Abstract
Background
Long noncoding RNAs (lncRNAs) have been reported to play crucial roles in breast cancer. This study aimed to determine the clinical significance and biological functions of lncRNA AFAP1-AS1 in breast cancer.
Methods
The expression of AFAP1-AS1 in breast cancer tissue and adjacent normal tissue from 160 patients and breast cancer cell lines were determined by qRT-PCR. The clinical characteristics of patients were collected to analyse the correlation between AFAP1-AS1 expression and malignancy status. Kaplan–Meier and Cox proportional hazards model were used to analyze whether AFAP1-AS1 expression impacted prognosis. To assess the effect of AFAP1-AS1 on MCF-7 cells proliferation, cell viability, EdU incorporation and colony formation assays were conducted after AFAP1-AS1 knockdown by siRNA. The apoptosis was detected by Caspase-3 activity, cell cycle analysis, Bcl-2 and Bax protein expression. Wound scratch assay and EMT-related protein expression (E-cadherin, N-cadherin and Vimentin) were conducted to evaluate the metastasis ability. To further determine the effect of AFAP1-AS1 on AFAP1, the mRNA and protein expression of AFAP1 and subsequent actin filament integrity were measured after AFAP1-AS1 knockdown.
Results
The expression of AFAP1-AS1 was up-regulated in human breast cancer tissue and associated with malignancy status, high expression of AFAP1-AS1 had a poor prognosis in breast cancer patients. AFAP1-AS1 expression was up-regulated in 4 breast cancer cell lines (MCF-7, SK-RB-3, MDA-MB-231and MDA-MB-468) compared with normal breast cell line HBL-100. MCF-7, the most up-regulation cancer cell, was used for following studies. AFAP1-AS1 knockdown can inhibit the proliferation, metastasis and promote apoptosis of MCF-7. However, the AFAP1 expression and actin filament integrity was not affected after AFAP1-AS1 knockdown.
Conclusion
Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis in breast cancer patients and regulated the breast cancer cells proliferation, apoptosis and metastasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer remains to be the most common cancer and the most frequent cause of cancer death in females worldwide. It represents a highly heterogeneous disease in terms of clinical outcomes and biological behavior and thus can be classified with various methods into different subtypes [1]. Novel concepts in genetics biology are leading to improved and higher resolution of the underlying events and increasing complexity in breast cancer biology [2]. Recently, growing evidence has demonstrated that long noncoding RNAs (lncRNAs) comprise noncoding RNAs longer than 200 nucleotides in length, are pervasively transcribed in the genome [3]. Aberrantly expressed lncRNAs have been observed in a serial of cancers, such as up-regulated PTENP1 in breast cancer [4], up-regulated PCAT1 in prostate cancer [5], and down-regulated GAS5 in colorectal cancer [6]. Moreover, lncRNAs act as either oncogene or tumor suppressor gene through regulating cancer cellular processes including cellular proliferation, apoptosis, cell cycle, and metastasis [7].
Actin filament associated protein1 antisense RNA1 (AFAP1-AS1), derived from the antisense strand at the AFAP1 coding gene locus, has been reported to regulate actin filament integrity [8]. High expression of AFAP1-AS1 correlates with poor prognosis, and promotes tumorigenesis in colorectal cancer [9], pancreatic ductal adenocarcinoma [10], non-small cell lung cancer [11], nasopharyngeal carcinoma [12]. Inhibiting its expression can decrease cellular proliferation, migration, invasion, along with increased apoptosis. Therefore, AFAP1-AS1 is considered to be a potential diagnostic and prognostic biomarker, as well as a therapeutic target in these cancers. However, to the best of our knowledge, study on the role of AFAP1-AS1 in breast cancer was not clear. Recently, Yang et al. analyzed the expression profile of lncRNAs in seven pairs of HER-2-enriched subtype breast cancer and paired adjacent normal tissue, and identified more than 1300 lncRNAs were significantly differently expressed [13]. Among these lncRNAs, AFAP1-AS1 was the most aberrantly expressed lncRNA, indicating AFAP1-AS1 may play a key role in breast cancer.
The present study, therefore, was aimed to investigate the expression and potential functional roles of AFAP1-AS1 in breast cancer. We sought to determine the different expression of AFAP1-AS1 between cancer tissues and paired adjacent normal tissues derived from breast cancer patients. Further in vitro studies revealed the effect of AFAP1-AS1 on cellular proliferation, colony formation, apoptosis and metastasis. This study suggested the critical role of AFAP1-AS1 in breast cancer, which might hopefully work as a novel biomarkers or targeted therapies of breast cancer.
Materials and methods
Patient samples
A total of 160 paired breast cancer tissue and adjacent normal tissue samples were obtained from patients that were histologically diagnosed as breast cancer and underwent surgical resections at the First Hospital of Lanzhou University between 2013 and 2016. All of the patients did not receive chemotherapy or radiotherapy before surgery and signed informed consent. Clinical characteristics of patients including age, family history, tumor grade, tumor-node-metastasis (TNM) stage, lymph node status, estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor-2 (HER2) were collected and summarized in Table 1. The information of family history was collected from patient’s medical record. Lymph node metastasis, Tumor grade, TNM stage, ER status, PR status and HER2 status were collected from the corresponding pathology report of the Gynecological Pathology Division, and all of these clinical characteristics were assigned according to Elston and Ellis [14]. Immunohistochemical analyses for ER, PR and HER2 were performed on 4-μm-thick sections according to standard procedures. ER or PR was considered negative if there were fewer than 1% immune-reactive cells (tumor cell nuclear staining) and positive otherwise, in line with recent guidelines [15]. HER2 was assessed through IHC exam or fluorescence in hybridization (FISH) test, positive HER2 was defined as 3+ [16]. The samples obtained in surgical resection were stored immediately in liquid nitrogen for qRT-PCR.
Quantitative real-time PCR (qRT-PCR)
Trizol reagent (Invitrogen) was used to extract total RNA. RNA (0.2–0.5 µg) was subjected to reverse transcription reaction using the PrimeScript reverse transcription reagent Kit (DRR037S; TaKaRa, Dalian, China). The expressions of AFAP1-AS1 in tissue samples and cultured cells were measured using SYBR Premix Ex TagTM II (Takara, Dalian, China) via ABI 7500 System (Applied Biosystems, Carlsbad, CA, USA) according to manufacturer instruction. The Primers sequence was designed by Primer Premier 5.0 according to the target gene target gene sequence and verified the specificity by dissolution curve of PCR products. The Primers sequence as follows: AFAP1-AS1, Forward 5′-TCGCTCAATGGAGTGACGGCA-3′; Reverse 5′-CGGCT GAGACCGCTGAGAACTT-3′. AFAP1, Forward 5′-CCGTGCATCAACGGCTCG CTC-3′; Reverse 5′-TTCACAACAGCCGCGGGATCC-3′. Internal control β-actin, Forward 5′-AACGGATTTGGTCGTATTGGG-3′; Reverse 5′-CCTGGAAGATGGTG ATGGGAT-3′. PCR cycling protocol was as following: initiate incubation at 95 °C for 15 s, followed by 40 amplification cycles of melting at 95 °C for 5 s, annealing at 60 °C for 31 s. Data analysis was performed by 2−ΔΔCt method using the ABI software.
Cell culture
The normal human mammary epithelial cell line MCF-10A and 4 breast cancer cell lines (MCF-7, SK-RB-3, MDA-MB-231and MDA-MB-468) were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China), Cells were cultured at 37 °C under 5% CO2 in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. The culture methods were consistent with the suggestion of ATCC.
AFAP1-AS1 siRNA transfection
MCF-7 cells grown to 50–60% confluence were transfected with either 50 nM AFAP1-AS1 siRNA or nontarget scramble control (NC) siRNA (Ribobio, Guangzhou, China) using Lipofectamine™ 2000 (Ribobio). After 24 h of transfection, qRT-PCR was used to measure the expression of AFAP1-AS1 for evaluating transfection efficiency. The detail protocol and siRNA segment sequences were achieved by reference to previous study [12]. The sequences of the AFAP1-AS1 targeting siRNAs were 5′-GGGCTTCAATTTACAAGCATT-3′ (segment 1), 5′-CCTA TCTGGTCAACACGTATT-3′ (segment 2) and 5′-GGGCTTCAATTTACAAGCATT-3′ (segment 3).
Cell viability assay
A cellTiter 96® AQueous one solution cell proliferation assay kit (MTS, Promega, USA) was used to detect cell viability. Briefly, after AFAP1-AS1 knockdown, 20 µL of reagent was added into each well (96-well plate) and then incubated at 37 °C for 2 h. The OD value, correlated to active cell number, was determined at 490 nm by using a colorimeter. The detailed protocol was conducted according to the manufacturer’s instructions as previously described [17].
EdU incorporation assay
The proliferation ability of MCF-7 cell was measured by 5-ethynyl-20-deoxyuridine (EdU) incorporation assay kit (Ribobio, Guangzhou China). Simple protocol as follow: after AFAP1-AS1 knockdown, each well (96-well plate) was added 100 µL of 50 µM EdU medium diluent for 3 h. The cells were fixed using 4% paraformaldehyde and incubated with 100 µL of the 0.5% Troxin X-100 into each well for 10 min followed by 100 µL of 1 × Apollo® 488 fluorescent staining reaction liquid for 30 min at 37 °C. After that, DAPI were used to stain cell nuclei and protected from light. The result presented as the ratio of EdU positive cells (green cells) to total DAPI positive cells (blue cells).
Colony formation
After AFAP1-AS1 knockdown, MCF-7 cells were plated into six-well plates and cultured in media containing 10% FBS for 14 days. Colonies were fixed with 4% paraformaldehyde and then stained with 0.5% crystal violet. The visible colonies were photographed and manually counted. The detail protocol was conducted according to previous study [18].
Caspase-3 activity assay
The activity of Caspase-3 was determined using a Caspase-3 activity kit (Beyotime Institute of Biotechnology, China). The total protein of cells was obtained through lysis buffer. 40 µg of protein was diluted to 50 µL of final volume that was then mixed with 75 µL of caspase-3 substrate for 3 h, the hydrolysis of Ac-DEVD-pNA by caspase-3 released free pNA (yellow formazan product) that was detected at 405 nm. Caspase-3 activity was expressed as the fold of enzyme activity compared to that of synchronized cells.
Cell cycle analysis
After AFAP1-AS1 knockdown, MCF-7 cells were harvested and washed three times with PBS. Staining with Annexin V-fluorescein isothiocyanate propidium iodide (FITC/PI) apoptosis detection kit (Life Technologies, USA) according to the manufacturer’ s recommendations. The cells were analyzed for their distribution in different phases of cell cycle on FACScalibur low cytometer using CellQuestPro software (Becton Dickinson, USA). The detailed protocol was conducted according to the manufacturer’s instructions as previously described [19].
Wound scratch assay
MCF-7 cells were grown to 80% confluence in six-well culture plates. The scratch on cell layer was created using a 200 µL pipet tip and photographed immediately (0 h) after wounding and 24 h after wounding. The wound closure was monitored by microscopy. An ocular ruler was used to verify the wound sizes.
Phalloidin staining
MCF-7 cells were fixed in 4% paraformaldehyde for 30 min. Subsequently, 0.2 µg/mL phalloidin (Invitrogen, USA) was diluted with preparation solution (0.1% NaN3, 3% BSA, 0.3% Triton X-100 in 0.01 M PBS). After washing with PBS, the cells were incubated with phalloidin solution overnight. The images were captured under a fluorescence confocal microscope. The detail protocol was conducted according to the manufacturer’s instructions as previously described [20].
Statistical analysis
A paired Wilcoxon signed-rank test was employed to evaluate AFAP1-AS1 expression in breast cancer tissues vs. adjacent normal tissues. Correlations between AFAP1-AS1 expression and clinical features were performed by a Person’s χ2 test. The optimal cut-off value of AFAP1-AS1 in tumor/normal was determined by a receiver operating characteristic (ROC) curve analysis. The Kaplan–Meier method was used to estimate survival curves, the log-rank test was used to compare between groups. The prognostic value of AFAP1-AS1 expression was tested accordingly to the high- AFAP1-AS1 vs. low- AFAP1-AS1 categorical definition. Hazard ratios (HR) and 95% confidence interval (95% CI) were calculated with the Cox proportional hazard regression model, adjusted for clinico-pathological prognostic factors. All the statistical tests were two-sided and considered significant when P < 0.05. Data in Table 1 was presented as number (percentage). The data of cell experiments were performed by One-way analysis of variance (ANOVA) with a Newman–Keuls post hoc test for multiple comparisons, these data were presented as means ± SEM. P < 0.05 was considered to be significant.
Results
The expression of AFAP1-AS1 was up-regulated in human breast cancer tissue and associated with malignancy status
To confirm the aberrant expression of AFAP1-AS1 in breast cancer, AFAP1-AS1 expression in 160 paired breast cancer tissue and adjacent normal tissue samples was measured by qRT-PCR. Results showed that AFAP1-AS1 expression was up-regulated in breast cancer tissue as compared with adjacent normal tissue (Fig. 1a). Moreover, the result of receiver operating characteristic (ROC) curve analysis indicated that AFAP1-AS1 was a good candidate to discriminate breast cancer tissue from adjacent normal tissue (sensitivity: 74%, specificity: 69%). The area under ROC curve was 0.736 (95% CI 0.64–0.83, P < 0.01; Fig. 1b). In addition, to further explore the association of AFAP1-AS1 expression with clinical characteristics, breast cancer patients were categorized as high or low expression group according to the optimal cut-off value of fold expression of AFAP1-AS1.
The expression of AFAP1-AS1 in breast cancer tissues and breast cancer cells. a AFAP1-AS1 expression in breast cancer tissues and adjacent normal tissues. b Optimal cutoff value of AFAP1-AS1 expression determined by ROC curve for 3-years overall survival. c Kaplan–Meier 3-years overall survival curves according to AFAP1-AS1 expression. d AFAP1-AS1 expression in 4 breast cancer cell lines (MCF-7, SK-RB-3, MDA-MB-231and MDA-MB-468) and normal human mammary epithelial cell line MCF-10A. Data are mean ± SEM. n = 160 (a, b) and 3 (c). **P < 0.01 vs. Adjacent tissues; ##P < 0.01 vs. MCF-10A
As shown in Table 1, the association between AFAP1-AS1 expression and clinical characteristics in 160 breast cancer patients were analyzed, and found high AFAP1-AS1 expression was remarkably correlated with TNM stage and lymph node metastasis, but not correlated with patient’s age, family history, and tumor grade, as well as ER, PR, and HER2 status. The result of Kaplan–Meier analysis using the log-rank test showed that the 3-years overall survival of the high AFAP1-AS1 group was significantly lower than that of low AFAP1-AS1 group, suggested that high expression of AFAP1-AS1 had a poor prognosis in breast cancer patients (Log Rank = 6.163, P = 0.0130; Fig. 1c). Significant factors in univariate analyses (AFAP1-AS1 expression, tumor grade, TNM stage, and lymph node metastasis) were further included in a multivariate Cox regression analysis. The result showed that AFAP1-AS1 expression was an independent prognostic factor of 3-years overall survival in breast cancer patients (hazard ratio 4.74, 95% CI 2.47–9.38, P < 0.001; Table 2). Collectively, these results suggested that up-regulated AFAP1-AS1 expression was associated with the malignancy status of breast cancer.
AFAP1-AS1 knockdown inhibits the proliferation and colony formation of breast cancer cell
To further verified the aberrantly expression of AFAP1-AS1 in breast cancer cell lines, AFAP1-AS1 expression in four breast cancer cell lines (MCF-7, SK-RB-3, MDA-MB-231and MDA-MB-468) and normal human mammary epithelial cell line MCF-10A were measured, and found the AFAP1-AS1 expression was up-regulated in breast cancer cells compared with MCF-10A (Fig. 1d). Among them, most up-regulation was detected in MCF-7, which was chosen to knockdown the expression of AFAP1-AS1 in the following studies.
Malignant proliferation and colony formation ability were the important pathological features of breast cancer cell. In view of the up-regulated AFAP1-AS1 expression in breast cancer, therefore, siRNA was transfected into MCF-7 to knockdown AFAP1-AS1 and then evaluated the proliferation and colony formation. To screen the best interference efficiency siRNA, three siRNA segments were designed according to the AFAP1-AS1 gene sequence and transfected into MCF-7. All of three siRNAs segments decreased the AFAP1-AS1 expression, while the third segment showed the best interference efficiency (Fig. 2a), which was used for following studies. As shown in Fig. 2b–d, AFAP1-AS1 knockdown decreased the cell viability and EdU positive cell percentage of MCF-7, suggesting the inhibiting role of AFAP1-AS1 knockdown on breast cancer cell proliferation. The colone number in MCF-7 was reduced after AFAP1-AS1 siRNA transfection (Fig. 2e, f), indicating AFAP1-AS1 knockdown can inhibit the colony formation of breast cancer cell.
Effect of AFAP1-AS1 knockdown on the proliferation and colony formation of MCF-7. The effect of AFAP1-AS1 siRNA on a AFAP1-AS1 expression, b cell viability, c, d EdU positive cell percentage, e, f colony formation in MCF-7. Data are mean ± SEM. n = 3. *P < 0.05, **P < 0.01 vs. control; #P < 0.05, ##P < 0.01 vs. si-NC
AFAP1-AS1 knockdown promotes apoptosis of breast cancer cell
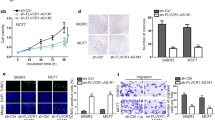
In addition to proliferation and colony formation, the effect of AFAP1-AS1 knockdown on breast cancer cell apoptosis was also explored. Caspase-3 activity, Bax and Bcl-2 were the important apoptosis-related marker. Results showed that AFAP1-AS1 knockdown decreased the Bcl-2 protein level and Caspase-3 activity in MCF-7, while increased the Bcl-2 protein level (Fig. 3a–c). The flow cytometry was used to determine the cell cycle, found that AFAP1-AS1 knockdown significantly induced G2/M phase arrest in MCF-7 (Fig. 3d). These results suggested AFAP1-AS1 knockdown promotes the apoptosis of breast cancer cell.
AFAP1-AS1 knockdown inhibited metastasis of breast cancer cell
Enhanced cell metastasis was another key pathological feature in breast cancer.
Wound healing assay, therefore, was used to investigate cell motility. As shown in Fig. 4a, b, the gap distance in si-AFAP1-AS1 was longer than si-NC, suggesting AFAP1-AS1 knockdown decreased the cell motility of MCF-7. Epithelial–mesenchymal transition (EMT) was a process characteristic by reprogramming of epithelial cells into a mesenchymal cells phenotype. It has been reported that EMT played a crucial role in breast cancer cell mobility, which finally led to metastasis. Thus, the EMT-related protein expression was measured, and found AFAP1-AS1 knockdown increased the E-cadherin protein expression, while decreased the N-cadherin and Vimentin protein expression (Fig. 4c, d). These results demonstrated that AFAP1-AS1 knockdown inhibited metastasis of breast cancer cell.
AFAP1-AS1 knockdown exhibited no effects on AFAP1 expression and actin filament integrity
The AFAP1-AS1 is derived from the antisense strand of the AFAP1. To determine the correlation between expression levels of AFAP1-AS1 and AFAP1 in breast cancer cell, we further examined the expression of AFAP1 after AFAP-AS1 knockdown. As shown in Fig. 5a, b, the all of the mRNA and protein expression of AFAP1 were not affected by AFAP-AS1 knockdown. Moreover, the actin filament integrity was not destroyed after AFAP-AS1 knockdown (Fig. 5c), which may attribute to the key role of AFAP1 in maintaining actin filament integrity.
Discussion
Although less than 2% of the human genome encodes protein-coding genes, at least 90% of the genome, including abundant pseudogenes and comparably numerous non-coding RNAs (ncRNAs), is actively transcribed. General conventions divide ncRNAs into small ncRNAs (< 200 nt in length) and long noncoding RNAs (lncRNAs, ≥ 200 nt in length) on the basis of size [21]. The regulating effect of lncRNAs on physiological and pathological functions is shown to be exerted via their interactions with microRNAs (miRNAs), mRNAs, proteins and genomic DNA [22]. Recently, more and more evidence has been demonstrated that lncRNAs are aberrantly expressed in breast cancer and associated with the poor prognosis of breast cancer patients [3]. Xu et al. constructed breast cancer-related lncRNA libraries using microarray. Microarray expression profiling suggests 1427 lncRNAs (log fold-change > 2.3, including 790 up-regulated and 637 down-regulated lncRNAs) were differently expressed between breast cancer tissues and its paired adjacent tissues [23]. In addition, Shen et al. identified 1758 lncRNAs that were differentially expressed (≥ twofold change) in triple-negative breast carcinomas [24]. Recently, Yang et al. sequenced seven pairs of HER-2-enriched subtype breast cancer and normal tissue and identified more than 1300 dysregulated expressed lncRNAs. Notable, AFAP1-AS1 was the most up-regulated lncRNA [13]. In the present study, we further measured the expression of AFAP1-AS1 in 110 paired breast cancer tissues and adjacent normal tissues and found AFAP1-AS1 expression was significantly up-regulated in breast cancer as compared with adjacent normal tissues. Moreover, we evaluated the potential correlations between AFAP1- AS1 expression and patients’ clinical characteristics. The results showed that there was a significant relationship between high AFAP1-AS1 expression and TNM stage, as well as lymph node metastasis, but high AFAP1-AS1 expression was not correlated with patient’s age, family history, and tumor grade, as well as ER, PR, and HER2 status. These results suggested that AFAP1-AS1 may be a novel diagnostic and prognostic indicator for breast cancer.
The expression of AFAP1-AS1 has been reported to be up-regulated and correlate with poor prognosis in several human cancers, such as hepatocellular carcinoma [25], non-small cell lung cancer [11], colorectal cancer [19], nasopharyngeal carcinoma [26], pancreatic ductal adenocarcinoma [10] and esophageal squamous cell carcinoma [27]. In the present study, we also found that breast cancer patients with high AFAP1-AS1 expression had a poor prognosis. It has been reported that AFAP1-AS1 acts as an oncogene to regulate tumorigenesis and progression through promoting cell proliferation, invasion, migration and inhibiting cell apoptosis. For example, AFAP1-AS1 was up-regulated in gastric cancer cells and regulated the gastric cancer cell proliferation and apoptosis [28]. Silencing of AFAP1-AS1 in hepatocellular carcinoma cell significantly reduced cell proliferation, clonal growth, cell migration, and invasion and increased apoptosis [18]. However, the function of AFAP1-AS1 in breast cancer cell biology remains unknown. Here, we measured the expression of AFAP1-AS1 in four breast cancer cell lines (MCF-7, SK-RB-3, MDA-MB-231and MDA-MB-468) and normal human mammary epithelial cell line MCF-10A. Results showed that the AFAP1-AS1 expression was up-regulated in breast cancer cell compared with MCF-10A cell. Among these breast cancer cells, most up-regulation was detected in MCF-7. AFAP1-AS1 acted as the most dysregulated lncRNA in HER-2-enriched subtype breast cancer [13], however, the present study found that high AFAP1-AS1 expression was not correlated with ER, PR, and HER2 status in breast cancer patients. Therefore, knockdown of AFAP1-AS1 in the following studies was conducted in MCF-7, in spite of its HER-2 (−). Further studies found AFAP1-AS1 knockdown inhibited the proliferation, colony formation and induced the apoptosis in MCF-7, suggesting AFAP1-AS1 served as an oncogene in breast cancer.
The AFAP1-AS1 is derived from the antisense strand of the AFAP1 (Actin Filament Associated Protein) gene. There are overlapping and complementary regions with the second AFAP1-AS1 exon and the 14, 15, and 16 exons of the AFAP1 transcripts [29]. AFAP1 is an adaptor protein of cSrc that binds to filamentous actin and regulates the activity of this tyrosine kinase to affect changes to the organization of the actin cytoskeleton. In breast cancer cells, AFAP1 has been shown to regulate cellular responses requiring actin cytoskeletal changes such as adhesion, invadopodia formation and invasion [30]. These studies urged us to evaluate the effect of AFAP1-AS1 on breast cancer cell migration and expression of EMT-related genes. Results showed that AFAP1-AS1 knockdown decreased migration and EMT-related genes expression in breast cancer cell. It has been documented that EMT is closely associated with triple negative subtype [31]. Therefore, AFAP1-AS1 may play a key role in regulating mesenchymal-like or mesenchymal stem-like phenotype in triple negative subtype, which deserved further study.
Usually, the antisense RNA regulates expression of its cognate sense gene, however, Wu et al. demonstrated that AFAP1-AS1 knockdown inhibited proliferation and colony forming ability, induced apoptosis, and reduced esophageal adenocarcinoma cell migration and invasion without altering the expression of AFAP1 [32]. Zeng et al. suggested that AFAP1-AS1 inhibited the AFAP1 mRNA translation, but did not affect the AFAP1 mRNA transcription without AFAP1 mRNA degradation in lung cancer cell [33]. In the present study, we found that AFAP1-AS1 knockdown did not affect AFAP1 mRNA and protein expression in breast cancer cells. Accordingly, the actin filament integrity was not affected after AFAP1-AS1 knockdown. Therefore, AFAP1-AS1 may not bind to its sense cognate gene, its effects may involve an AFAP1-independent mechanism during development or progression of breast cancer.
In conclusion, we confirmed that AFAP1-AS1 expression was significantly up-regulated in breast cancer tissues. The expression of AFAP1-AS1 was correlated with the malignancy status and patients with high AFAP1-AS1 expression had a poor prognosis in breast cancer. Furthermore, knockdown of AFAP1-AS1 suppressed proliferation, colony formation, metastatic ability and induced apoptosis in breast cancer cell. The present study suggested that AFAP1-AS1 acted as an oncogene in breast cancer, which may be a novel diagnostic and prognostic indicator for breast cancer. To determine whether the AFAP1-AS1 can be a therapeutic target for breast cancer, animal study and more preclinical study should be performed in further investigation.
References
Bartsch R, Bergen E. ASCO 2017: highlights in breast cancer. Memo. 2017;10:228–32.
Low SK, Zembutsu H, Nakamura Y. Breast cancer: the translation of big genomic data to cancer precision medicine. Cancer Sci. 2017;109:497
Kumar M, DeVaux RS, Herschkowitz JI. Molecular and cellular changes in breast cancer and new roles of lncRNAs in breast cancer initiation and progression. Prog Mol Biol Transl Sci. 2016;144:563–86.
Li RK, Gao J, Guo LH, Huang GQ, Luo WH. PTENP1 acts as a ceRNA to regulate PTEN by sponging miR-19b and explores the biological role of PTENP1 in breast cancer. Cancer Gene Ther. 2017;24:309–15.
Xu W, Chang J, Du X, Hou J. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother. 2017;95:1112–8.
Zheng Y, Song D, Xiao K, Yang C, Ding Y, Deng W, et al. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:83727–34.
Hui H, Zhai Y, Ao L, Cleveland JC Jr, Liu H, Fullerton DA, et al. Klotho suppresses the inflammatory responses and ameliorates cardiac dysfunction in aging endotoxemic mice. Oncotarget. 2017;8:15663–76.
Liu FT, Xue QZ, Zhu PQ, Luo HL, Zhang Y, Hao T. Long noncoding RNA AFAP1-AS1, a potential novel biomarker to predict the clinical outcome of cancer patients: a meta-analysis. Onco Targets Ther. 2016;9:4247–54.
Han X, Wang L, Ning Y, Li S, Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biol Res. 2016;49:36.
Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y, et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137.
Deng J, Liang Y, Liu C, He S, Wang S. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacother. 2015;75:8–11.
Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–18.
Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761–72.
Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–10.
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010;134:e48–72.
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43.
Li WQ, Li XH, Wu YH, Du J, Wang AP, Li D, et al. Role of eukaryotic translation initiation factors 3a in hypoxia-induced right ventricular remodeling of rats. Life Sci. 2016;144:61–8.
Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L, et al. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9699–707.
Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152–9.
Wang L, Cao J, Shi Z, Fan W, Liu H, Deng J, et al. Experimental study on the neurotoxic effect of beta-amyloid on the cytoskeleton of PC12 cells. Int J Mol Med. 2018;41:2764–70.
Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219–51.
Giroud M, Scheideler M. Long non-coding RNAs in metabolic organs and energy homeostasis. Int J Mol Sci. 2017;18:2578
Xu N, Wang F, Lv M, Cheng L. Microarray expression profile analysis of long non-coding RNAs in human breast cancer: a study of Chinese women. Biomed Pharmacother. 2015;69:221–7.
Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D, et al. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015;6:21730–9.
Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q, Wu JY, et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590–8.
Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y, et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001–11.
Zhou XL, Wang WW, Zhu WG, Yu CH, Tao GZ, Wu QQ, et al. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog. 2016;55:2095–105.
Guo JQ, Li SJ, Guo GX. Long Noncoding. RNA AFAP1-AS1 promotes cell proliferation and apoptosis of gastric cancer cells via PTEN/p-AKT pathway. Dig Dis Sci. 2017;62:2004–10.
Baisden JM, Gatesman AS, Cherezova L, Jiang BH, Flynn DC. The intrinsic ability of AFAP-110 to alter actin filament integrity is linked with its ability to also activate cellular tyrosine kinases. Oncogene. 2001;20:6607–16.
Cunnick JM, Kim S, Hadsell J, Collins S, Cerra C, Reiser P, et al. Actin filament-associated protein 1 is required for cSrc activity and secretory activation in the lactating mammary gland. Oncogene. 2015;34:2640–9.
Lv ZD, Kong B, Liu XP, Jin LY, Dong Q, Li FN, et al. miR-655 suppresses epithelial-to-mesenchymal transition by targeting Prrx1 in triple-negative breast cancer. J Cell Mol Med. 2016;20:864–73.
Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956 e4–66 e4.
Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–37.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consent
All patients had signed the informed consent before surgery.
Ethics approval
Clinicopathological follow-up information from breast cancer patients was available. All clinical investigations in the present study were approved by the Human Ethics Committee in the First Hospital of Lanzhou University (Lanzhou, China) and conducted according to the principles expressed in Declaration of Helsinki.
Conflict of interest
The authors declare that they have no conflict of interest.
About this article
Cite this article
Ma, D., Chen, C., Wu, J. et al. Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis and promotes carcinogenesis of breast cancer. Breast Cancer 26, 74–83 (2019). https://doi.org/10.1007/s12282-018-0891-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-018-0891-3