Abstract
Soil-borne diseases are closely related to rhizosphere microecosystem. While, plant species and genotypes are important factors affected rhizosphere microecosystem. In this study, the rhizosphere soil microbial community and metabolites of susceptible and resistant tobacco cultivars were investigated. The results showed that there were significant differences in the rhizosphere microbial community and metabolites between susceptible cultivar Yunyan87 and resistant cultivar Fandi3. Furthermore, the rhizosphere soil of Fandi3 showed a higher microbial diversity than that of Yunyan87. The abundance of R. solanacearum was much higher in the rhizosphere soil of Yunyan87 than in the rhizosphere soil of Fandi3, resulting in a higher disease incidence and index. While the abundance of beneficial bacteria in the rhizosphere soil of Fandi3 were higher than that of Yunyan87. Additionally, there were significant differences in metabolites between Yunyan87 and Fandi3 cultivars, and 4-hydroxybenzaldehyde, 3-hydroxy-4-methoxybenzoic acid, vamillic aldehyde, benzoic acid, 4-hydroxybenzyl alcohol, p-hydroxybenzoic acid and phthalic acid were notably high in Yunyan87. Redundancy analysis (RDA) indicated that the rhizosphere microbial community of Fandi3 and Yunyan87 were highly correlated with various environmental factors and metabolites. Overall, susceptible and resistant tobacco cultivars had different impact on rhizosphere microbial community and metabolites. The results expand our understanding of the roles of tobacco cultivars in plant-micro-ecosystem interactions, and provide a basis for the control of tobacco bacterial wilt.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil-borne pathogens have become one of the potential threats to agriculture production, causing great losses to the yield of many kinds of crops (Raaijmakers et al., 2009). Tobacco bacterial wilt (TBW) caused by Ralstonia solanacearum is one of the most serious soil-borne diseases, which caused huge yield and economic loss to tobacco production in China (Jiang et al., 2017). It is very necessary to find effective and sustainable measures to control TBW.
The rhizosphere soil is a special microecosystem, which includes soil physicochemical properties, microorganism and plant (Sun et al., 2013). Rhizosphere soil is considered critical to the maintenance of soil health and quality (Garbeva et al., 2004). Several reports have shown that soil-borne disease is associated with rhizosphere soil microbial community (Wang et al., 2017; Xiong et al., 2017). The interaction between the rhizosphere microorganisms and plants has an important impact on plant life activities, such as promoting plant growth, preventing microbial pathogens and so on (Carrión et al., 2019; Durán et al., 2018; Kwak et al., 2018). Rhizosphere microorganisms can also transform and decompose nutrients that are difficult for plants to absorb and utilize, thereby improving the efficiency of plant utilization of nutrients (Zhang et al., 2019). At the same time, plant root exudates can provide metabolites necessary for rhizosphere bacteria to recruit beneficial bacteria, and these metabolites can also inhibit the enrichment of some bacteria in plant roots (Huang et al., 2019; Yuan et al., 2018). Plant species and genotypes are also important factors that affect the rhizosphere microecosystem (Huang et al., 2019; Zhang et al., 2019). Plants with different genotypes have different responses to the environment, and their rhizosphere microecosystem may also be different (Zhang et al., 2019). Therefore, studying the role of plant genotypes in rhizosphere soil microecosystem is helpful to control soil-borne diseases.
Our previous study has analyzed gene-expression profiles of susceptible and resistant tobacco varieties (Yunyan87 and Fandi3) at different stages after R. solanacearum infection, and revealed the molecular mechanism of tobacco resistance to R. solanacearum (Li et al., 2021). However, there is still a lack of systematic analysis of the rhizosphere soil microecosystem of susceptible and resistant cultivars, and the suppressing of TBW.
In this study, the rhizosphere soil microbial community and its metabolites of susceptible and resistant tobacco cultivars were analyzed. The objective was to understand the role of tobacco cultivars in the rhizosphere soil microecosystem. Comparing the rhizosphere microbial community and metabolites between susceptible and resistant tobacco cultivars will help us to understand the mechanism of genotype on rhizosphere soil microecosystem in suppressing soil-borne diseases. This study hypothesized that there were significant differences between susceptible and resistant tobacco cultivars in rhizosphere soil microecosystem, which were related to the resistance of tobacco cultivars to TBW.
Materials and Methods
Field Experiment
The field trial was carried out in tobacco fields with 15 years of continuous cropping tobacco fields in Xuanen County (109° 26′ 20″ E, 29° 59′ 55″ N), Hubei province, China. The trial field was divided into three blocks according to the experimental design, each 220 m2 in size. Each block was divided into two plots of 110 m2, representing two plantation systems: (1) Yunyan87 (susceptible to TBW, YY87); (2) Fandi3 (resistant to TBW, FD3). There were 360 tobacco plants per block, 180 plants in each plot. Tobacco cultivars Yunyan87 and Fandi3 were provided by the Tobacco Research Institute of Hubei, Wuhan, China. The seeds of Yunyan87 and Fandi3 were surface sterilized in 10% NaClO for 5 min, rinsed with sterile distilled water three times to remove the disinfectant residue, then cultured in floating polystyrene trays in a greenhouse. When seedlings grew to the 4–5 leaf stage, the susceptible and resistant cultivars were transplanted to the trail field. The planting density of Yunyan87 and Fandi3 were the same, and all plantation systems were randomly placed in the field.
Disease Occurrence in the Fields
Symptoms of TBW across Yunyan87 and Fandi3 plantation systems were monitored from 45 to 105 days post-transplantion. The disease incidence (I) and disease index (DI) of TBW based on a severity scale of 0–9 was described by Chen et al. (2020). Briefly, “0” represents plants without visible symptoms; “1” represents the presence of occasional chlorotic spots on stems, or less than half of the leaves wilted on unilateral stems; “3” represents the presence of a black streak less than half the height of the stem, or between half to two-thirds of the leaves wilted on unilateral stems; “5” represents the presence of a black streak over half the length of the stem, but not reaching the top of the stem, or more than two-thirds of the leaves wilted on unilateral stems; “7” represents the presence of a black streak going the top of the stem, or all leaves wilted; and “9” represents the dead plant. Based on the number of plants in each rating scale, I and DI of TBW were calculated as follows formula: I = n′/N × 100%, and DI = ∑(r × n)/(N × 9) × 100, where n′ is the total number of infected tobacco plants, N is the total number of plants, r is the rating scale of disease severity, and n is the number of infected tobacco plants with a rating of r.
Rhizosphere Soil Sampling and Physicochemical Properties Analysis
Rhizosphere soil were collected by five-point sampling method at 45 days, 75 days, and 105 days tobacco post-transplantation. For each treatment, a total of 15 plants were selected randomly, and each replicate consisted of a mixture of rhizosphere soil from five plants. The loosely attached soil around the root area was shaken and collected separately into the plastic bags and transported to the laboratory in an ice box. Then the soil samples crushed and sifted through a sterile 2 mm sieve, partitioned into two subsamples, one was stored at − 80 °C for microbiological and metabolome analysis, and the other subsamples were air-dried for physicochemical properties analysis. The measurement of soil pH, alkali-hydrolyzed nitrogen (AN), available phosphorus (AP), available potassium (AK), organic matter (OM) was performed according to Hu et al. (2021).
Soil DNA Extraction
Soil microbial genomic DNA was extracted from 0.5 g rhizosphere soil using the FastDNA Spin Kit (MP Biomedicals) following the manufacture’s protocol. The integrity of DNA samples was determined by 1% agarose gel electrophoresis. Then the concentration and purity of the DNA were determined using a Nanodrop ND-1000 Spectrophotmeter (Nanodrop Tchenologies).
Microbial rRNA Gene Amplification and Illumina Sequencing (PCR Amplification and Sequencing)
The extracted soil genomic DNA was used as template to amplify 16S rRNA and ITS rRNA genes, respectively. The V4 regions of 16S rRNA gene were amplified using primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Wu et al., 2016), and the ITS1 regions of ITS rRNA gene were amplified using primers ITS5-1737F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2-2043R (5′-GCTGCGTTCTTCATCGATGC-3′) (Zhang et al., 2017). The library was sequenced on an Illumina HiSeq platform (Novogene Bioinformatics Technology Co., Ltd). The sequence quality was statistically analyzed by CASAVA1.8. The raw sequence data was preliminarily filtrated using the FASTX Toolkit 0.0.13 software package, removing the low mass base at the tail of the sequence (Q value less than 20) and the sequences with lengths less than 35 bp. Finally, the length of the valid reads was approximately 250 bp. All effective tags of all samples were clustered using Uparse software (V7.0.1001, http://drive5.com/uparse/). Sequences with ≥ 99.5% identity for 16S rDNA and sequences with ≥ 97% identity for ITS were assigned to the same OTUs (operational taxonomic units). The OTUs, Chao1, and Shannon index were calculated with QIIME (Version 1.7.0) to evaluate the richness and diversity of soil microbial community (Hill et al., 2003).
Quantification of R. solanacearum by Real-Time PCR
The abundance of R. solanacearum was determined by quantitative real-time PCR analysis of lpxC gene of R. solanacearum strain LF-17. Each reaction was performed in a 10 μl volume containing 5 μl of SYBR Green Supermix, 1 μl sample DNA, 3 μl ddH2O, and 0.5 μl of each primer. The primers used for the amplification were as follows: 759 (5′-GTCGCCGTCAACTCACTTTCC-3′) and 760 (5′-GTCGCCGTCAGCAATGCGGAATCG-3′) (Opina et al., 1997). Real-time PCR was performed on a BIO-RAD C1000 Touch™ Thermal Cycler with CFX96 real-time system. Quantitative PCR were performed under the following conditions: 95 °C for 3 min and 40 cycles of 95 °C for 10 s, 54 °C for 20 s, and 72 °C for 1 min. Standard curves were generated with serial dilution series of quantified plasmid DNA (pMD 19-T vector, Takara). Three independent quantitative PCRs were performed for each sample.
Soil Metabolite Extraction
The ground soil (1 g) was extracted in 5 mL 80% (v/v) methanol (10 min, 20 °C) using sonicator. The residue was extracted twice with the same procedure and the total combined supernatant was filtered through Whatman filter paper (125 mm). Then, the supernatant was placed in a glass derivative bottle for vacuum draining.
The dried extracts were added to 60 μl of a methoxyamination hydrochloride (20 mg/ml in pyridine), and incubated for 30 min at 80 °C. Then, 80 μl of the BSTFA regent (1% TMCS, v/v) was added, and incubated for 1.5 h at 70℃. All samples were analyzed by gas chromatograph system coupled with a Pegasus HT time-of-flight mass spectrometer (Lykogianni et al., 2020) (GC-TOF–MS).
GC-TOF–MS Analysis and Data Preprocessing
GC-TOF–MS analysis was performed using an Agilent 7890 gas chromatograph system (Agilent Technologies Inc., USA) coupled with a Pegasus HT time-of-flight mass spectrometer (LECO Corporation). The system utilized a DB-5MS capillary column coated with 5% diphenyl cross-linked with 95% dimethylpolysiloxane (30 m × 250 μm inner diameter, 0.25 μm film thickness; J&W Scientific). The samples (1 μl) were injected in splitless mode. Helium was used as the carrier gas, the front inlet purge flow was 3 ml/min, and the gas flow rate through the column was 1 ml/min. The initial temperature of the oven was 50 °C for 1 min, raised to 310 °C at a rate of 20 °C/min, then kept for 6 min at 310 °C. The injection, transfer line, and ion source temperatures were 280, 280, and 250 °C, respectively. The energy was -70 eV in electron impact mode. The mass spectrometry data were acquired in full-scan mode with 50–500 Da mass (scan rate of 12.5 scans per second) after a solvent delay of 4.78 min (Lykogianni et al., 2020).
Chroma TOF 4.3X software (LECO Corporation) and the LECO-Fiehn Rtx5 database were used for raw peaks exacting, data baselines filtering and calibration, peak alignment, deconvolution analysis, peak identification and integration of the peak area. Both of mass spectrum match and retention index match were considered in metabolites identification.
Statistical Analysis
Statistically significant differences (p < 0.05) in disease incidence, disease index, soil physicochemical properties, microbial alpha-diversity, the abundance of R. solanacearum, and root exudate compound abundance between Yunyan87 and Fandi3 systems were evaluated by Student’s t test using SPSS version 18.0 (IBM). Principal coordinate analysis (PCoA) based on the OTUs to explore the differences in bacterial and fungal community composition. The difference of root exudate composition between Yunyan87 and Fandi3 systems was analyzed by principal component analysis (PCA). Redundancy analysis (RDA) based on the relative abundances of bacterial and fungal genera, soil physicochemical properties (pH, AN, AP, AK, and OM), and metabolites. Based on the Sørensen’s distance of disease index, soil physicochemical properties, metabolites and microbial community, the Spearman correlation coefficient and significance were calculated with the R package Hmisc.
Results
The Disease Incidence and Disease Index of TBW
The symptoms of TBW in Yunyan87 (susceptible cultivar) and Fandi3 (resistant cultivar) were recorded at 45 days, 75 days and 105 days post-transplantation, and the disease incidence (I) and index (DI) of TBW were calculated (Table 1). From 45 to 105 days post-transplantation, the I and DI of TBW in Fandi3 were significantly lower than that in Yunyan87 (p < 0.05). The results indicated that the rhizosphere ecosystem of Fandi3 could more effectively inhibit the incidence and severity of TBW than that of Yunyan87.
Physicochemical Properties in Rhizosphere Soil
Five physicochemical properties of the rhizosphere soil of Yunyan87 and Fandi3 at 45 days, 75 days and 105 days post-transplantation were analyzed (Table 2). From 45 to 105 days post-transplantation, except alkali-hydrolyzed nitrogen (AN), there was significant difference in available phosphorous (AP), available potassium (AK), pH and organic matter (OM) between Yunyan87 and Fandi3. The OM content in Yunyan87 system was observed higher (p < 0.05) than that in Fandi3 systems. While, the content of AP, AK and pH in Yunyan87 system was lower (p < 0.05) than in Fandi3 system, respectively. It implied that soil acidification may be inhibited in the rhizosphere soil of Fandi3 to prevent the incidence of TBW.
Bacterial Diversity and Community Structure in Rhizosphere Soil
In total, 1,316,690 high-quality raw sequences with an average length of 251 bp for bacteria were obtained from rhizosphere soil samples after removing low-quality reads. The OTUs number, Chao1 and Shannon index were used to assess and compare the diversity and richness of bacterial community between susceptible and resistant tobacco varieties (Table 3). From 45 to 105 days post-transplantation, the OTUs number and Chao1 were both significantly higher in Fandi3 system than in Yunyan87 system. Analysis by shannon, a higher richness of bacteria was also found in Fandi3 system. These results indicated that Fandi3 system has a higher bacteria diversity and richness than Yunyan87 system.
The result of PCoA (principal co-ordinates analysis) with the weighted UniFrac distance showed that PC1 and PC2 explained 43.21% and 23.55% of the variations in the bacterial community variations, respectively (Fig. 1A). YY87 (YY87-45, YY87-75, YY87-105) and FD3 (FD3-45, FD3-75, FD3-105) treatments were separated from each other at PC1 axis. Therefore, the results of the PCoA suggested that the structure of soil bacterial community were different between Yunyan87 and Fandi3. The two-way PERMANOVA analysis showed that both growth stage (R2 = 0.196, p = 0.002) and cultivar (R2 = 0.566, p = 0.001) had significant effects on the diversity of bacterial with cultivar playing a bigger role (Table S1).
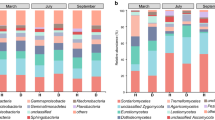
Soil bacterial community in Fandi3 and Yunyan87 at 45 days, 75 days, and 105 days post-transplanted, respectively. A Principal coordinate analysis (PCoA) of soil bacterial community with permutational MANOVA was conducted to analysis the bacterial community were significantly different between Fandi3 and Yunyan87. B The relative abundance of bacterial phyla in soil samples. C Hierarchical cluster analysis of predominant bacterial genera
A total of 59 bacterial phyla were identified from all soil samples. The top ten abundant bacterial phyla were selected to compare the changes of bacterial communities in rhizosphere soil between Yunyan87 and Fandi3 systems (Fig. 1B). From 45 to 105 days post-transplantation, Proteobacteria was dominant (31.61–58.81%), followed by Acidobacteria (8.40–21.63%), Actinobacteria (4.42–9.54%), Bacteroidetes (4.59–7.02%), Gemmatimonadetes (3.92–8.29%), Chloroflexi (2.05–6.24%), Verrucomicrobia (1.27–3.84%), Firmicutes (0.59–3.23%), Thaumarchaeota (0.36–8.07%), and Cyanobacteria (0.26–1.33%). The relative abundance of Chloroflexi and Firmicutes were lower in Fandi3 than Yunyan87. The result indicated that the soil bacterial community composition of Yunyan87 and Fandi3 were different.
The Heatmap analysis of the top 30 genera with hierarchical clusters was used to identify the different composition of bacterial community structure between Yunyan87 and Fandi3 systems (Fig. 1C). Yunyan87 and Fandi3 systems were divided into two broad categories, suggesting there were distinction of bacterial community structure between Yunyan87 and Fandi3 systems. A statistical comparison of the top 30 abundant bacterial genera was conducted to better understand the difference of relative abundance at the genus level between susceptible and resistant tobacco varieties (Fig. 1C). Fourteen of the 30 bacterial genera showed significantly different between the two varieties. The relative abundances of Massilia, Bryobacter, and Ralstonia in Fandi3 were significantly lower than that in Yunyan87. However, the relative abundances of Ramlibacter, Paenibacillus, Novosphingobium, Rhodococcus, Haliangium, Sphingobium, Flavobacterium, Sphingomonas, Granulicella, Solirubrobacter and Bacillus in Fandi3 were more abundant than that in Yunyan87 (Fig. 1C).
Fungal Diversity and Community Structure in Soil
All rhizosphere soil samples consist of 966,546 high-quality raw sequences for fungal. The difference of the OTUs number, Chao1 and Shannon index of fungal community between susceptible and resistant tobacco varieties were also analyzed (Table 3). The OTUs number, Chao1 and Shannon index in Fandi3 were also higher than those in Yunyan87, indicating Fandi3 system has a higher fungal diversity and richness than Yunyan87 system.
According to PCoA analysis, PC1 and PC2 explained 68.49% of the total fungal community (Fig. 2A). The fungal community of Yunyan87 and Fandi3 systems were separated from each other at PC1 axis, and different tobacco growth stages within one treatment showed close distances, indicating the fungal community between Yunyan87 and Fandi3 systems was different. The two-way PERMANOVA analysis showed that both growth stage (R2 = 0.211, p = 0.002) and cultivar (R2 = 0.319, p = 0.001) had significant effects on the diversity of fungal with cultivar playing a bigger role (Table S1).
Soil fungal community in Fandi3 and Yunyan87 at 45 days, 75 days, and 105 days post-transplanted, respectively. A Principal coordinate analysis (PCoA) of soil fungal community with permutational MANOVA was conducted to analysis the fungal community were significantly different between Fandi3 and Yunyan87. B The relative abundance of fungal phyla in soil samples. C Hierarchical cluster analysis of predominant fungal genera
10 main known fungal phyla were identified from all soil samples (Fig. 2B), including Ascomycota (6.19–75.45%), followed by Chytridiomycota (0.60–22.62%), Basidiomycota (2.07–9.66%), Mortierellomycota (1.93–5.66%), Olpidiomycota (0–3.79%), Rozellomycota (0.02–0.36%), Mucoromycota (0.04–0.28%), Glomeromycota (0.02–0.22%) and Monoblepharomycota (0–0.04%), Aphelidiomycota (0–0.02%). The relative abundance of Ascomycota decreased from 45 to 75 days post-transplantation, and increased from 75 to 105 days post-transplantation. Additionally, Ascomycota in Fandi3 system were abundant than that in Yunyan87 system.
In the Heatmap for fungal community structures (Fig. 2C), two broad categories were divided, and same treatments in different period were clustered together, indicating there were distinction of fungal community structure in different cultivars. Comparison of the relative abundances of the top 30 fungal genera showed significant variations between Yunyan87 and Fandi3 systems (Fig. 2C). The relative abundance of Fusarium in Fandi3 was lower than that in Yunyan87, whereas the relative abundances of Chaetomium, Conlarium, Cercophora, Aspergillus, and Trichoderma in Fandi3 system were more abundant than that in Yunyan87 system.
The Abundance of R. solanacearum
The abundance of R. solanacearum in the rhizosphere soil of Yunyan87 and Fandi3 at different growth periods was analyzed by real-time PCR. The abundance of R. solanacearum increased from 45 to 105 days. The variation trend of the abundance of R. solanacearum in Yunyan87 and Fandi3 was similar. However, the abundance of R. solanacearum in Yunyan87 was significantly higher than Fandi3 from 75 to 105 days (Fig. 3).
Metabolomics Analysis
In order to explore how susceptible and resistant cultivars alters the soil microbiome, soil metabolites of Yunyan87 and Fandi3 at different tobacco growth periods were collected and analyzed by GC-TOF–MS. 5,893 and 4,497 features were detected from positive and negative modes, respectively. A total of 606 metabolites were obtained and assigned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Human Metabolome Database (HMDB). PCA was applied to understand the clustering features of Yunyan87 and Fandi3 metabolites at different tobacco growth periods. The first components (PC1) showed 36.29% difference in variation, and PC2 explained 20.25% of the variance (Fig. 4A). The metabolites of Yunyan87 and Fandi3 cultivars were separated from each other, indicating the overall exudation patterns from the roots of Yunyan87 were distinct from that of Fandi3. The two-way PERMANOVA analysis showed that growth stage (R2 = 0.037, p = 0.016) and cultivar (R2 = 0.642, p = 0.001) had significant effects on the diversity of metabolites with cultivar playing a bigger role. The 132 of the 606 metabolites could be placed into broad categories in which there were their chemical nature, namely lipids and lipid-like molecules (LLMs, 33 compounds), organic oxygen compounds (OOCs, 25 compounds), organic acids and derivatives (OADs, 24 compounds), benzenoids (15 compounds), organoheterocyclic compounds (ORCs, 10 compounds), phenylpropanoids and polyketides (PHPs, 8 compounds), organic nitrogen compounds (ONCs, 5 compounds), nucleosides (3 compounds), hydrocarbons (2 compounds), and others (7 compounds). After evaluating differences of these metabolites between two cultivars, it was found that LLMs, OADs, and benzenoids in Fandi3 were significantly lower than that in Yunyan87, while OOCs, ORCs, and ONCs in Fandi3 were significantly higher than that in Yunyan87 (Fig. 4B). The Heatmap with hierarchical clusters demonstrated that 30 metabolites exhibited significant variations between Yunyan87 and Fandi3 systems (Fig. 4C). Moreover, 7 of 30 metabolites (4-hydroxybenzaldehyde, vamillic aldehyde, benzoic acid, 3-hydroxy-4-methoxybenzoicacid, 4-hydroxybenzyl alcohol, p-hydroxybenzoic acid, phthalic acid) showed higher abundance than that in Yuyan87, while the remaining (cerotinic acid, sedoheptulose, etc.) showed the converse tendency (Fig. 4C). These results indicated that root exudates in Fandi3 had an effect on the prevention of soil acidification and the incidence of TBW.
Principal component analysis (PCA) of root exudates of Fandi3 and Yunyan87 at 45 days, 75 days and 105 days post-transplanted, respectively (A). The permutational MANOVA analysis was conducted to evaluate the difference of metabolites between Fandi3 and Yunyan87. Cumulative peak area of compound categories (B). Heatmap analysis (left) and VIP score plot (right) of root exudates changes of Fandi3 and Yunyan87 (C). Statistical analysis is by t test (*p < 0.05, **p < 0.01, ***p < 0.001). ONCs organic nitrogen compounds, PHPs phenylpropanoids and polyketides, ORCS organoheterocyclic compounds, OADs organic acids and derivatives, OOCs organic oxygen compounds, LLMs lipids and lipid-like molecules
Relationships Among Disease Index, Microbial Community, Soil Physicochemical Properties and Metabolites
All the disease index, soil metabolites and soil physicochemical properties show significantly positive correlation with bacterial and fungi diversity (Table S2). The RDA was used to investigate the special relationships among microbial genera, soil physicochemical properties and metabolites (Figs. 5, 6). The correlation analysis of 14 dominant bacterial genera, soil physicochemical factors, and metabolites is shown in Fig. 5. It is clear that the bacterial community of Yunyan87 and Fandi3 cultivars were separated from each other. Additionally, the relative abundances of 11 bacteria (Ramlibacter, Bacillus, Sphingomonas, Haliangium, Novosphingobium, Sphingobium, Paenibacillus, Rhodococcus, Granulicella, Flavobacterium and Solirubrobacter) in Fandi3 were positively correlated with pH, available potassium (AK), and available phosphorous (AP). While, the relative abundances of Massilia, Ralstonia, and Bryobacter in Yunyan87 were positively correlated with organic matter (OM), alkali-hydrolyzed nitrogen (AN), 4-hydroxybenzyl alcohol, vamillic acid, 4-hydroxybenzaldehyde, benzoic acid, 3-hydroxy-4-methoxybenzoci acid, phthalic acid, and p-hydroxybenzoic (Fig. 5). For the fungi, the correlation analysis of 6 dominant fungal genera, soil physicochemical factors, and metabolites is shown in Fig. 6. In the first axis, Fusarium was related to OM, AK, phthalic acid, p-hydroxybenzoic, and 4-hydroxybenzaldehyde. The relative abundances of 4 fungi (Conlarium, Aspergillus, Trichoderma, and Cercophora) were positively correlated with AN, pH, and AP (Fig. 6).
Redundancy analysis (RDA) of 14 dominant bacterial genera and soil properties changes (A). RDA of 14 dominant bacterial genera and metabolites changes (B). The soil properties and metabolites are indicated with arrows, including soil pH, available nitrogen (AN), available phosphorous (AP), available potassium (AK), organic matter (OM), 4-hydroxybenzaldehyde, 3-hydroxy-4-methoxybenzoicacid, vamillic aldehyde, benzoic acid, 1, 4-discarboxybenzene, p-hydroxybenzoic acid and phthalic acid
Redundancy analysis (RDA) of 6 dominant fungal genera and soil properties changes (A). RDA of 6 dominant fungal genera and metabolites changes (B). The soil properties and metabolites are indicated with arrows, including soil pH, available nitrogen (AN), available phosphorous (AP), available potassium (AK), organic matter (OM), 4-hydroxybenzaldehyde, 3-hydroxy-4-methoxybenzoicacid, vamillic aldehyde, benzoic acid, 1,4-discarboxybenzene, p-hydroxybenzoic acid and phthalic acid
Discussion
Rhizosphere is a special micro-ecosystem of plant microorganism interactions. Rhizosphere microbial community plays an important role in inhibiting soil-borne diseases and promoting plant growth. Plant roots release a variety of compounds into the surrounding soil. The composition of rhizosphere soil metabolites is affected by the plant species and genotypes (Siciliano et al., 1998; Sun et al., 2013). Rhizosphere soil metabolites create unique environments for rhizosphere microorganisms. In this study, rhizosphere microbial community and metabolites of susceptible cultivar Yunyan87 and resistant cultivar Fandi3 were observed. We found that there were significant differences in the rhizosphere microbial community and metabolites between Yunyan87 and Fandi3.
Changes in the rhizosphere microbial community structure are closely related to factors such as soil physicochemical properties, plant species, and genotypes. Some studies have shown that rhizosphere microbial community structure in natural systems responds to plant species and genotype (An et al., 2011; Sun et al., 2013). In this study, resistant cultivar Fandi3 showed a higher microbial diversity and richness than that of susceptible cultivar Yunyan87 (Table 3). Diverse microbial community is less vulnerable to pathogens than simple microbial community (van Elsas et al., 2012). Therefore, Fandi3 is more resistant to pathogens than Yunyan87. The PCoA results suggest that there are differences in the rhizosphere microbial community structure between Yunyan87 and Fandi3 cultivars (Figs. 1A, 2A). Cultivar had significant effects on the diversity of microbial community (Table S1). Similar to previous study, plant genotype had a significant impact on the soil microbial community composition (Yao & Wu, 2010). It was also testified by the Heatmap, showing the difference of microbial community structure between Yunyan87 and Fandi3. Moreover, the genera Ramlibacter, Paenibacillus, Novosphingobium, Rhodococcus, Haliangium, Sphingobium, Flavobacterium, Sphingomonas, Granulicella, Solirubrobacter, Bacillus, Chaetomium, Conlarium, Cercophora, Aspergillus, and Trichoderma were more abundant in the Fandi3 (Figs. 1C, 2C). Most of these genera were reported to be beneficial microorganisms, which are beneficial to soil nutrient cycling, promote plants growth, and protect plants from pathogens (Badri et al., 2009; Pattee, 1969; Raza et al., 2016; Silva et al., 2019). The genus Bacillus is a beneficial bacteria in the soil, which can participates in many processes in the soil (Yang et al., 2011). Some Bacillus species can produce volatile organic compounds which can inhibit the growth of R. solanacearum (Raza et al., 2016). Some Bacillus species can fix nitrogen and increase the tolerance of plants to diseases (Awasthi et al., 2011). There were also some Bacillus species can kill harmful bacteria, arthropods or nematodes in rhizosphere soil (Huang et al., 2005; Kayalvizhi & Gunasekaran, 2010; Kramarz et al., 2007). The genera Rhodococcus and Bacillus were reported as phosphate solubilizing bacteria, which can promote phosphorus cycling in rhizosphere soil (Chen et al., 2006). The genera Granulicella and Solirubrobacter were found to use different substances as carbon sources and participate in carbon cycling in the soil (Mannisto et al., 2012; Sakai et al., 2014). It has been well documented that the genera Paenibacillus, Novosphingobium, Haliangium, Sphingobium, Sphingomonas, Flavobacterium, Bacillus, Aspergillus and Trichoderma as antagonistic microorganisms can directly interact with plant roots to produce bioactive substances, promote plant growth and resist biotic and abiotic stresses, so as to inhibit pathogenic bacteria (Badri et al., 2009; Pattee, 1969; Ramesh et al., 2009; Raza et al., 2016; Silva et al., 2019; Xue et al., 2013). In addition, the abundances of pathogens Massilia, Bryobacter, Ralstonia, and Fusarium were significant lower in Fandi3 than Yunyan87 (Figs. 1C, 2C). The abundances of beneficial bacteria were higher in Fandi3, while pathogens were lower, indicating that resistant cultivar Fandi3 could promote the growth of beneficial bacteria and inhibit the growth of pathogens. More and more evidences show that root exudates play an important role in plant disease resistance. The root exudates of resistant cultivar had inhibitory activity against pathogens, while the exudates of susceptible cultivar had promoting effects on pathogens (Wu et al., 2006; Zhang et al., 2020).
The interaction between plant root exudates, soil and microbes can significantly change soil physicochemical properties, which in turn alter the microbial community in the rhizosphere (Haichar et al., 2014; Huang et al., 2014; Nihorimbere et al., 2011). Our study showed that the contents of AP, AK, and pH in the rhizosphere soil of resistant cultivar Fandi3 were higher than those of susceptible cultivar Yunyan87 (Table 2). The content of OM in the rhizosphere soil of Fandi3 was lower than that of Yunyan87 (Table 2). One possible explanation is that root exudates change soil physicochemical properties and microbial community. Studies have shown that phosphorus can increase the soil microbial diversity, and the increased phosphorus supply significantly decreased the relative density of R. Solanacearum (Leff et al., 2015; Yang et al., 2018). Soil pH has been reported as an environmental factor can regulate the soil microbial community, and the increased pH is important for inhibiting R. Solanacearum (Chen et al., 2020). Root exudates play an important role in regulating the interactions between microorganisms and plants (Huang et al., 2014; Wu et al., 2015). In current study, the Heatmap showed that there were significant differences in metabolites between Yunyan87 and Fandi3 cultivars. Metabolites 4-hydroxybenzaldehyde, 3-hydroxy-4-methoxybenzoic acid, vamillic aldehyde, benzoic acid, 4-hydroxybenzyl alcohol, p-hydroxybenzoic acid and phthalic acid were found notably high in Yunyan87 (Fig. 4). This similar to previous research which indicated that the content of oxalic acids in root exudates of susceptible cultivar was significantly higher than that of resistant cultivar (Wu et al., 2015). Additionally, organic acids could significantly increase the recruitment of R. solanacearum to tobacco root (Wu et al., 2015). As an important component of root exudates, organic acids can stimulate the growth of pathogenic microorganisms and aggravate the occurrence of soil-borne diseases (Haichar et al., 2014; Li et al., 2017; Liu et al., 2015). Previous studies have been shown that organic acids benzoic acid and p-hydroxybenzoic acid from tobacco root exudates can inhibit the growth of pant and simulate the growth of R. solanacearum (Li et al., 2017; Liu et al., 2015; Wu & Wang, 2006). Hasegawa et al. (2019) showed several aromatic acids secreted by plants are chemoattractants of R. solanacearum. The susceptibility of Yunyan87 may be related to the organic acids exuded by roots to attract R. solanacearum.
After RDA, the relationships among microbial genera, soil physicochemical properties and metabolites showed that pH, AK, AP, OM, AN, 4-hydroxybenzyl alcohol, vamillic acid, 4-hydroxybenzaldehyde, benzoic acid, 3-hydroxy-4-methoxybenzoci acid, phthalic acid and p-hydroxybenzoic acid played major roles in the shaping of soil microbial community (Figs. 5, 6). In accordance with other studies, soil physicochemical properties and metabolites were correlated with microbial abundance (Song et al., 2020; Wang et al., 2017). Besides, the distribution of changed some beneficial microbial were positively correlated with pH, AK, and AP. Some pathogens were positively correlated with OM, AN, and some organic acids (Figs. 5, 6). In accordance with other researches, plant root exudates could change soil physicochemical properties, provide necessary metabolites for rhizosphere bacteria to recruit beneficial bacteria, and inhibit the accumulation of certain pathogens in plant roots (Sun et al., 2013; Yuan et al., 2018; Zhalnina et al., 2018).
The present study showed a range of correlations between disease index, microbial community, soil physicochemical properties and metabolites. We conducted PLSPM analyses (Fig. 7) to profile the complex relationship between rhizosphere microecosystem and disease index. Soil properties had strong effect on bacteria diversity (positively, 0.7424) and disease index (negatively, − 0.4857). Additionally, soil properties can also affect metabolites indirectly through bacteria diversity (positively, 0.8344).
Conclusion
Our findings indicate that there were significant differences in rhizosphere microbial community and metabolites between Yunyan87 and Fandi3 cultivars. The rhizosphere microbial community of resistant cultivar Fandi3 was significantly higher than that of Yunyan87. Additionally, the root exudates of Fandi3 were distinct from those of Yunyan87. These results indicated that tobacco cultivars had a significant impact on the rhizosphere microecosystem, which might be the reason for the differences in tobacco resistance to TBW.
Data availability
All data generated or analysed in this study are included in the published article.
References
An, M. J., Zhou, X. G., Wu, F. Z., Ma, Y. F., & Yang, P. (2011). Rhizosphere soil microorganism populations and community structures of different watermelon cultivars with differing resistance to Fusariumoxysporum f. sp. Niveum. Canadian Journal of Microbiology, 57, 355–365.
Awasthi, A., Bharti, N., Nair, P., Singh, R., Shukla, A. K., Gupta, M. M., Darokar, M. P., & Kalra, A. (2011). Synergistic effect of Glomus mosseae and nitrogen fixing Bacillus subtilis strain Daz26 on artemisinin content in Artemisia annua L. Applied Soil Ecology, 49, 125–130.
Badri, D. V., Chaparro, J. M., Zhang, R. F., Shen, Q. R., & Vivanco, J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. Journal of Biological Chemistry, 288, 4502–4512.
Badri, D. V., Weir, T. L., van der Lelie, D., & Vivanco, J. M. (2009). Rhizosphere chemical dialogues: Plant–microbe interactions. Current Opinion in Biotechnology, 20, 642–650.
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., de Hollander, M., Ruiz-Buck, D., Mendes, L. W., van Ijcken, W. F. J., Gomez-Exposito, R., Elsayed, S. S., et al (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science, 366, 606–612.
Chen, S., Qi, G. F., Ma, G. Q., & Zhao, X. Y. (2020). Biochar amendment controlled bacterial wilt through changing soil chemical properties and microbial community. Microbiological Research, 231, 126–373.
Chen, Y. P., Rekha, P. D., Arun, A. B., Shen, F. T., Lai, W. A., & Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Applied Soil Ecology, 34, 33–41.
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., & Hacquard, S. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell, 175, 973–983.
Garbeva, P., Veen, J. A., & Elsas, J. D. (2004). Microbial diversity in soil: Selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annual Review of Phytopathology, 42, 43–270.
Haichar, F. E. Z., Santaella, C., Heulin, T., & Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biology & Biochemistry, 77, 69–80.
Hasegawa, T., Kato, Y., Okabe, A., Itoi, C., Ooshiro, A., Kawaide, H., & Natsume, M. (2019). Effect of secondary metabolites of tomato (Solanum lycopersicum) on chemotaxis of Ralstonia solanacearum, pathogen of bacterial wilt disease. Journal of Agricultural and Food Chemistry, 67, 1807–1813.
Hill, T. C., Walsh, K. A., Harris, J. A., & Moffett, B. F. (2003). Using ecological diversity measures with bacterial communities. FEMS Microbiology Ecology, 43, 1–11.
Hu, Y., Li, Y. Y., Yang, X. Q., Li, C. L., Wang, L., Feng, J., Chen, S. W., Li, X. H., & Yang, Y. (2021). Effects of integrated biocontrol on bacterial wilt and rhizosphere bacterial community of tobacco. Scientific Reports, 11, 2653.
Huang, A. C., Jiang, T., Liu, Y. X., Bai, Y. C., Reed, J., Qu, B., Goossens, A., Nützmann, H. W., Bai, Y., & Osbourn, A. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science, 364, eaau6389.
Huang, X. F., Chaparro, J. M., Reardon, K. F., Zhang, R. F., Shen, Q. R., & Vivanco, J. M. (2014). Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany, 92, 267–275.
Huang, X. W., Tian, B. Y., Niu, Q. H., Yang, J. K., Zhang, L. M., & Zhang, K. Q. (2005). An extracellular protease from Brevibacillus laterosporus G4 without parasporal crystals can serve as a pathogenic factor in infection of nematodes. Research in Microbiology, 156, 719–727.
Jiang, G. F., Wei, Z., Xu, J., Chen, H. L., Zhang, Y., She, X. M., Macho, A. P., Ding, W., & Liao, B. S. (2017). Bacterial wilt in China: History, current status, and future perspectives. Frontiers in Plant Science, 8, 1549–1558.
Kayalvizhi, N., & Gunasekaran, P. (2010). Purifification and characterization of a novel broad spectrum bacteriocin from Bacillus licheniformis MKU3. Biotechnology and Bioprocess Engineering, 15, 365–370.
Kramarz, P. E., De Vauflfleury, A., Zygmunt, P. M. S., & Verdun, C. (2007). Increased response to cadmium and Bacillus thuringiensis maize toxicity in the snail Helix aspersa infected by the nematode Phasmarhabditis hermaphrodita. Environmental Toxicology and Chemistry, 26, 73–79.
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., Lee, P. A., Choi, S. Y., Seo, M., Lee, H. J., et al (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nature Biotechnology, 36, 1100–1109.
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., & Firn, J. L. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proceedings of the National Academy of Sciences of the USA, 112, 10967–10972.
Li, S. L., Xu, C., Wang, J., Guo, B., Yang, L., Chen, J. N., & Ding, W. (2017). Cinnamic, myristic and fumaric acids in tobacco root exudates induce the infection of plants by Ralstonia solanacearum. Plant and Soil, 412, 381–395.
Li, Y. Y., Wang, L., Sun, G. W., Li, X. H., Chen, Z. G., Feng, J., & Yang, Y. (2021). Digital gene expression analysis of the response to Ralstonia solanacearum between resistant and susceptible tobacco varieties. Scientific Reports, 11, 3887.
Liu, Y. X., Li, X., Cai, K., Cai, L., Lu, N., & Shi, J. X. (2015). Identification of benzoic acid and 3-phenylpropanoic acid in tobacco root exudates and their role in the growth of rhizosphere microorganisms. Applied Soil Ecology, 93, 78–87.
Lykogianni, M., Papadopoulou, E. A., Sapalidis, A., Tsiourvas, D., Sideratou, Z., & Aliferisa, K. A. (2020). Metabolomics reveals differential mechanisms of toxicity of hyperbranched poly(ethyleneimine)-derived nanoparticles to the soil-borne fungus Verticillium dahliae Kleb. Pestic. Biochemical and Physics, 165, 104535.
Mannisto, M. K., Rawat, S., Starovoytov, V., & Haggblom, M. M. (2012). Granulicella arctica sp nov., Granulicella mallensis sp nov., Granulicella tundricola sp nov and Granulicella sapmiensis sp nov., novel acidobacteria from tundra soil. International Journal of Systematic and Evolutionary Microbiology, 62, 2097–2106.
Nihorimbere, V., Ongena, M., Smargiassi, M., & Thonart, P. (2011). Benefificial effect of the rhizosphere microbial community for plant growth and health. Biotechnology, Agronomy and Society, 15, 327–337.
Opina, N., Tavner, F., Hollway, G., Wang, J. F., Li, T. H., Maghirang, R., Fegan, M., Hayward, A. C., Krishnapillai, V., Hong, W. F., Holloway, B. W., & Timmis, J. N. (1997). A novel method for development of species and strain- specific DNA probes and PCR primers for identifying Burkholderia Solanacearum (Formerly Pseudomonas Solanacearum). Asia-Pacific Journal of Molecular Biology and Biotechnology, 5, 19–30.
Pattee, H. E. (1969). Production of aflatoxins by Aspergillus flavus cultured on flue-cured tobacco. Applied Microbiology, 18, 952.
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., & Moënne-Loccoz, Y. (2009). The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant and Soil, 321, 341–361.
Ramesh, R., Joshi, A. A., & Ghanekar, M. P. (2009). Pseudomonads: Major antagonistic endophytic bacteria to suppress bacterial wilt pathogen, Ralstonia solanacearum in the eggplant (Solanum melongena L.). World Journal of Microbiology and Biotechnology, 25, 47–55.
Raza, W., Ling, N., Yang, L. D., Huang, Q. W., & Shen, Q. R. (2016). Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Scientific Reports, 6, 24856.
Sakai, M., Hosoda, A., Ogura, K., & Ikenaga, M. (2014). The growth of Steroidobacter agariperforans sp nov., a novel agar-degrading bacterium isolated from soil, is enhanced by the diffusible metabolites produced by bacteria belonging to rhizobiales. Microbes and Environments, 29, 89–95.
Siciliano, S. D., Theoret, C. M., de Freitas, J. R., Hucl, P. J., & Germida, J. J. (1998). Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Canadian Journal of Microbiology, 44, 844–851.
Silva, R. N., Monteiro, V. N., Steindorff, A. S., Gomes, E. V., & Ulhoa, C. J. (2019). Trichoderma/pathogen/plant interaction in pre-harvest food security. Fungal Biology, 123, 565–583.
Song, Y., Li, X. N., Yang, X. L., & Jiang, X. (2020). Correlations between soil metabolomics and bacterial community structures in the pepper rhizosphere under plastic greenhouse cultivation. Science of the Total Environment, 728, 138439.
Sun, J. B., Peng, M., Wang, Y. G., Li, W. B., & Xia, Q. Y. (2013). The effects of different disease-resistant cultivars of banana on rhizosphere microbial communities and enzyme activities. FEMS Microbiology Letters, 345, 121–126.
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., Elhottovā, D., Krištůfek, V., & Salles, J. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proceedings of the National Academy of Sciences of the USA, 109, 1159–1164.
Wang, R., Zhang, H. C., Sun, L. G., Qi, G. F., Chen, S., & Zhao, X. Y. (2017). Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Scientific Reports, 7, 343.
Wu, B., Wang, X., Yang, L., Yang, H., Zeng, H., Qiu, Y., Wang, C. J., Yu, J., Li, J. P., Xu, D. H., He, Z. L., & Chen, S. W. (2016). Effects of Bacillus amyloliquefaciens ZM9 on bacterial wilt and rhizosphere microbial community of tobacco. Applied Soil Ecology, 103, 1–12.
Wu, F. Z., Han, X., & Wang, X. Z. (2006). Allelopathic effect of root exudates of cucumber cultivars on Fusarium oxysporum. Allelopathy Journal, 18, 163–172.
Wu, F. Z., & Wang, X. Z. (2006). Effect of p-hydroxybenzoic and cinnamic acid on soil fungi (Fusarium oxysporum f.sp. cucumerinim) growth and microbial population. Allelopathy Journal, 18, 129–139.
Wu, K., Yuan, S. F., Xun, G. H., Shi, W., Pan, B., Guan, H. L., Shen, B., & Shen, Q. R. (2015). Root exudates from two tobacco cultivars affect colonization of Ralstonia solanacearum and the disease index. European Journal of Plant Pathology, 141, 667–677.
Xiong, W., Li, R., Ren, Y., Liu, C., Zhao, Q. Y., Wu, H. S., Jousset, A., & Shen, Q. R. (2017). Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biology & Biochemistry, 107, 198–207.
Xue, Q. Y., Ding, G. C., Li, S. M., Yang, Y., Lan, C. Z., Guo, J. H., & Smalla, K. (2013). Rhizocompetence and antagonistic activity towards genetically diverse Ralstonia solanacearum strains an improved strategy for selecting biocontrol agents. Applied Microbiology and Biotechnology, 97, 1361–1371.
Yang, T. J., Han, G., Yang, Q. J., Friman, V. P., Gu, S. H., Wei, Z., Kowalchuk, G. A., Xu, Y. C., Shen, Q. R., & Jousset, A. (2018). Resource stoichiometry shapes community invasion resistance via productivity-mediated species identity effects. Proceedings of the Royal Society B: Biological Sciences, 285, 20182035.
Yang, X. P., Wang, S. M., Zhang, D. W., & Zhou, L. X. (2011). Isolation and nitrogen removal characteristics of an aerobic heterotrophic nitrifying-denitrifying bacterium, Bacillus subtilis A1. Bioresource Technology, 102, 854–862.
Yao, H. Y., & Wu, F. Z. (2010). Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt. FEMS Microbiology Ecology, 72, 456–546.
Yuan, J., Zhao, J., Wen, T., Zhao, M. G., Li, R., Goossens, P., Huang, Q. W., Bai, Y., Vivanco, J. M., Kowalchuk, G. A., Berendsen, R. L., & Shen, Q. R. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome., 6, 156.
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S. J., Cho, H. J., Karaoz, U., Loqué, D., Bowen, B. P., Firestone, M. K., Northen, T. R., & Brodie, E. L. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nature Microbiology, 3, 470–480.
Zhang, C. S., Feng, C., Zheng, Y. F., Wang, J., & Wang, F. L. (2020). Root exudates metabolic profiling suggests distinct defense mechanisms between resistant and susceptible tobacco cultivars against black shank disease. Frontiers in Plant Science, 11, 559775.
Zhang, C. S., Lin, Y., Tian, X. Y., Xu, Q., Chen, Z. H., & Lin, W. (2017). Tobacco bacterial wilt suppression with biochar soil addition associates to improved soil physiochemical properties and increased rhizosphere bacteria abundance. Applied Soil Ecology, 112, 90–96.
Zhang, J. Y., Liu, Y. X., Zhang, N., Hu, B., Jin, T., Xu, H. R., Qin, Y., Yan, P. X., Zhang, X. N., Guo, X. X., et al (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nature Biotechnology, 37, 676–684.
Acknowledgements
This work was financially supported by the key technology project of CNTC (110202101047 [LS-07]), the key technology project of Hubei tobacco company (No. 027Y2022-023), and the science and technology research project of the education department of Hubei province (D20201003), and Wuhan key research and development project (2022023102015166).
Author information
Authors and Affiliations
Contributions
YH and YY conceived and designed the experiments. WZ and YL performed the experiments. WZ and CY analyzed the data. YH and YY wrote and revised the paper. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, W., Li, Y., Yang, C. et al. Rhizosphere Microbial Community and Metabolites of Susceptible and Resistant Tobacco Cultivars to Bacterial Wilt. J Microbiol. 61, 389–402 (2023). https://doi.org/10.1007/s12275-023-00012-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00012-0