Seed zinc content in pigeonpea was also influenced by soil zinc content in a genotype-dependent manner. The higher zinc content in the seeds of C. cajan has been reported to inhibit the pod borer herbivory (Kaur et al. 2014; Cabot et al. 2019). In the present study, two pod borer susceptible genotypes (C. cajan acc. ICP-28 and C. cajan acc. ICP-26) have comparatively lower Zn content in its seeds (11.08 ± 0.42 and 11.51 ± 0.33 µg g−1) at par with susceptible control (10.95 ± 0.4 µg g−1). Whereas the two resistant genotypes (C. scarabaeoides acc. ICPW-94 and C. scarabaeoides acc. ICPW-90), primarily used as the genetic resources for the introgression of pod borer resistance allele, had higher seed Zn content (16.37 ± 0.57 and 15.65 ± 0.61 µg g−1) than susceptible genotypes tested. This finding was also well corroborated with the earlier report on Zn content in pod borer resistant genotypes in pigeonpea (Kaur et al. 2014). Zn-deficient soybean plants also showed increased aphid colonization, and was attributed to a higher accumulation of amino acids caused by reduced protein synthesis under zinc deficiency (Helfenstein et al. 2015; Cabot et al. 2019). Similarly, low Zn status inhibit the expression of defence-related genes, including PR1, through evolutionary conserved Zn sensing mechanism with respect to plant growth vis-à-vis defence (Bouain et al. 2018). The reproductive leaves in dicots play a crucial role in phloem unloading of micronutrients in the seed through pod wall and maternally derived seed coat (Garcia and Grusak 2015) and are also valid for the legumes; thus, it influences the seed nutrition composition. Even the seed development in legumes accomplished by networks of regulatory and metabolic genes of several ontological pathways, which affect its size and composition (Weber et al. 2005), and among them, Zinc finger (CCHC and C2H2) type proteins were associated with seed size (Radkova et al. 2019). Thus, the expression analysis of CcZIP genes was carried out in two different plant parts (reproductive leaves and matured green pods) of four different genotypes, with contrasting host responses to pod borer and heterogeneous seed zinc content, and their involvement in seed zinc accumulation and pod borer herbivory has been hypothesized.
The qRT-PCR analysis revealed that expression of seven CcZIP genes (CcZIP1, CcZIP2, CcZIP4, CcZIP8, CcZIP12, CcZIP14 and CcZIP16) were relatively up-regulated both in the reproductive leaves and matured green pods of susceptible genotypes, whereas CcZIP17 was up-regulated in both tissues of resistant genotypes (Fig. 6). The expression of CcZIP3 and CcZIP15 were up-regulated in the reproductive leaves whereas down-regulated in the matured green pods of resistant genotypes (Fig. 7). In contrast, the expression of CcZIP6 and CcZIP13 was induced in the matured green pods of the resistant genotypes compared to their reproductive leaves (Fig. 7). On comparison of the relative expression of CcZIPs between two tissues of the resistant genotypes, it has been revealed that six CcZIPs (CcZIP4, CcZIP7, CcZIP8, CcZIP15, CcZIP17 and CcZIP19) in reproductive leaves, and 11 CcZIPs (CcZIP1, CcZIP2, CcZIP3, CcZIP6, CcZIP9, CcZIP10, CcZIP11, CcZIP12, CcZIP13, CcZIP14 and CcZIP18) in matured green pods showed higher expression. Similarly, among the susceptible genotypes four CcZIPs (CcZIP4, CcZIP6, CcZIP7 and CcZIP8) showed higher expression in reproductive leaves, whereas 10 CcZIPs (CcZIP1, CcZIP2, CcZIP3, CcZIP9, CcZIP10, CcZIP12, CcZIP14, CcZIP15, CcZIP16 and CcZIP18) in matured green pods. Similar heterogeneous expression of the ZIP genes has already been reported in several plants, including A. thaliana and P. vulgaris (Astudillo et al. 2013; Jain et al. 2013), and this might be attributed to different stress and growth responsive CREs and transcription factors with regards to different genotypes. In P. vulgaris, seven genes were characterized among two genotypes (G19833 and DOR364) under different zinc treatments, and four of the genes (PvZIP12, PvZIP13, PvZIP16 and PvbZIP1) showed differential expression depending upon the type of tissues (roots, leaves and pods), genotype and zinc treatment. PvZIP12 and PvZIP13 showed more expression in G19833 genotype than DOR 364, and PvZIP01, PvZIP12 and PvZIP16 showed maximum expression under zinc deficit treatment in pods, vegetative leaves and reproductive leaves, respectively. Further, Astudillo et al. (2013) recommended PvZIP12 as a good candidate gene for enhancing seed zinc concentration through their genetic mapping studies. A similar kind of genotype-specific heterogeneous expression of the zinc transporters genes has also been reported in maize and rice, the strategy II plants (Lee et al. 2010a, b; Ishimaru et al. 2011; Mondal et al. 2014). In A. thaliana three members of ZIP family (AtZIP4, AtZIP9 and AtZIP12) also showed higher expression in roots and shoots under Zn deficient seedlings and they were suppressed with the availability of Zn (Jain et al. 2013). During this study, CcZIP3 and CcZIP15 (containing ZDRE in its promoter region) were significantly up-regulated in the reproductive leaf tissues and down-regulated in matured green pods of the resistant genotypes with higher zinc content. Thus the role of these two CcZIP genes could be attributed to the translocation (cellular movement) of Zn from leaves to pod walls (Garcia and Grusak 2015). Alternatively, CcZIP6, and CcZIP13 were up-regulated in the matured green pods, and down-regulated reproductive leaf tissues of the genotypes with higher zinc content; thus these CcZIP genes might have played a subsequent role in phloem unloading and accumulation in the seeds (Garcia and Grusak 2015). This kind of tissue specific expression of ZIP genes was reported in several dicots and monocots (Astudillo et al. 2013; Jain et al. 2013; Mondal et al. 2014). Therefore, CcZIP3, CcZIP6, CcZIP13 and CcZIP15 might be considered probable candidate genes for higher accumulation seed zinc content in the reproductive leaves and matured green pods of the pod borer resistant genotypes atleast used in this study, which is quite relevant for bio-fortification and genetic improvement to inhibit the pod borer herbivory in pigeonpea. Differential tissue-specific expression of these CcZIP genes and other allied gene families, cis- regulatory elements in the promoter of these genes and their regulatory transcription factors in pigeonpea under both Zn-abundant and Zn-deficient conditions, as well as various plant defence-related enzymes activities are being investigated to support the findings.
Relative expression of eight CcZIPs in two tissues (reproductive leaves and matured green pods) among the four genotypes of Cajanus spp. (on X-axis) with contrasting host response to pod borer and seed zinc content. The expression normalized by measuring ΔCT value for each of the CcZIP gene with respect to the endogenous control ‘TUB6’ and was represented as 2−ΔCT on Y-axis. The error bars represent the standard deviation from three biological replicates
Differential expression of four CcZIPs (CcZIP3, CcZIP6, CcZIP13 and CcZIP15) in reproductive leaves and matured green pods among the genotypes of Cajanus spp. (on X-axis) with contrasting host response to pod borer and seed zinc content. The expression normalized by measuring ΔCT value for each of the CcZIP gene with respect to the endogenous control ‘TUB6’ and was represented as 2−ΔCT on Y-axis. The error bars represent the standard deviation from three biological replicates
Conclusion
This study on identification, characterization and analysis of zinc transporters, including the members of ZIP gene family in C. cajan could be used in functional genomic studies on Zn biofortification, stress modulation and plant development. Gene structure characterization and expression analysis showed the possible role of CcZIP3 and CcZIP15, having ZDRE in their promoter region, on the mobilization of Zn from leaves to pods in the pod borer resistant genotypes. In addition, the higher expression of CcZIP6 and CcZIP13 in matured green pods of both the resistant genotypes compared to reproductive leaves indicates their possible involvement in phloem unloading, leading to higher seed Zn content in the resistant genotypes. Differential expression of these CcZIPs along with other allied gene families and transcription factors could delineate the strategy not only in Zn bio-fortification but also for the genetic improvement to inhibit the pod borer herbivory in pigeonpea.
Abbreviations
- ABRE:
-
ABA responsive elements
- ARE:
-
Auxin responsive elements
- CDF:
-
Cation diffusion facilitator
- CRE:
-
Cis Regulatory elements
- ERE:
-
Ethylene responsive elements
- GRAVY:
-
Grand average of hydropathicity
- HMA:
-
Heavy metal ATPase
- HMM:
-
Hidden Markov model
- LRE:
-
Light regulatory elements
- LTRE:
-
Low-temperature responsive elements
- MRE:
-
Metal regulatory element
- MUSCLE:
-
Multiple sequence comparison by log-expectation
- PCR:
-
Plant cadmium resistance
- pI:
-
Isoelectric point
- PLH:
-
Pore line helices
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- ROS:
-
Reactive oxygen species
- TMH:
-
Trans-membrane helices
- VIT:
-
Vacuolar iron transporter
- ZDRE:
-
Zinc deficiency-related elements
- ZIP:
-
Zinc-regulated transporter (ZRT) and iron-regulated transporter (IRT) like protein
References
Alagarasan G, Dubey M, Aswathy KS, Chandel G (2017) Genome-wide identification of orthologous ZIP genes associated with zinc and iron translocation in Setaria italica. Front Plant Sci 8:775
Allagulova CR, Gimalov FR, Shakirova FM, Vakhitov VA (2003) The plant dehydrins: structure and putative functions. Biochem Mosc 68:945–951
Assunção AGL, Herrero E, Lin YF et al (2010) Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA 107:10296–10301. https://doi.org/10.1073/pnas.1004788107
Astudillo C, Fernandez A, Blair M, Cichy K (2013) The Phaseolus vulgaris ZIP gene family: identification, characterization, mapping, and gene expression. Front Plant Sci 4:286
Astudillo-Reyes C, Fernandez AC, Cichy KA (2015) Transcriptome characterization of developing bean (Phaseolus vulgaris L.) pods from two genotypes with contrasting seed zinc concentrations. PLOS ONE 10:e0137157
Bailey TL, Boden M, Buske FA et al (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Bashir K, Ishimaru Y, Nishizawa NK (2012) Molecular mechanisms of zinc uptake and translocation in rice. Plant Soil 361:189–201. https://doi.org/10.1007/s11104-012-1240-5
Bashir K, Rasheed S, Kobayashi T et al (2016) Regulating subcellular metal homeostasis: the key to crop improvement. Front Plant Sci 7:1192
Bouain N, Satbhai SB, Korte A, Saenchai C et al (2018) Natural allelic variation of the AZI1gene controls root growth under zinc-limiting condition. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007304
Cabot C, Martos S, Llugany M et al (2019) A role for zinc in plant defense against pathogens and herbivores. Front Plant Sci 10:1171. https://doi.org/10.3389/fpls.2019.01171
Ćalić I, Koch J, Carey D, et al. (2017) Genome-wide association study identifies a major gene for beech bark disease resistance in American beech (Fagus grandifolia Ehrh.). BMC Genomics 18:547. https://doi.org/10.1186/s12864-017-3931-z
Chapman B, Chang J (2000) Biopython: Python tools for computational biology. SIGBIO Newsletter 20:15–19. https://doi.org/10.1145/360262.360268
Chen WR, Feng Y, Chao YE (2008) Genomic analysis and expression pattern of OsZIP1, OsZIP3, and OsZIP4 in two rice (Oryza sativa L.) genotypes with different zinc efficiency. Russ J Plant Physiol 55:400–409. https://doi.org/10.1134/S1021443708030175
Clemens S (2014) Zn and Fe biofortification: the right chemical environment for human bioavailability. Plant Sci 225:52–57
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004
D’Ovidio R, Raiola A, Capodicasa C et al (2004) Characterization of the complex locus of bean encoding polygalacturonase- inhibiting proteins reveals sub-functionalization for defense against fungi and insects. Plant Physiol 135:2424–2435. https://doi.org/10.1104/pp.104.044644
de Castro E, Sigrist CJA, Gattiker A et al (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. https://doi.org/10.1093/nar/gkl124
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113. https://doi.org/10.1186/1471-2105-5-113
El-Gebali S, Mistry J, Bateman A et al (2018) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. https://doi.org/10.1093/nar/gky995
Eng BH, Guerinot ML, Eide D, Saier MH Jr (1998) Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol 166:1–7. https://doi.org/10.1007/s002329900442
FAOSTAT (2017) FAOSTAT. In: Food and Agriculture Organization of the United Nations (FAO). http://www.fao.org/faostat/en/#data/QC/visualize. Accessed 28 Nov 2020
Floden EW, Tommaso PD, Chatzou M et al (2016) PSI/TM-Coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res 44:W339–W343. https://doi.org/10.1093/nar/gkw300
Fu XZ, Zhou X, Xing F, Ling LL, Chun CP, Cao L, Aarts MGM, Peng L-Z (2017) Genome-wide identification, cloning and functional analysis of the Zinc/Iron-regulated transporter-like protein (ZIP) gene family in Trifoliate Orange (Poncirus trifoliata L. Raf.). Front Plant Sci 8:588. https://doi.org/10.3389/fpls.2017.00588
Garcia C, Grusak M (2015) Mineral accumulation in vegetative and reproductive tissues during seed development in Medicago truncatula. Front Plant Sci 6:622
Grotz N, Fox T, Connolly E et al (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA 95:7220–7224. https://doi.org/10.1073/pnas.95.12.7220
Grover A, Pental D (2003) Breeding objectives and requirements for producing transgenics for major field crops of India. Curr Sci 84:310–320
Guerinot M, lou, (2000) The ZIP family of metal transporters. Biochim Biophys Acta Biomembr 1465:190–198
Hambidge M (2000) Human zinc deficiency. J Nutr 130:1344S-1349S. https://doi.org/10.1093/jn/130.5.1344S
Helfenstein J, Pawlowski ML, Hill C et al (2015) Zinc deficiency alters soybean susceptibility to pathogens and pests. J Plant Nutr Soil Sci 178:896–903. https://doi.org/10.1002/jpln.201500146
Hu B, Jin J, Guo A-Y et al (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Ishimaru Y, Bashir K, Nishizawa NK (2011) Zn uptake and translocation in rice plants. Rice 4:21–27. https://doi.org/10.1007/s12284-011-9061-3
Jain A, Sinilal B, Dhandapani G, Meagher RB, Shai SV (2013) Effects of deficiency and excess of zinc on morphological traits and spatiotemporal regulation of zinc responsive genes reveal incidence of cross talk between micro- and macronutrients. Environ Sci Technol 47(10):5327–5335. https://doi.org/10.1021/es400113y
Kaur R, Gupta AK, Taggar GK (2014) Zinc as an important factor determining resistance against Helicoverpa armigera herbivory in pigeon pea (Cajanus cajan L.). Curr Sci 107:1871–1875. https://doi.org/10.18520/cs/v107/i11/1871-1875
Kawakami Y, Bhullar NK (2018) Molecular processes in iron and zinc homeostasis and their modulation for biofortification in rice. J Integr Plant Biol 60:1181–1198. https://doi.org/10.1111/jipb.12751
Krishna TPA, Maharajan T, Roch GV et al (2020) Structure, function, regulation and phylogenetic relationship of ZIP family transporters of plants. Front Plant Sci 11:1–18. https://doi.org/10.3389/fpls.2020.00662
Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Edited by F. Cohen. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kundu SS, Podder R, Bett KE et al (2017) Optimizing seed sample size for zinc and iron analysis of wild and cultivated lentil. Commun Soil Sci Plant Anal 48:1584–1594. https://doi.org/10.1080/00103624.2017.1374397
Lawrence SD, Novak NG, Jones RW et al (2014) Herbivory responsive C2H2 zinc finger transcription factor protein StZFP2 from potato. Plant Physiol Biochem 80:226–233. https://doi.org/10.1016/j.plaphy.2014.04.010
Lee S, Jeong HJ, Kim SA et al (2010a) OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol Biol 73:507–517. https://doi.org/10.1007/s11103-010-9637-0
Lee S, Kim SA, Lee J et al (2010b) Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol Cells 29:551–558. https://doi.org/10.1007/s10059-010-0069-0
Li S, Zhou X, Huang Y et al (2013) Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol 13:114. https://doi.org/10.1186/1471-2229-13-114
Li XB, Suo HC, Liu JT, Wang L, Li CC, Liu WW (2020) Genome-wide identification and expression analysis of the potato ZIP gene family under Zn-deficiency. Biol Plant 64: 845–855. https://doi.org/10.32615/bp.2020.125
Lilay GH, Castro PH, Guedes JG et al (2020) Rice F-bZIP transcription factors regulate the zinc deficiency response. J Exp Bot 71:3664–3677. https://doi.org/10.1093/jxb/eraa115
Lira-Morales JD, Varela-Bojorquez N, Montoya-Rojo MB, Sanudo-Barajas JA (2019) The role of ZIP proteins in zinc assimilation and distribution in plants: Current challenges. Czech J Genet Plant Breed 55:45–54. https://doi.org/10.17221/54/2018-CJGPB
Livak KJ, Scmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and 2 (-ΔΔ C(T)) method. Methods 25:402-408. https://doi.org/10.1006/Meth.2001.1262
Maksaev G, Shoots JM, Ohri S, Haswell ES (2018) Nonpolar residues in the presumptive pore-lining helix of mechanosensitive channel MSL10 influence channel behaviour and establish a nonconducting function. Plant Direct 2:e00059. https://doi.org/10.1002/pld3.59
Malviya N, Gupta S, Singh VK et al (2015) Genome-wide in silico characterization of Dof gene families of pigeonpea (Cajanus cajan (L) Millsp.). Mol Biol Rep 42:535–552. https://doi.org/10.1007/s11033-014-3797-y
Marchler-Bauer A, Derbyshire MK, Gonzales NR et al (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. https://doi.org/10.1093/nar/gku1221
Mondal TK, Ganie SA, Rana MK, Sharma TR (2014) Genome-wide analysis of zinc transporter genes of maize (Zea mays). Plant Mol Biol Report 32:605–616. https://doi.org/10.1007/s11105-013-0664-2
Moretti S, Armougom F, Wallace IM et al (2007) The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res 35:645–648. https://doi.org/10.1093/nar/gkm333
Nugent T, Ward S, Jones DT (2011) The MEMPACK alpha-helical transmembrane protein structure prediction server. Bioinformatics 27:1438–1439. https://doi.org/10.1093/bioinformatics/btr096
Olsen L, Palmgren M (2014) Many rivers to cross: the journey of zinc from soil to seed. Front Plant Sci 5:30
Olsen LI, Hansen TH, Larue C et al (2016) Mother-plant-mediated pumping of zinc into the developing seed. Nat Plants 2:16036. https://doi.org/10.1038/nplants.2016.36
Pazhamala LT, Purohit S, Saxena RK et al (2017) Gene expression atlas of pigeonpea and its application to gain insights into genes associated with pollen fertility implicated in seed formation. J Exp Bot 68:2037–2054. https://doi.org/10.1093/jxb/erx010
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Pita-Barbosa A, Ricachenevsky FK, Wilson M et al (2019) Transcriptional plasticity buffers genetic variation in zinc homeostasis. Sci Rep 9:19482. https://doi.org/10.1038/s41598-019-55736-0
Quevillon E, Silventoinen V, Pillai S et al (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. https://doi.org/10.1093/nar/gki442
Radkova M, Revalska M, Kertikova D, Iantcheva A (2019) Zinc finger CCHC-type protein related with seed size in model legume species Medicago truncatula. Biotechnol Biotechnol Equip 33:278–285. https://doi.org/10.1080/13102818.2019.1568914
Ricachenevsky FK, Menguer PK, Sperotto RA, Fett JP (2015) Got to hide your Zn away: molecular control of Zn accumulation and biotechnological applications. Plant Sci 236:1–17
Schweizer F, Bodenhausen N, Lassueur S et al (2013) Differential contribution of transcription factors to Arabidopsis thaliana defense against Spodoptera littoralis. Front Plant Sci 4:13. https://doi.org/10.3389/fpls.2013.00013
Shahmuradov IA, Solovyev V (2015) Nsite, NsiteH and NsiteM computer tools for studying transcription regulatory elements. Bioinformatics 31:3544–3545. https://doi.org/10.1093/bioinformatics/btv404
Sharma O P, Gopali J B, Yelshetty S, et al. (2010) Pests of pigeonpea and their management. NCIPM, IARI, New Delhi
Shivashankar S, Sumathi M, Krishnakumar NK, Rao VK (2015) Role of phenolic acids and enzymes of phenylpropanoid pathway in resistance of chayote fruit (Sechium edule) against infestation by melon fly, Bactrocera cucurbitae. Ann Appl Biol 166:420–433. https://doi.org/10.1111/aab.12194
Sinclair SA, Krämer U (2012) The zinc homeostasis network of land plants. Biochim Biophys Acta Mol Cell Res 1823:1553–1567. https://doi.org/10.1016/j.bbamcr.2012.05.016
Singh A, Singh PK, Sharma AK, et al. (2019) Understanding the role of the WRKY gene family under stress conditions in pigeonpea (Cajanus cajan l.). Plants 8:214. https://doi.org/10.3390/plants8070214
Sinha P, Singh VK, Suryanarayana V et al (2015) Evaluation and validation of housekeeping genes as reference for gene expression studies in Pigeonpea (Cajanus cajan) under drought stress conditions. PLoS ONE 10:e0122847. https://doi.org/10.1371/journal.pone.0122847
Stephens BW, Cook DR, Grusak MA (2011) Characterization of zinc transport by divalent metal transporters of the ZIP family from the model legume Medicago truncatula. Biometals 24:51–58. https://doi.org/10.1007/s10534-010-9373-6
Thingholm TE, Rönnstrand L, Rosenberg PA (2020) Why and how to investigate the role of protein phosphorylation in ZIP and ZnT zinc transporter activity and regulation. Cell Mol Life Sci 77:3085–3102
Tiong J, McDonald G, Genc Y et al (2015) Increased expression of six ZIP family genes by zinc (Zn) deficiency is associated with enhanced uptake and root-to-shoot translocation of Zn in barley (Hordeum vulgare). New Phytol 207:1097–1109. https://doi.org/10.1111/nph.13413
Vallee BL, Falchuk KH (1993) The biochemical basis of zinc physiology. Physiol Rev 73:79–118. https://doi.org/10.1152/physrev.1993.73.1.79
Varshney RK, Chen W, Li Y et al (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89. https://doi.org/10.1038/nbt.2022
Vigani G, Hanikenne M (2018) Metal homeostasis in plant mitochondria. In: Annual plant reviews online. Wiley, Chichester, pp 111–142
Wang Z, Cheng K, Wan L et al (2015) Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16:1–15. https://doi.org/10.1186/s12864-015-2258-x
Wang X, Wang Y, Liu P et al (2017) TaRar1 is involved in wheat defense against stripe rust pathogen mediated by YrSu. Front Plant Sci 8:156
Weber H, Borisjuk L, Wobus U (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56:253–279. https://doi.org/10.1146/annurev.arplant.56.032604.144201
Wongpia A, Lomthaisong K (2010) Changes in the 2DE protein profiles of chilli pepper (Capsicum annuum) leaves in response to Fusarium oxysporum infection. Sci Asia 36:259–270. https://doi.org/10.2306/scienceasia1513-1874.2010.36.259
Yamasaki S, Sakata-Sogawa K, Hasegawa A et al (2007) Zinc is a novel intracellular second messenger. J Cell Biol 177:637–645. https://doi.org/10.1083/jcb.200702081
Zang D, Li H, Xu H et al (2016) An Arabidopsis zinc finger protein increases abiotic stress tolerance by regulating sodium and potassium homeostasis, reactive oxygen species scavenging and osmotic potential. Front Plant Sci 7:1272
Zaun HC, Shrier A, Orlowski J (2012) N-myristoylation and Ca2+ binding of calcineurin B homologous protein CHP3 are required to enhance Na+/H+ exchanger NHE1 half-life and activity at the plasma membrane. J Biol Chem 287:36883–36895. https://doi.org/10.1074/jbc.M112.394700
Acknowledgements
The authors are grateful to the Department of Biotechnology, Govt. of India, New Delhi for financial assistance to the corresponding author (JP) through R&D project (F. No. BT/PR13468/AGR/02/702/2010). The authors are also thankful the Vice Chancellor of Central University of Rajasthan and the Vice chancellor of Berhampur University for providing necessary laboratory facilities to carry out this work.
Funding
Department of Biotechnology, Govt. of India (Grant No. F. BT/PR13468/AGR/02/702/2010).
Author information
Authors and Affiliations
Contributions
JP and AN conceived and designed the experiments; AN, SKM, KG, ND performed the experiments and bioinformatics analysis; JP, KG and AN analysed the data and interpreted the results; AN, KG, SKM and JP wrote the manuscript. All authors have reviewed the manuscript and have given consent to the final version submitted.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nag, A., Gupta, K., Dubey, N. et al. Genomic characterization of ZIP genes in pigeonpea (CcZIP) and their expression analysis among the genotypes with contrasting host response to pod borer. Physiol Mol Biol Plants 27, 2787–2804 (2021). https://doi.org/10.1007/s12298-021-01111-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01111-1