Abstract
To analyze the risk and reason of false-negative HBV DNA results of NAT reagents among blood donations of China and discuss the necessity of two amplification targets for HBV DNA tests among donations. In this study, samples that showed discordant results on two commercially available assay platforms were further detected by established in-house methods based on conserved regions of the HBV genome. The HBV concentration of these samples was determined using two commercially available reagents. The samples with high titers of HBV were detected by an in-house method. The samples showing high Ct differences between two regions in the in-house method were further sequenced and aligned with primers and probes. The results showed that the established method has a good detection performance. The mismatch between reverse primers and sample sequences led to decreased detection capacity of S and C regions by the in-house method, but it could be compensated by another region. Among the false-negative samples detected by commercial reagents, most were because of low titers; however, there were 7 samples with HBV DNA concentrations much higher than the LOD of the commercial reagents, which may be due to mismatch of the primer or probe. This study highlights the potential risk of HBV false-negative detection by commercial NAT reagents. The dual-target assay may be helpful for HBV screening and reduce the risk of false-negative detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nucleotide acid technology (NAT) is an important part of blood screening, which plays critical roles in preventing the transmission of blood-borne viruses through transfusion and improves blood safety. Generally, it is used for screening of hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus (HIV).
HBV is highly prevalent globally and there are a total of 350 million HBV carriers, a one-third of whom live in China [1]. An epidemiological survey has shown an HBV surface antigen (HBsAg) prevalence of 7.18% among Chinese individuals [2]. The meta-analysis results showed an HBsAg prevalence of 1.085% among Chinese blood donors, which poses risk of transfusion-transmitted HBV infection and thus threatens blood safety [3]. To improve blood safety, China started to implement NAT as a mandatory tool for blood screening in addition to serological screening from 2010. The prevalence of HBV DNA among Chinese HBsAg-negative blood donations was 1/1482 from 2010 to 2015 [4].
HBV transmission routes includes sexual transmission, mother to child transmission, and transfusion transmission. This study aimed to address the strategies for reducing the risk of transfusion transmission. The presence of HBV at low concentrations in blood donations, especially the occult hepatitis B virus infections (OBI), demands the requirement of highly sensitive NAT reagents. HBV titers lower than the limit of detection (LOD) of NAT reagents pose a risk of false-negative results.
Another factor which may result in “false negative” screening is the mismatch between HBV sequence and primer/probes in NAT reagents. HBV is a highly variable DNA virus that replicates through its pre-genomic RNA. The mutation frequency of HBV was 10 times higher than that of other DNA viruses [5, 6]. Mutations like drug-resistant mutations caused by antiviral drugs and vaccine escape mutations further increase the complexity of HBV DNA mutations [7, 8]. These factors result in a high risk of HBV DNA detection by real-time PCR assays. Previous studies report underestimation of HBV quantitation due to the mismatch between primer/probe and samples [9, 10].
Among the viruses screened by NAT, HIV is a genetically diverse retrovirus. Blood infected with HIV is undetected by NAT reagents due to mutations in primer/probe binding regions, leading to the transmission of HIV in recipients [13]. The mono-target HIV NAT reagents are reportedly more vulnerable to sequence variations than dual-target reagents [11, 12].
There are very few studies reporting false-negative screening of HBV DNA. This study aimed to discuss the risk of false-negative screening of HBV using NAT. Most commercial NAT reagents target the surface “S”, pre-Core, and Core “C” region of the HBV genome. In this study, we developed an in-house method for HBV DNA screening based on the conserved S and C region of HBV. The risk of “false negative” screening of single “S” or “C” region was analyzed. Considering the high mutation rate of HBV DNA and high prevalence of HBsAg in Chinese individuals and blood donors, we speculated that mono-target NAT assays may result in “false negative” screening because of the mismatch between primer/probe and HBV sequence. The necessity of two different targets of HBV DNA for NAT screening was also explored.
Materials and Methods
Samples
A total of 372,318 HBsAg-negative blood donations were screened by COBAS® TaqScreen MPX test, version 2 (Cobas) nucleic acid test (NAT) reagent in 7 blood banks of China from July, 2014 to July, 2016. There were 372 HBV DNA reactive blood donations. A total of 70,387 HBsAg-negative blood donations were screened by Haoyuan NAT reagent in 8 blood banks. There were 86 HBV DNA reactive blood donations. Procedures were conducted according to the manufacturer’s instructions. Among the HBV-reactive blood donations, 314 Cobas-positive and 70 Haoyuan-positive blood donations were sent to the National Center for Clinical Laboratories (NCCL) of China.
HBV-Reactive Blood Donations Detected by Commercial Kits in NCCL
The 314 Cobas-positive blood donations in blood banks were further confirmed by Haoyuan NAT reagent in NCCL. Samples which were negative by Haoyuan were further evaluated by COBAS® TaqScreen MPX test, version 2 (Cobas) nucleic acid test (NAT) reagent. The samples which were positive by Cobas but negative by Haoyuan were tested by an in-house method and also for HBV DNA titer detection. The 70 Haoyuan-positive blood donations in blood banks were further confirmed by COBAS® TaqScreen MPX test, version 2 (Cobas) nucleic acid test (NAT) reagent in NCCL. Samples which were negative by Cobas were tested by Haoyuan NAT reagent. The samples which were positive by Haoyuan but negative by Cobas were tested by an in-house method and also for HBV DNA titer detection. Testing was done according to the manufacturer’s (NAT reagents) instructions. In addition, blood donations with Cycle Threshold (Ct) below 32.5 detected by Haoyuan were also tested by an in-house method. Haoyuan and Cobas NAT reagents targeted S and C region of the HBV genome, respectively.
Establishment of the In-house Method
Based on the conserved S and C regions, an in-house method was developed for HBV DNA screening using primers and probes described previously [10]. Quantitative polymerase chain reaction (qPCR) was performed separately using primers and probes for S and C region. The HBV samples which were traced to WHO HBV DNA Second International Standard (97/750) were diluted to 50 IU/mL, 20 IU/mL, 10 IU/mL, 5 IU/mL, 2.5 IU/mL, and 1 IU/mL. LOD was determined by testing 20 replicates by the in-house method. Specificity was determined by testing 10 samples each of HCV-positive, HIV-positive, human genome, and HBV-negative normal plasma.
Blood Donations Detected by the In-house Method
Each sample (4 mL) was concentrated by centrifugation at 20,000g and 4°C for 1 h. After centrifugation, 3.2 mL supernatant was removed and the pellet was reconstituted in the remaining 800 µL supernatant. DNA was extracted from 500 μL of reconstituted pellet using an mSample Preparation System DNA extraction kit (Abott, USA) on a m2000sp nucleotide acid extraction machine (Abott, USA). The sequences of designed primers and probes as described previously [10] were (5′–3′) S-F: GATGTGTCTGCGGCGTTTTA; S-R:GCAACATACCTTGATAGTCCAGAAGAA, S-P:Fam-CCTCTICATCCTGCTGCTATGCCTCA-BHQ1; C-:TTCCGGAAACTACTGTTGTTAGAC, C-R:ATTGAGATTCCCGAGATTGAGA, C-Probe:Fam-CCCTAGAAGAAGAACTCCCTCGCCTC-BHQ1.
We added S-F (concentration: 0.8 μmol/μL), S-R (concentration: 0.8 μmol/μL), S-P (concentration: 0.3 μmol/μL) and C-F (concentration: 0.3 μmol/μL), C-R (concentration: 0.3 μmol/μL) and C-P (concentration: 0.3 μmol/μL) into two different qPCR tubes to target S and C regions of the HBV genome, respectively. In-house qPCR was performed using 5 × baorui master mix (baorui, Zhuhai, China) and ABI7500 Real-Time PCR System (Applied Biosystems, USA). Volume of the qPCR was 50 μL with 36 μL DNA template. The PCR program comprised the following steps: 50 °C for 2 min, 95 °C for 1 min, 95 °C for 5 s, 55 °C for 40 s, 60 cycles, 25 °C, 1 min. A weak positive and negative external control were included in the assay as a quality check.
HBV Viral Load Testing
The HBV viral load of the samples was determined by Abbott Real Time HBV Assay and Cobas TaqMan HBV test, version 2, which targeted the S and C region of HBV, respectively.
Sequence and Alignment
Samples that showed very high differences between the Ct values of S and C regions by the in-house method were sequenced as described previously [10]. The sequences were aligned with the primer/probes of S and C regions from the in-house method.
Results
The Results of HBV Reactive Blood Donations Detected by Commercial Kits in NCCL
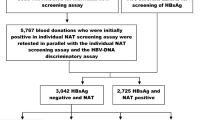
Most of HBV-reactive blood donations were detected as positive by both Cobas and Haoyuan NAT reagent methods. However, 21 Cobas-positive/Haoyuan-negative and 19 Haoyuan-positive/Cobas-negative blood donations showed conflicting results between Cobas and Haoyuan reagents. In addition, 12 blood donations were detected by Haoyuan with a Ct below 32.5 (Fig. 1).
LOD and Specificity of the In-house Method
Results showed that the 95% LOD was 5 IU/mL for S and C regions of the in-house method. Results of the 10 cases of HCV-positive, HIV-positive, human genome, and HBV-negative normal plasmas detected by an in-house method were negative, demonstrating good specificity of the in-house method.
Results of Blood Donations Detected by the In-house Method
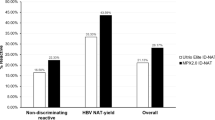
Nineteen Haoyuan-positive/Cobas-negative, 21 Cobas-positive/Haoyuan-negative and 12 Cobas-positive/Haoyuan-positive samples were tested by the in-house method. Results from the in-house method confirmed and detected the S and C region of all the Cobas-positive/Haoyuan-positive samples as positive. Of the 19 Haoyuan-positive/Cobas-negative samples, the S region of 7 samples and C region of 7 samples was positive, and both S and C regions of 10 samples were negative (Table 1). Thus, 9/19 (47.4%) Haoyuan-positive/Cobas-negative blood samples were also positive by the in-house method using two primers. Among the 21 Cobas-positive/Haoyuan-negative samples, the S region of 16 samples was positive, C region of 19 samples was positive, and both S and C regions of 2 samples were negative (Table 2). 19/21 (90.4%) Cobas-positive/Haoyuan-negative blood samples were positive by the in-house method.
HBV Viral Load Testing
The HBV viral load of all the Haoyuan-positive/Cobas-negative and Haoyuan-negative/Cobas-positive samples was determined by Abbott Real Time HBV Assay and Cobas TaqMan HBV test, version 2. Results showed that among the Haoyuan-positive/Cobas-negative samples, the HBV viral load of only one sample was above the limit of quantitation (LOQ) of Abbott Real Time HBV Assay. Among the Haoyuan-negative/Cobas-positive samples, the HBV viral load of 6 samples was above the LOQ of Abbott Real Time HBV Assay. Of the 6 samples tested, the highest HBV DNA viral load was 139 IU/mL and 97 IU/mL, determined by Abbott Real Time HBV Assay and Cobas TaqMan HBV test, version 2, respectively (Table 3).
Sequence and Alignment
Among the samples detected by the in-house method, two samples showed very high difference in Ct values of the S and C region (Table 4). Sequencing results showed the presence of a mutation in the 3′ region of reverse primers of S and C regions (Fig. 2).
Discussion
Hepatitis B virus (HBV), a DNA virus, seriously endangers human health through blood transfusion, one of the most important transmission routes. Although HBV is highly prevalent in China, with the development of serology assays and NAT reagents, the risk of transfusion transmitted HBV was reduced. However, because of the complex HBV infection models, limitations of the LOD of serological and NAT assays, and highly variant HBV DNA, a part of HBV-positive samples remained undetected by current blood screening reagents.
In the current blood screening strategies, NAT is the last method of detection for blood donations and the result of NAT is critical to prevent transfusion-transmitted HBV and improve blood safety. NAT could detect blood donations in a window period or OBI stage [13, 14]. The HBV load of part of the OBI blood donations were below 5 IU/mL, which could lead to undetectable results because of limitations of the LOD of NAT reagents [15, 16]. In this study, the viral load of most of the Haoyuan-positive/Cobas-negative, Haoyuan-negative/Cobas-positive and in-house method-positive samples were below the LOQ of Abbott Real Time HBV Assay (10 IU/mL) and Cobas TaqMan HBV test, version 2 (20 IU/mL). These results showed that samples with low HBV titer detected by NAT reagents could be undetectable. The undetectable samples with low titer detected by NAT reagents compose a big part of “false negative” blood donations. This demonstrated the necessity to improve the sensitivity of NAT reagents.
In this study, an in-house method was developed targeting the S and C regions of the HBV genome to avoid false-negative results due to mismatches in the primer/probe binding region of mono-targets to some extent. The in-house method showed good specificity. The samples were detected by an in-house method to show that the blood donations were positive for HBV DNA by another method. If one blood donation was HBV DNA-positive in blood banks and NCCL by one commercial NAT reagent and in-house methods, but negative by another commercial reagent, it is a case of false-negative screening. To understand the cause of false-negative results, we performed HBV DNA quantitation detection of selected samples in this study. Most of them were undetected or under the limit of quantitation of commercial reagents for HBV DNA quantitation. This implies to a large part of false-negative results which was because of the low titer of HBV. Among the Haoyuan-positive/Cobas-negative samples, the HBV DNA titer of only one sample was 14 IU/mL determined by Abbott Real Time HBV Assay, while undetectable by Cobas TaqMan HBV test, version 2. The titer of the sample was 6 times higher than that of the LOD of Cobas (2.3 IU/mL). This may have occurred because of mismatches in the region targeted by Cobas NAT reagents and also may be due to the fluctuations of HBV DNA quantification when HBV titer was low. Among the Cobas-positive/Haoyuan-negative samples, the HBV DNA titer of 6 samples was higher than 10 IU/mL, ranged from 12 to 139 IU/mL which was much higher than the LOD of Haoyuan NAT reagents (6.3 IU/mL). Especially, one sample was determined 139 IU/mL by Abbott Real Time HBV Assay and 97 IU/mL by Cobas TaqMan HBV test, version 2. The false-negative results by Haoyuan for this sample were because of the mismatch of primer/probe with sequence of NAT reagents and were independent of the low titer of HBV DNA. However, as the sequences of primer/probe of commercial NAT reagents were not disclosed by the manufacturer, it becomes difficult to check for mismatch by alignment. Though the proportion of mismatch samples among blood donations was low, the false-negative result of donations with high titer of HBV would easily lead to HBV transmission through blood transfusion. These results demonstrated a risk of false-negative screening by commercial NAT reagents with mono-targets among Chinese blood donations.
The in-house method detected samples on the basis of different Ct values for S and C region of two samples. The targeted S and C region were sequenced and aligned with the primer/probe sequence. In the samples whose Ct value of the S region were higher than that of the C region by 9.75, there was a mutation in the 3′ region of the reverse primer binding region of the S region. In the sample whose Ct value of the C region were higher than that of the S region by 7.96, there was a mutation in the 3′ region of the reverse primer binding region of the C region. This demonstrated that mismatch of primer/probe sequences with samples leads to decreased detection capacity and even undetectable results. Study also demonstrated decreased HBV titer due to the occurrence of mutants in the binding region of the primer or probe in the C region of HBV [10, 17]. Therefore, NAT reagents with mono-target may pose the risk of false-negative results of samples with titers higher than the LOD due to mismatch of primer/probe with the sequence of samples. In the in-house method of this study, results indicated that the use of dual-targets for screening could compensate the mismatch and decrease the risk of false-negatives of mono-target reagents. However, the dual-target NAT reagents used for screening may lead to increased costs of blood donation screening.
Conclusions
NAT plays a critical role in preventing HBV transmission and improving blood transfusion safety. NAT reagents with mono-target screening may pose “false negative” risk. The samples with low HBV titer were undetected by commercial NAT reagents, as the concentration of HBV was lower than LOD of reagents. However, there were still several undetected samples whose HBV titer was much higher than the LOD, which may be attributable to the mismatch of primer/probe with HBV DNA sequence. In the in-house method, mono-target screening decreased detection capacity due to the mismatch of primer with sequence and addition of another target region could compensate for the mismatched target. This demonstrated that there remains a risk of HBV false-negative screening by commercial NAT reagents. Dual-target may be helpful for HBV screening and reduce the risk of false-negative screening.
References
Custer B, Sullivan SD, Hazlet TK, Iloeje U, Veenstra DL, Kowdley KV (2004) Global epidemiology of hepatitis B virus. J Clin Gastroenterol 38:S158–S168
Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, Zhang Y, Liu J, Gong X, Chen Y, Wang F, Zheng H, Wang F, Guo J, Jia Z, Ma J, Wang H, Luo H, Li L, Jin S, Hadler SC, Wang Y (2009) Epidemiological serosurvey of hepatitis B in China—declining HBV prevalence due to hepatitis B vaccination. Vaccine 27:6550–6557
Liu GC, Sui GY, Liu GY, Zheng Y, Deng Y, Gao YY, Wang L (2013) A Bayesian meta-analysis on prevalence of hepatitis B virus infection among Chinese volunteer blood donors. PLoS ONE 8:e79203
Liu C, Chang L, Ji H, Guo F, Zhang K, Lin G, Zhang R, Li J, Wang L (2016) Prevalence of HBV DNA among 20 million seronegative blood donations in China from 2010 to 2015. Sci Rep. 6:36464
Echevarría JM, Avellón A (2006) Hepatitis B virus genetic diversity. J Med Virol 78(Suppl):S36–S42
Girones R, Miller RH (1989) Mutation rate of the hepadnavirus genome. Virology 170:595–597
Tipples GA, Ma MM, Fischer KP, Bain VG, Kneteman NM, Tyrrell DL (1996) Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology 24:714–717
Dos Santos MIMA, Pacheco SR, Stocker A, Schinoni MI, Paraná R, Reis MG, Silva LK (2017) Mutations associated with drug resistance and prevalence of vaccine escape mutations in patients with chronic hepatitis B infection. J Med Virol 89:1811–1816
Lindh M, Hannoun C, Malmström S, Lindberg J, Norkrans G (2006) Lamivudine resistance of hepatitis B virus masked by coemergence of mutations in probe region of the COBAS AMPLICOR assay. J Clin Microbiol 44:2587–2589
Liu C, Chang L, Jia T, Guo F, Zhang L, Ji H, Zhao J, Wang L (2017) Real-time PCR assays for hepatitis B virus DNA quantification may require two different targets. Virol J 14:94
Chudy M, Weber-Schehl M, Pichl L, Jork C, Kress J, Heiden M, Funk MB, Nübling CM (2012) Blood screening nucleic acid amplification tests for human immunodeficiency virus Type 1 may require two different amplification targets. Transfusion 52:431–439
Shah K, Ragupathy V, Saga A, Hewlett I (2016) High sensitivity detection of HIV-1 using two genomic targets compared with single target PCR. J Med Virol 88:1092–1097
Tsoi WC, Lelie N, Lin CK (2013) Enhanced detection of hepatitis B virus in Hong Kong blood donors after introduction of a more sensitive transcription-mediated amplification assay. Transfusion 53:2477–2488
Taira R, Satake M, Momose S, Hino S, Suzuki Y, Murokawa H, Uchida S, Tadokoro K (2013) Residual risk of transfusion-transmitted hepatitis B virus (HBV) infection caused by blood components derived from donors with occult HBV infection in Japan. Transfusion 53:1393–1404
Larralde O, Dow B, Jarvis L, Davidson F, Petrik J (2013) Hepatitis B escape mutants in Scottish blood donors. Med Microbiol Immunol 202:207–214
Allain JP, Mihaljevic I, Gonzalez-Fraile MI, Gubbe K, Holm-Harritshøj L, Garcia JM, Brojer E, Erikstrup C, Saniewski M, Wernish L, Bianco L, Ullum H, Candotti D, Lelie N, Gerlich WH, Chudy M (2013) Infectivity of blood products from donors with occult hepatitis B virus infection. Transfusion 53:1405–1415
Andonov A, Osiowy C, Borlang J, Swidinsky K (2016) Sequence variability of the Cobas taqman assay target region impacts accurate HBV DNA detection. Vox Sang 111:S58
Acknowledgements
The authors thank the staff at the participating blood banks who are engaged in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethical Statement
The study was approved by the institutional review board of the National Center for Clinical Laboratories. Written informed consent was obtained from all subjects participating in this research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, C., Chang, L. & Wang, L. HBV DNA Test Among Blood Donations May Require Two Amplification Targets. Indian J Hematol Blood Transfus 35, 544–550 (2019). https://doi.org/10.1007/s12288-018-01064-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12288-018-01064-8