Abstract
Extracellular vesicles (EVs) derived from mouse bone marrow mesenchymal stem cells (mBMSCs) convey the CAV1 protein, influencing the TGF-β1/SMAD2/c-JUN pathway and thus the molecular mechanisms underlying myocardial fibrosis (MF) post-myocardial infarction (MI). Through various experimental methods, including transmission electron microscopy, Nanosight analysis, Western blot, ELISA, and qRT-PCR, we isolated, purified, and identified EVs originating from mBMSCs. Bioinformatics and experimental findings show a reduced expression of CAV1 in myocardial fibrosis tissue. Furthermore, our findings suggest that mBMSC-EVs can deliver CAV1 to cardiac fibroblasts (CFs) and that silencing CAV1 in mBMSC-EVs promotes CF fibrosis. In vivo studies further corroborated these findings. In conclusion, mBMSC-EVs mitigate myocardial fibrosis in MI mice by delivering the CAV1 protein, inhibiting the TGF-β1/SMAD2/c-JUN pathway.
Graphical Abstract

Molecular mechanism of mBMSC-EVs-CAV1-mediated TGF-β1/SMAD2/c-JUN axis in inhibiting cardiac fibroblast differentiation to improve MF after MI. mBMSC-EVs deliver CAV1 protein to CFs where the protein expression of CAV1 is upregulated upon hypoxia conditions. The TGF-β1/SMAD2 signaling pathway downstream of CAV1 is consequently inactivated, the transcription of c-JUN is inhibited, and transcription of SMAD2/c-JUN transcription complex target genes α-SMA and Collagen I is reduced. By this mechanism, CF fibrosis and apoptosis are suppressed in vitro and MF is ameliorated in MI mice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myocardial infarction (MI) is a disorder with high morbidity and mortality worldwide, which occurs when part or all of the blood supply to the heart is interrupted [1]. MI is a consequence of the blockade of the coronary artery, which results in oxygen deficit in the myocardium, thus inducing the death of cardiomyocytes and non-cardiomyocytes as well as the remodeling of maladaptive left ventricular [2]. The degree of cell death (infarct size) mainly depends on the duration of ischemia, the size of the ischemic or danger zone, and the extent of coronary collateral blood flow and coronary microvascular impairment [3]. Cardiac fibroblasts (CFs) are crucial in the myocardium and MI wound healing [4]. Fibroblasts participate in the excessive production of collagen, thus causing myocardial fibrosis (MF) [5]. Hyperactive MF may contribute to adverse remodeling in patients with MI, triggering the pathogenesis of heart failure [6]. Therefore, delineating the molecular mechanism underlying MF in MI will enable the development of potential targets to curb the consequences of MI.
It has been proposed that BMSCs facilitate the repair of cardiomyocytes following MI due to their anti-fibrotic activity and differentiation into cardiomyocyte-like cells and endothelial cells [7]. Of note, almost every type of cell can release extracellular vesicles (EVs) [8]. Moreover, prior research elucidated that BMSC-derived EVs (BMSC-EVs) could facilitate blood flow recovery, decrease infarct size, and maintain cardiac systolic and diastolic performance in MI rats [9]. EVs mainly originate from endosomes and encase many microRNA, mRNA, and proteins transmitted from donor cells to recipient cells [10]. For instance, castration-resistant prostate cancer–derived EVs containing caveolin-1 (CAV1) could accelerate radio- and chemo-resistance of recipient cells [11]. Based on this, the role of CAV1 in MI is significant for exploring the mechanism of BMSC-EVs in MI.
Notably, the anti-fibrotic potential of CAV1, an integral membrane protein, has been documented [12]. A prior work reported that CAV1 knockout fostered cardiac interstitial fibrosis by inducing M2 macrophage activation in MI mice [13]. More importantly, another research revealed that CAV1 functioned as a repressor of the TGF-β1/SMAD2 pathway to attenuate mouse liver fibrosis [14]. TGF-β1/SMAD2 pathway activation caused the elevation of c-JUN to exacerbate MF and heart failure in rats [15]. Interestingly, the research of Qiu et al. illustrated that the blocked TGF-β1/SMAD2 pathway alleviated atrial interstitial fibrosis in post-MI rats [16].
In this context, we speculated that BMSC-EVs might deliver CAV1 to reduce MF following MI via the TGF-β1/SMAD2/c-JUN axis. Therefore, we treated CFs with mouse BMSC-derived EVs (mBMSC-EVs) to observe the impact of BMSC-EVs on MF in MI. The role of BMSC-EV-enclosed CAV1 in the TGF-β1/SMAD2/c-JUN axis of CFs was also researched.
Materials and Methods
Ethics Statement
Animal experimentations were authorized by the Animal Ethics Committee of Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences; Guangdong Cardiovascular Institute) (No. KY-7.-2022-2317-01). Appropriate measures had been taken to avoid any unnecessary distress to the animals. This study was approved by the Ethics Committee of Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences; Guangdong Cardiovascular Institute), and the approved guidelines carried out the methods. All the patients have been informed and signed informed consent before the experiments.
In Silico Analysis
MI-related array-based dataset GSE24519 was downloaded from the Gene Expression Omnibus database based on the GPL2895 platform, which contains whole blood samples from 17 patients with MI and two healthy individuals. After two replicates for each patient, 34 AMI samples and four control samples were yielded for analysis. GEO2R tool [17] was applied for differential analysis of the GSE24519 dataset to identify the differentially expressed genes with |log2FC| > 1 and p value < 0.05 as the threshold. The genes contained in MSC-EVs were obtained from the ExoCarta database [18]. Next, the association between genes and disease was analyzed by the Phenolyzer tool [19]. Protein-protein interaction (PPI) of critical factors was analyzed, and the network was constructed using the STRING database [20], followed by visualization using Cytoscape 3.5.1 software [21]. The network nodes and edges were analyzed using the built-in tool Network Analyzer, and the number of edges for each gene (Degree value) was sorted. KEGG enrichment analysis of related genes was conducted using the Network Analyst tool [22].
Construction of Left Anterior Descending Artery Acute MI (LAD AMI) Mouse Model
Thirty-two specific pathogen-free (SPF)–grade C57BL/6 male mice aged 6–8 weeks, weighing 18–25 g, were procured from Beijing Biotech-HD Co. Ltd. (Beijing, China) and bred in an SPF-grade animal room. Mice were anesthetized through intraperitoneal injection of phenobarbital sodium (60 mg/kg) and ventilated by inserting endotracheal intubation connected to a rodent ventilator. The left anterior descending artery was ligated utilizing 8–0 nylon suture. The success of MI modeling was determined by observing the whitening of the myocardium at the damaged site. Mice were randomized into the sham group (mice were only sutured by thoracotomy without ligation), the phosphate buffer saline (PBS) group (injection of 25-μL PBS), the EVs-overexpression (oe)-negative control (NC) group (injection of 25-μL EVs-oe-NC derived from mBMSCs) and the EVs-oe-CAV1 group (injection of 25-μL EVs-oe-CAV1 derived from mBMSCs), with eight mice in each group. EV injection was performed 30 min after ligation, and EVs were injected into multiple sites around MI [23]. Every 20-g mouse was injected with 200-μg EVs [24]. After 28 days of modeling, the mice were euthanized for a follow-up study.
Isolation, Culture, and Identification of mBMSCs
Isolation and Culture of mBMSCs
C57BL/6 mice (aged 6–8 weeks) were executed by cervical dislocation. The hind limbs of mice were disinfected with 70% ethanol, followed by skin and muscle tissue removal utilizing tweezers. Then, the bone marrow was taken out and subjected to filtration employing a 70-mm filter screen for discarding the muscle and cell mass. The harvested cells were cultured in DMEM (11965092, Gibco-Life Technologies, Grand Island, NY) replenishing 10% fast calcification solution penicillin (100 IU/mL) and streptomycin (100 μg/mL) in a 5% CO2 incubator at 37 °C, followed by removal of non-adherent cells after 3 h of culturing. The culture medium was renewed every 3–4 days. Upon 80% cell density, 0.25% trypsin was used for subculture. The surface markers were identified when mBMSCs were subcultured to the third generation.
Identification of mBMSCs
mBMSCs (1 × 104) were probed with primary antibodies to anti-CD44 (1/100, ab51037, Abcam, Cambridge, UK), anti-CD151 (1/100, 750814, BD Biosciences, Franklin Lakes, NJ), anti-CD73 (1/100, 561544, BD Biosciences), anti-CD34 (1/100, ab81289, Abcam), and anti-CD133 (1/100, MAB11332, R&D Systems, Minneapolis, MN) at 4 °C for 30 min, respectively. After that, surface marker expression was analyzed using a FACSCalibur flow cytometer (BD Biosciences).
Isolation and Culture of CFs in Neonatal Mice
Isolation and culture of CFs in Neonatal Mice
The heart tissue of newborn C57BL/6 mice within 24 h was cut into small pieces of 0.5–1 mm3 utilizing surgical scissors. The tissue was detached with 0.25% type IV collagenase-trypsin-EDTA (T4049, Sigma-Aldrich, St Louis, MO) solution preheated to 37 °C. After that, the tissue solution was filtered through 150-μm and 74-μm stainless steel cell filters (GongLu Co. Ltd., Hangzhou, Zhejiang, China) to obtain cells. Then, the cells were placed in DMEM complete medium replenishing 10% FBS and incubated in a 37 °C, 5% CO2 incubator. The attained adherent cells after 72 h of culture were identified by trypan blue staining. The expression of vimentin was quantified utilizing immunohistochemistry to identify the purity of cardiac fibroblasts (CFs). CFs needed to maintain 70–80% density culture to prevent its spontaneous trans-differentiation. CFs were treated with TGF-β1 (ZY124Bo011, HZbscience, varying concentration: 0, 1, 5, 10, or 20 ng/mL) or TGF-β1 + TGF-β1 inhibitor SB431542 (S4317, 2.5 μM, Sigma-Aldrich).
The shRNA (purchased from Hanheng Biotechnology) was transfected into logarithmic growth phase CFs using Lipofectamine 2000 (11668-019, Invitrogen, New York, California, USA) according to the manufacturer’s instructions. After transfection, the cells were incubated in a CO2 incubator at 37 °C. After 6 h, the transfection reagent medium was replaced, and the cells were further cultured. After 48 h, the CFs were collected for subsequent experiments. CFs were divided into the sh-NC group, sh-c-JUN group, sh-NC group, sh-NC + TGF-β1 group, and sh-c-JUN + TGF-β1 group. The sequence of sh-c-JUN was 5′-CGGCTACAGTAACCCTAAGAT-3′.
Construction of MF Cell Models In Vitro
CFs were cultured under normoxia and hypoxia conditions, respectively. CFs were cultured under hypoxia conditions to construct an in vitro MF cell model to simulate myocardial injury. Primary CFs under normoxia conditions were cultured in a 5% CO2 incubator, while those under hypoxia conditions were in an incubator with 1% O2 and 5% CO2 at 37 °C. The cells were grouped into the normoxia conditions, normoxia conditions + TGF-β1, hypoxia conditions, and hypoxia conditions + TGF-β1 groups.
Cell Culture, Transfection, and Transduction
HEK-293T cell (CRL-11268G-1, American Type Culture Collection, VA) was cultured in DMEM complete medium replenishing 10% FBS and 1% penicillin-streptomycin. The plasmid (Hanbio Biotechnology Co., Ltd) was transfected into logarithmically growing 293T cells using the Lipofectamin 2000’s manual (11668-019, Invitrogen). After 6 h of culturing, the medium replenishing transfection medium was discarded, and the medium was changed to continue the culture. The fresh virus solution was collected after 48 h. The 293T cells were grouped into the sh-NC, sh-CAV1, oe-NC, and oe-CAV1 groups.
mBMSCs (1 × 106) were seeded in 6-well plates with 2-mL medium per well. Under 50% confluence, cell transduction was implemented with a mixture of 800-μL fresh virus solution, 800-μL complete culture medium, and Polybrene (6 μg/mL, TR-1003-G, Sigma-Aldrich). Afterward, the cells were cultured in the incubator under 5% CO2 and 37 °C. After 48 h of transduction, the cells were cultured in DMEM replenishing puromycin (1 μg/mL, A1113803, Thermo Fisher Scientific Inc., Waltham, Massachusetts) for identifying stably transfected cell lines. The cells were harvested when they no longer died in the puromycin-containing medium.
mBMSCs were cultured until 80% confluency and then treated with the exosome secretion inhibitor GW4869 (D1692, Sigma-Aldrich Corporation, St. Louis, MO, USA) at a final concentration of 10 μM. After 24 h, the medium was changed to a serum-free culture medium containing 10 μM GW4869, and the cells were further cultured for 24 h before isolating exosomes. The control group received 1% DMSO (D2650, Sigma-Aldrich Corporation, St. Louis, MO, USA), and exosomes were isolated after 48 h for subsequent analysis. In the following in vitro cell experiments, the exosome concentration was 20 μM [25]. mBMSCs were divided into the following groups: DMSO, GW4869; control, sh-NC, sh-CAV1, sh-NC + TGF-β1, sh-c-JUN + TGF-β1, oe-NC, oe-CAV1. The sequence for sh-NC was 5′-GCTTTGTGATTCAATCTGTAA-3′, sh-CAV1 was 5′-GACGTGGTCAAGATTGACTTT-3′, and sh-c-JUN was 5′-GGTGCCTACGGCTACAGTAAC-3′ [26].
Isolation, Culture, and Identification of EVs of mBMSCs
Isolation of EVs was implemented employing a differential ultracentrifugation method with the L-80XP ultracentrifuge (Beckman Coulter Life Sciences, Brea, CA) [27]. The concentration of EVs was assessed by Pierce™ bicinchoninic acid (BCA) kit (23225, Thermo Fisher Scientific).
Identification of mBMSC-EVs
The EV protein markers CD9, CD81, CD63, Calnexin, and Tsg101 were analyzed by Western blot. Additionally, EVs were photographed by a H7650 transmission electron microscope (TEM, Hitachi Ltd., Tokyo, Japan). Then, the particle size distribution of EVs was checked utilizing a NanoSight nanoparticle tracking analyzer (Malvern Instruments, Ltd., Malvern, UK).
EV Uptake by CFs
EVs secreted by mBMSCs were labeled with PKH67 green fluorescence (HR8569, Beijing Biolab Technology Co., Ltd., Beijing, China). Firstly, 4-μL PKH67 was added to the 10 μg/mL EVs for incubation for 15 min. Then, 1 mL of 5% BSA was supplemented for reaction termination. Subsequently, the EVs were ultracentrifuged at 4 °C with 110,000×g for 1 h. The EVs were resuspended with pre-cooled PBS. The labeled EVs were treated with CFs for 24 h and fixed with 4% paraformaldehyde. The nuclei were stained with 10 μg/mL of DAPI staining solution for 10 min. The uptake of labeled EVs by CFs was observed with the help of a Nikon Eclipse fluorescence microscope (Nikon, Tokyo, Japan).
RT-qPCR
Total RNA was extracted from cells and mouse cardiac tissue using the Trizol reagent kit (15596018, Invitrogen, Thermo Fisher, USA). The quality and concentration of RNA were assessed using ultraviolet-visible spectrophotometry (ND-1000, Nanodrop, USA). Reverse transcription was performed using the PrimeScript™ qRT-PCR kit (RR047A, TaKaRa, Japan). Sample preparation was carried out using SYBR (RR820A, TaKaRa, Japan), and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed on a LightCycler 480 system (Roche Diagnostics, Pleasanton, CA, USA). The housekeeping gene GAPDH was used as an internal control for mRNA analysis. The primer sequences for amplification were designed and provided by Shanghai Universal Biotech Co., Ltd. The primer sequences can be found in Table S1. The fold change in target gene expression between the experimental group and the control group was determined using the 2−ΔΔCt method, where ΔΔCt = ΔCt experimental group − ΔCt control group, and ΔCt was calculated by subtracting the cycle threshold (Ct) of the housekeeping gene from the Ct of the target gene.
Immunoblotting
Immunoblotting of the target protein was implemented with diluted primary antibodies from Cell Signaling Technology (CST, Beverly, MA) to α-SMA (rabbit monoclonal antibody, 19245, 1/1000), Collagen I (rabbit monoclonal antibody, 91144, 1/1000), CAV1 (rabbit polyclonal antibody, 3238, 1/1000), c-JUN (rabbit monoclonal antibody, 9165, 1/1000), total-SMAD2 (rabbit monoclonal antibody, 5339, 1/1000), and p-SMAD2 (rabbit monoclonal antibody, 18338) and antibodies from Abcam to TGF-β1 (rabbit monoclonal antibody, ab215715, 1/1000), CD9 (rabbit polyclonal antibody, 1/1000, ab223052), CD81 (rabbit monoclonal antibody, 1/1000, ab109201), CD63 (rabbit monoclonal antibody, 1/1000, ab134045), Tsg101 (mouse monoclonal antibody, 1/1000, ab83), Calnexin (rabbit polyclonal antibody, 1/1000, ab22595), and GAPDH (mouse monoclonal antibody, ab8245, 1/10,000) as well as HRP-labeled goat anti-mouse secondary antibodies (ab6808, 1/2000, Abcam) and goat anti-rabbit secondary antibodies (ab6721, 1/2000, Abcam) [28]. Afterward, an enhanced chemiluminescence reagent (42029053, EMD Millipore Inc., Billerica, Mass) was employed for result visualization. The grayscale of bands in Western blot images was quantified utilizing ImageJ analysis software.
Flow Cytometry
The cells were digested with pancreatin, collected in FACS tubes, centrifuged, and the supernatant was discarded. The cells were then washed three times with cold PBS, followed by centrifugation and removal of the supernatant. FITC Annexin V Apoptosis Detection Kit I (Catalog number: 556547, BD Biosciences) was used according to the manufacturer’s instructions, and Annexin-V-FITC, PI, and HEPES buffer were mixed in a ratio of 1:2:50 to prepare the Annexin-V-FITC/PI staining solution. Each 1 × 106 cell was resuspended, mixed with 100-μL staining solution, incubated at room temperature for 15 min, then 1-mL HEPES buffer and gentle shaking. Cell apoptosis was detected using the FACSCalibur System (BD Biosciences, San Jose, CA, USA) equipped with FACScan flow cytometry.
ChIP
ChIP assay was performed with a ChIP kit (17-295, Sigma-Aldrich). In short, the cells were fixed with 4% paraformaldehyde and incubated with glycine to a final concentration of 0.125M for 5 min. Then, the cells were resuspended in 1 mL sodium dodecyl sulfate lysis buffer, sonicated 30 times at an interval of 30 s with 30 s each time (output H), and centrifuged at 14,000 ×g and 4 °C. Next, the lysate was immunoprecipitated with magnetic protein A beads bound with antibodies. The cells in the NC group were probed with rabbit IgG (ab171870, Abcam). The cells in the Anti-SMAD2 group were probed with SMAD2 antibodies (5339, 1/1000, CST). Finally, the precipitated DNA was quantified employing RT-qPCR. The primer sequence of α-SMA was F: 5′-GCTGTCGTCTTATCTCCA CCA-3′, R: 5′-GCAGGAGTCTAGCAGAAGTTC-3′. The primer sequence of Collagen I was F: 5′-TCAGGACCTCTAGAAGAGCAG-3′, R: 5′-CTTCCTTCCTTCCTTCCTTCC-3′.
Echocardiography
After 28 days of modeling, the echocardiography system evaluated the left ventricular (LV) systolic function (Siemens Sequoia 512, Mountain View, CA). The mice were kept in the supine position after anesthesia. The transducer frequency was set at 10 MHz. Left ventricular internal diastolic diameter (LVIDd), left ventricular end-systolic internal dimension (LVIDs), ejection fraction (EF), end-systolic chamber volume (ESCV), and left ventricular internal diameter (LVID) were recorded. Left ventricular end-systolic volume (LVESV) and left ventricular end-diastolic volume (LVEDV) were measured according to 7.0 × LVIDs3/(2.4 + LVIDs) and 7.0 × LVIDd3/(2.4 + LVIDd). Fractional shortening (FS, %) = (Left ventricular end-systolic internal dimension (LVEDD) − left ventricular end-systolic diameter (LVESD))/ LVEDD × 100. Left ventricular ejection fraction (LVEF) was calculated utilizing the Teich method.
ELISA
Mouse serum or cell culture supernatant was harvested and centrifuged at 3000 r/min at 4 °C for 15 min with the supernatant harvested. Total protein was quantified utilizing Pierce™ BCA Kit. The culture supernatant was harvested after 48 h. The expression of the corresponding protein was detected using ELISA kit TGF-β1 (MB100B, R&D Systems), TNF-α (ab208348, Abcam), and IL-1β (ab197742, Abcam), respectively.
Immunohistochemistry
Paraffin sections of myocardial tissue were placed in an incubator and stored at 60 °C for 2 h. The sections were dewaxed with xylene, hydrated with gradient ethanol (100%, 95%, 85%, 70%), soaked in citric acid buffer (0.01 mol/L, pH 6.0), and subjected to antigen retrieval. Then, the sections were treated utilizing 0.5% TritonX 100 (30 min) and probed with rabbit anti-CAV1 (3238, 1/250, CST) at 4 °C overnight.
CF identification
The sterilized and dried cell slides were coated with polylysine for 30 min. After drying, the slide was placed into 6-well culture plates. The cells were treated with trypsin and counted again. After that, 2-mL cell suspension was added to each well. After 48 h, the slides were fixed with 4% paraformaldehyde (30 min) and cleared with 0.5% TritonX 100 (30 min) at ambient temperature. Then, the slides were blocked with a 5% BSA blocking solution and probed with rabbit anti-Vimentin (5741, 1/100, CST).
Afterward, the slides were re-probed with HRP-labeled goat anti-rabbit IgG secondary antibodies (1/500, Life Technologies, Carlsbad, CA) for 1 h at ambient temperature. Then, the slides were treated with diaminobenzidine (DAB) solution for 3–5 min, re-stained in hematoxylin for 1–3 min, dehydrated, and sealed with neutral balm. Brown staining indicated a positive immune response. The images were visualized utilizing the Nikon ECLIPSE Ti microscope system (Fukasawa, Japan). The CAV1 and vimentin-positive cells were counted in randomly selected five fields of view from each section, with three sections randomly selected from each mouse.
Masson’s Trichrome Staining
In this assay, 8–12 cross-sections were taken to measure the left ventricular infarct size (one section per 200 μm tissue). The sections (4 μm thick) were dewaxed, hydrated, stained with hematoxylin (5–10 min), and hydrolyzed with acid ethanol (5–15 s). The sections were blued in Masson blue solution (DC0032, Leagene Biotechnology Co. Ltd., Beijing, China) (3–5 min), stained with ponceau-acid fuchsin solution (5–10 min), washed with phosphomolybdic acid solution (1–2 min), and stained in aniline blue solution (1–2 min). Next, the sections were dehydrated, cleared, and sealed for observation. Images were captured at 200 times magnification under an Olympus BX51 microscope (Olympus, Tokyo, Japan). Images were analyzed employing the Image-Pro Plus 6.0 image analysis system. Three sections were randomly taken from each mouse from the apex to the bottom of the heart, and five fields of view were randomly selected from each section and observed under a high-power microscope. Analysis was performed in a single-masked manner. The collagen volume fraction was evaluated by measuring the ratio of the blue staining area to the total atrial area.
Sirius Red Staining
The myocardial tissue of mice was cut into sections (6 μm thick), which were dewaxed, stained with hematoxylin (10–20 min), hydrolyzed with acidic differentiation solution (10 s), and stained with Sirius red (365548-5G, Sigma-Aldrich) (1 h). The sections were dehydrated, cleared, and finally sealed before observation. Three sections were randomly taken from each mouse, and five fields of view were randomly selected from each section and observed under a high-power microscope. Image-Pro Plus 6.0 software was employed to analyze the positive area for semi-quantitative analysis.
TUNEL Assay for Cell Apoptosis
Mouse myocardial tissues were fixed in 4% paraformaldehyde (15 min) and permeabilized with 0.1% Triton-X 100 in PBS for 3 min. Cardiomyocytes were stained with a TUNEL staining kit (C1091, Beyotime, Shanghai, China) [29]. The apoptosis ratio was calculated utilizing Image-Pro Plus 6.0 software.
Statistical Analysis
All data were processed using SPSS 21.0 statistical software (IBM Corp. Armonk, NY). Measurement data were described as mean ± standard deviation. Before conducting any statistical tests, all data were assessed for normality using the Shapiro-Wilk test. The experimental results demonstrated that the data followed a normal distribution. Independent-sample t test was adopted for data comparison between two groups; one-way analysis of variance for data comparison among groups, followed by Tukey’s post hoc test. p < 0.05 depicted statistically significant.
Results
mBMSC-EVs Limited the Expression of Fibrosis-Related Factors in Hypoxia-Exposed CFs
CFs were isolated from mice for analysis. After a 3-day culture, the isolated cells were swirled or radial-shaped under the optical microscope (Figure S1A). Immunohistochemistry depicted that more than 95% of the cells were positive for vimentin (Figure S1B).
Subsequently, CFs were induced by hypoxia conditions to simulate MF in vitro. Flow cytometry showed that CF apoptosis increased under hypoxia conditions (Fig. 1A). As reflected by ELISA results, the expression of inflammatory factors (TNF-α and IL-1β) elevated in the supernatant of CFs under hypoxia conditions (Fig. 1B). Additionally, the expression of MF markers (α-SMA and Collagen I) was enhanced in CFs cultured under hypoxic conditions, as detected using Western blot (Fig. 1C). Hypoxia conditions could substantially promote the expression of markers linked to CF fibrosis and induce their apoptosis.
mBMSC-EVs restrain the expression of fibrosis-related factors in hypoxia-exposed CFs. A Flow cytometry detected the apoptosis rate of CFs under normoxia and hypoxia conditions. B The expression of TNF-α and IL-1β in the supernatant of CFs assessed by ELISA. C The expression of α-SMA and Collagen I in CFs under normoxia and hypoxia conditions measured by Western blot analysis. *p < 0.05 vs. normoxic CFs. D The internalization of PKH-67 (green)-labeled mBMSC-EVs by CFs detected by immunofluorescence (× 400). E The apoptosis rate of CFs treated for 24 h with the CM of mBMSCs in the presence or absence of GW4869 evaluated by flow cytometry. F The expression of TNF-α and IL-1β in the supernatant of CFs treated for 24 h with the CM of mBMSCs in the presence or absence of GW4869 estimated by ELISA. G The expression of α-SMA and Collagen I in CFs treated for 24 h with the CM of mBMSCs in the presence or absence of GW4869 detected by using Western blot. In A–C: *p < 0.05 vs. normoxic CFs; in E–G *p < 0.05 vs. control CFs. #p < 0.05 vs. CFs treated with mBMSC-CM. All cell experiments were repeated three times
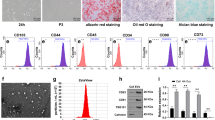
MSC-EVs effectively treat myocardial diseases and reduce oxidative stress and fibrosis in animal models [30,31,32]. Next, to verify whether MSC-EVs affect the fibrosis of hypoxia-exposed CFs, mBMSCs were extracted from the bone marrow of C57BL/6 mice. It was noted that mBMSCs were spindle-like fibroblasts under an optical microscope (Figure S2A). In addition, the mBMSC surfaces positive markers CD151 (100.0%), CD3 (99.1%), and CD44 (100%) and negative markers CD34 (0.78%) and CD133 (1%) were measured in mBMSCs at passage 3 by flow cytometry (Figure S2B), which suggested the successful isolation of mBMSCs.
Furthermore, the morphology of mBMSC-EVs was circular or oval membranous vesicles (Figure S2C) with the diameter of EVs in the range of 50–150 nm (Figure S2D). Western blot analysis documented that the expression of CD9, CD63, CD81, and TGS101 was elevated in mBMSC-EVs, while Calnexin was not expressed compared with cell lysate (Figure S2E), indicating the successful extraction of mBMSC-EVs. mBMSC-EVs were labeled with PKH-67 (green) and cultured with CFs for 24 h. Furthermore, the changes in extracellular vesicle concentration were determined using NTA after treatment of BMSCs with GW4869. The results showed a significant decrease in extracellular vesicle concentration per unit volume secreted by BMSCs after treatment with GW4869 compared to the control group (Figure S2F). The observation under the confocal microscopy depicted green fluorescence in CFs, which illustrated that mBMSC-EVs could be internalized by CFs (Fig. 1D).
To ascertain whether EVs functioned in MI, CFs were incubated with the conditioned medium (CM) of mBMSCs in the presence or absence of GW4869 (mBMSC-CM or mBMSCs + GW4869-CM). As manifested by the results of flow cytometry, ELISA, and Western blot analysis, treatment with mBMSC-CM decreased the apoptosis and TNF-α, IL-1β, α-SMA, and Collagen I expression in hypoxia-exposed. In contrast to treatment with mBMSC-CM, treatment with mBMSCs + GW4869-CM enhanced α-SMA, Collagen I, TNF-α, and IL-1β expression and the apoptosis in hypoxia-exposed CFs (Fig. 1E–G).
Conclusively, hypoxia conditions promoted the expression of fibrosis-related factors in hypoxia-exposed CFs, whereas mBMSC-EVs could repress those expressions.
mBMSC-EVs Carrying CAV1 Reduced the Expression of Fibrosis-Related Factors in Hypoxia-Exposed CFs
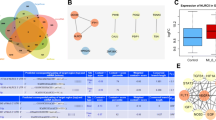
To investigate the specific molecular mechanism of MSC-EV involvement in myocardial infarction (MI), we performed differential gene expression analysis using the MI-related microarray dataset GSE24519. As a result, we identified 2102 genes that were upregulated and 734 genes that showed downregulation (Fig. 2A). Next, we compared these genes with the 938 genes present in the ExoCarta database of MSC-EVs and identified 22 candidate genes (KRT2, IGHA2, HSPA6, CCL7, SRI, SCAMP3, ATP2B1, IQGAP1, IGHA1, BPIFA1, AHSG, HSPD1, MARCKSL1, DSP, SFN, CAV1, RPL18, CD59, FLT1, QSOX1, RAB14, EHD4) (Fig. 2B).
CAV1 expression was poor in myocardial tissues. A Volcano map of differentially expressed genes in MI-related dataset GSE24519. B Venn map of the intersection of MSC-EVs gene in the GSE24519 and the ExoCarta database. C Correlation between 22 candidate factors and MI, abscissa is correlation score; ordinate is gene name. D The expression of CAV1 in control and MI samples in the GSE24519 dataset (control, n = 4, MI (myocardial infarction), n = 34). E RT-qPCR and Western blot detection of CAV1 protein expression in mBMSCs and EVs derived from mBMSCs. ***p < 0.001
Twenty out of the 22 candidate factors were found to be significantly correlated with MI using the Phenolyzer tool. Among them, Caveolin-1 (CAV1) demonstrated the highest correlation score with MI (Fig. 2C). Analysis results of the GSE24519 dataset indicated that CAV1 was significantly poorly expressed in MI tissues (Fig. 2D). Additionally, the expression of CAV1 was validated through Western blot and RT-qPCR. The results indicated a high expression of CAV1 in mBMSC-EVs, whereas the mRNA levels of CAV1 were significantly reduced in mBMSC-EV. This suggests that EVs primarily deliver CAV1 protein to target cells (Fig. 2E). Therefore, CAV1 might be a critical factor in MI.
To further determine whether mBMSC-EVs inhibited fibrosis by delivering CAV1 into CFs, we first detected the expression of CAV1 by RT-qPCR, which displayed the downregulated CAV1 in hypoxia-exposed CFs (Fig. 3A). In addition, mBMSC-EVs could depress hypoxia-exposed CF fibrosis. Therefore, mBMSC-EVs could transfer CAV1 into CFs to curtail fibrosis. Next, mBMSCs were treated with GW4869 and then treated with hypoxia-exposed CFs. As expected, CAV1 expression reduced evidently in hypoxia-exposed CFs treated with GW4869-treated mBMSCs (Fig. 3B), suggesting that the inhibition of the secretion of EVs by mBMSCs could lower CAV1 expression in hypoxia-exposed CFs.
mBMSC-EV-shuttled CAV1 represses the expression of fibrosis-related factors in hypoxia-exposed CFs. A Expression of CAV1 in CFs under normoxia and hypoxia conditions detected by RT-qPCR. *p < 0.05 vs. normoxic CFs. B The protein expression of CAV1 in hypoxia-exposed CFs treated with mBMSCs treated or with DMSO or GW4869 detected by using Western blot. *p < 0.05 vs. CFs treated with DMSO-treated mBMSCs. C The protein expression of CAV1 after sh-CAV1 transfection in mBMSCs detected by using Western blot. *p < 0.05 vs. mBMSCs treated with sh-NC. Hypoxia-exposed CFs were treated or not treated with mBMSC-sh-NC-EVs or mBMSC-sh-CAV1-EVs for 24 h. D The apoptosis rate of hypoxia-exposed CFs assessed by flow cytometry. E The expression of TNF-α and IL-1β in the supernatant of hypoxia-exposed CFs measured by ELISA. F The protein expression of α-SMA and Collagen I in hypoxia-exposed CFs detected by using Western blot. In D–F, *p < 0.05 vs. control (hypoxia-exposed CFs). #p < 0.05 vs. hypoxia-exposed CFs treated with mBMSC-sh-NC-EVs. All cell experiments were repeated three times
We then commenced to study the impacts of mBMSC-EVs-enclosed CAV1 on the expression of fibrosis-related factors in hypoxia-exposed CFs; CAV1 was knocked down in mBMSCs. A decline of CAV1 protein expression was witnessed in mBMSCs with sh-CAV1 transduction (Fig. 3C), indicating the successful construction of the CAV1 knockdown cell model. EVs derived from sh-CAV1-treated mBMSCs (mBMSC-sh-CAV1-EVs) were treated with hypoxia-exposed CFs. Flow cytometry results depicted that apoptosis of hypoxia-exposed CFs diminished following treatment with mBMSC-sh-NC-EVs, which was the opposite after treatment with mBMSC-sh-CAV1-EVs (Fig. 3D). In addition, augmented CAV1 expression and decreased TNF-α, IL-1β, α-SMA, and Collagen I expressions were witnessed in hypoxia-exposed CFs treated with mBMSC-sh-NC-EVs, while contrary results were observed after treatment with mBMSC-sh-CAV1-EVs (Fig. 3E, F).
Together, mBMSC-EVs could deliver CAV1 into hypoxia-exposed CFs, thereby limiting the expression of fibrosis-related factors in hypoxia-exposed CFs.
CAV1 Transferred by mBMSC-EVs Blocked the TGF-β1/SMAD2 Pathway in Hypoxia-Exposed CFs
For further predicting the downstream pathway of CAV1, the PPI relationship of key factor CAV1 was analyzed through the STRING database. The results showed higher degree values of EGF, EGFR, SRC, AKT1, INS, NOS3, FYN, HSP90AA1, TNF, and TGFB1 (Fig. 4A). KEGG enrichment analysis exhibited that these genes were mainly involved in FoxO signaling pathway, Insulin signaling pathway, EGFR tyrosine kinase inhibitor resistance, HIF-1 signaling pathway, MAPK signaling pathway, and TGF-β signaling pathway (Fig. 4B), in which TGF-β1, SMAD2, and TNF were critical factors of TGF-beta pathway.
mBMSC-EV-packaged CAV1 curtails the expression of fibrosis-related factors in hypoxia-exposed CFs by inactivating the TGF-β1/SMAD2 pathway. A PPI network diagram of CAV1. The degree value of the gene from small to large is indicated by the color scale orange to blue. B KEGG enrichment analysis diagram. The abscissa represents −log10 (p value), and the ordinate represents the pathway name. Hypoxia-exposed CFs were treated with mBMSC-oe-NC-EVs, mBMSC-oe-CAV1-EVs, or mBMSC-oe-CAV1-EVs + TGF-β1. C The expression of TGF-β1 protein and p-SMAD2/SMAD2 protein in hypoxia-exposed CFs measured by Western blot analysis. D The protein expression of α-SMA and Collagen I in hypoxia-exposed CFs assessed by Western blot analysis. E The expression of TNF-α and IL-1β in the supernatant of hypoxia-exposed CFs detected by ELISA. F The apoptosis rate of hypoxia-exposed CFs determined by flow cytometry. *p < 0.05 vs. hypoxia-exposed CFs treated with mBMSC-oe-NC-EVs. #p < 0.05 vs. hypoxia-exposed CFs treated with mBMSC-oe-CAV2-EVs. All cell experiments were repeated three times
Our next focus was the correlation of CAV1 with the TGF-β1/SMAD2 pathway. CAV1 was overexpressed in mBMSCs, from which EVs were isolated (mBMSC-oe-CAV1-EVs) and treated with hypoxia-exposed CFs, followed by supplementation of TGF-β1. Western blot analysis depicted that mBMSC-oe-CAV1-EVs increased CAV1 expression and decreased TGF-β1 protein expression and SMAD2 phosphorylation extent in hypoxia-exposed CFs, while the addition of TGF-β1 did not affect CAV1 expression but facilitated TGF-β1 protein expression and SMAD2 phosphorylation extent in hypoxia-exposed CFs treated with mBMSC-oe-CAV1-EVs (Fig. 4C).
Moreover, reduced TNF-α, IL-1β, α-SMA, and Collagen I expression was detected in hypoxia-exposed CFs after treatment with mBMSC-oe-CAV1-EVs, which was abolished by further TGF-β1 treatment (Fig. 4D, E). Flow cytometric data depicted that hypoxia-exposed CF apoptosis decreased following treatment with mBMSC-oe-CAV1-EVs, which was neutralized by additional TGF-β1 treatment (Fig. 4F).
In a word, CAV1 in mBMSC-EVs could repress the expression of fibrosis-related factors in hypoxia-exposed CFs by disrupting the TGF-β1/SMAD2 pathway.
TGF-β1 Elevated SMAD2 Expression to Upregulate c-JUN in Hypoxia-Exposed CFs
TGF-β1/SMAD2 pathway can upregulate AP-1 transcription factor subunit c-JUN [15]. To investigate the specific mechanism of TGF-β1/SMAD2/c-JUN axis involved in fibrosis of hypoxia-exposed CFs, CFs were cultured under hypoxic conditions and stimulated with TGF-β1 of varying concentrations for 24 h. As reflected, TGF-β1 stimulation boosted c-JUN expression (concentration-dependently), and there was a remarkable difference when TGF-β1 concentration was 10 ng/mL (Fig. 5A, B). Under hypoxia conditions, TGF-β1 enhanced c-JUN expression and SMAD2 phosphorylation extent (Fig. 5C, D).
TGF-β1 activates SMAD2/c-JUN axis in hypoxia-exposed CFs. A RT-qPCR was used to measure the effect of different concentrations of TGF-β1 on the mRNA expression of c-JUN. **p < 0.01; ***p < 0.001 vs. hypoxia-exposed CFs treated with TGF-β1 (0 ng/mL). B Western blot analysis was used to determine the effect of different concentrations of TGF-β1 on the protein expression of c-JUN. **p < 0.01; ***p < 0.001 vs. hypoxia-exposed CFs treated with TGF-β1 (0 ng/mL). C Detection of the effects of TGF-β1 on the mRNA expression of c-JUN under normoxic or hypoxic conditions by RT-qPCR. D Western blot analysis was used to detect the effect of TGF-β1 stimulation on the c-JUN protein expression and SMAD2 phosphorylation extent in CFs under normoxia or hypoxia conditions. *p < 0.05 vs. normoxic CFs; #p < 0.05 vs. hypoxia-exposed CFs. TGF-β1-stimulated hypoxia-exposed CFs were treated or not treated with SB431542. E RT-qPCR was used to assess the mRNA expression of c-JUN in hypoxia-exposed CFs. F c-JUN protein expression and SMAD2 phosphorylation extent in hypoxia-exposed CFs detected by using Western blot. *p < 0.05 vs. CFs treated with TGF-β1. All cell experiments were repeated three times
To evaluate the impacts of the TGF-β1/SMAD2 pathway on the c-JUN axis, TGF-β1 inhibitor SB431542 was supplemented to treat hypoxia-exposed CFs, followed by 48 h of TGF-β1 stimulation. We found that c-JUN expression and SMAD2 phosphorylation extent were diminished in TGF-β1-stimulated hypoxia-exposed CFs following treatment with SB431542 (Fig. 5E, F).
In brief, TGF-β1 could activate SMAD2 phosphorylation and upregulate c-JUN in hypoxia-exposed CFs.
TGF-β1/SMAD2 Pathway Activated Downstream α-SMA and Collagen I Transcription Dependent on c-JUN
It has been documented that c-JUN, as a co-transcriptional activator of SMAD2, forms a complex after binding to SMAD2, which can promote downstream gene transcription [33]. Consequently, we further elucidated the orchestrating influence of the TGF-β1/SMAD2/c-JUN axis on α-SMA and Collagen I in CFs.
First, c-JUN was silenced in CFs, and transfection efficiency was validated utilizing western blot analysis (Fig. 6A). In addition, Western blot analysis exhibited that TGF-β1 augmented SMAD2 phosphorylation extent and c-JUN, α-SMA, and Collagen I expression, which was counterweighed by silencing c-JUN without affecting SMAD2 phosphorylation extent (Fig. 6B, C).
TGF-β1/SMAD2 pathway facilitates α-SMA and Collagen I transcription in CFs via c-JUN. A The effect of sh-c-JUN sequence on the expression of c-JUN protein in CFs detected by using Western blot. *p < 0.05 vs. CFs treated with sh-NC. CFs were treated with sh-NC, sh-NC + TGF-β1, or sh-c-JUN + TGF-β1. B The protein expression of c-JUN, α-SMA, and Collagen I in CFs under hypoxia conditions measured by Western blot analysis. C SMAD2 phosphorylation extent in CFs under hypoxia conditions assessed by Western blot analysis. D ChIP experiment was used to detect the enrichment of SMAD2 in the α-SMA and Collagen I promoter regions in CFs under hypoxia conditions. *p < 0.05 vs. CFs treated with sh-NC; #p < 0.05 vs. CFs treated with sh-NC + TGF-β1. All cell experiments were repeated three times
Finally, ChIP experiments displayed that the enrichment of SMAD2 protein in α-SMA and Collagen I promoters enhanced under the stimulation of TGF-β1, which was counteracted after silencing of c-JUN (Fig. 6D).
Collectively, TGF-β1-activated α-SMA and Collagen I transcription were dependent on the SMAD2/c-JUN axis.
mBMSC-EVs Carrying CAV1 Improved MF in MI Mice
To further ascertain whether mBMSC-EV-CAV1 affected MF in mice, an MI animal model was established and injected with mBMSC-EVs at the myocardium (Fig. 7A). We discerned that TNF-α and IL-1β levels were enhanced in the serum of mice after MI modeling. Moreover, EVs-oe-NC resulted in a decline in those levels in the serum of MI mice versus PBS treatment, which was subsequently promoted to injection with EVs-oe-CAV1 (Fig. 7B).
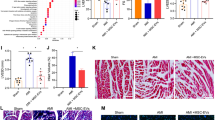
mBMSC-EV-enclosed CAV1 attenuates MF in MI mice by inactivating the TGF-β1/SMAD2/c-JUN axis. A Schematic diagram of intracardiac injection with mBMSC-EVs-CAV1 in a mouse model of MI. Sham-operated mice were used as control, and MI mice injected with PBS, EVs-oe-NC, or EVs-oe-CAV1. B The expression of inflammatory factors in the serum of mice evaluated by ELISA. C The protein expression of CAV1 in myocardial tissue of mice estimated by immunohistochemistry. D Sirius red staining of the myocardial infarction area of mice after 28-day injection. E Masson’s trichrome staining of the deposition of ventricular interstitial fibrosis in mice after 28-day injection (cardiomyocytes are red and interstitial collagen is blue-green). F Measurements of EF%, FS%, ESCV, and LVID of mice utilizing echocardiography. G TUNEL staining of cardiomyocyte apoptosis in MI mice. H Western blot analysis of TGF-β1, p-SMAD2/SMAD2, c-JUN proteins in myocardial tissues of mice. *p < 0.05 vs. sham-operated mice. #p < 0.05 vs. MI mice treated with PBS. $p < 0.05 compared with MI mice treated EVs-oe-NC. There were eight mice in each group
Immunohistochemistry manifested that the proportion of CAV1-positive cells diminished in mice after MI modeling. Additionally, the proportion of CAV1-positive cells was higher in EVs-oe-NC-treated MI mice than in PBS-treated MI mice, which was further facilitated in MI mice injected with EVs-oe-CAV1 (Fig. 7C, Figure S3A). It was found by Sirius red staining and Masson’s trichrome staining that the fibrotic deposition area was enlarged in mice following MI modeling. However, the fibrotic deposition area was reduced in MI mice after EVs-oe-NC treatment in contrast to PBS treatment and was further diminished in MI mice injected with EVs-oe-CAV1 (Fig. 7D, E, Figure S3B, C).
Additionally, MI mice exhibited lower EF and FS yet higher ESCV and LVID than the sham-operated mice. However, EVs-oe-NC treatment led to the opposite results. EF and FS were further augmented, and ESCV and LVID further narrowed in MI mice treated with EVs-oe-CAV1 (Fig. 7F), indicating that EVs-oe-CAV1 reduced left ventricular injury and improved cardiac systolic function.
As for TUNEL staining, cardiomyocyte apoptosis accelerated in MI mice but restricted in MI mice by EVs-oe-NC treatment versus PBS treatment, which was further facilitated in MI mice treated with EVs-oe-CAV1 (Fig. 7G, Figure S3D). Concerning Western blot analysis, TGF-β1 and c-JUN expression and SMAD2 phosphorylation extent in mice were augmented by MI modeling, and EVs-oe-NC triggered conflicting trends in MI mice compared to PBS. Besides, TGF-β1 and c-JUN expression and SMAD2 phosphorylation extent were further reduced in MI mice by EVs-oe-CAV1 (Fig. 7H).
Conclusively, mBMSC-EV-encapsulated CAV1 blocked the TGF-β1/SMAD2/c-JUN axis to improve MF in MI mice.
Discussion
Despite decades of therapeutic advances, MI remains one of the significant causes of death on a global scale [34]. Moreover, MSC-derived EVs have improved the infarcted cardiac function [35]. EVs carry different macromolecules, including proteins and non-coding RNAs and exert cardioprotective effects in MI [36, 37]. Returning to the initial speculation, we provided evidence in the current research that mBMSC-EV-shuttled CAV1 disrupted the TGF-β1/SMAD2 pathway, thus repressing c-JUN transcription and finally attenuating MF in MI mice.
While our study reveals the critical role of CAV1 carried by BMSC EVs in CF target cells, it should be emphasized that EVs solely serve as carriers for regulating proteins [38, 39]. Previous studies have also shown the direct involvement of CAV-1 protein in cardiac fibroblasts [40, 41], suggesting that CAV1 protein may exert its influence on myocardial fibrosis after myocardial infarction through the TGF-β1/SMAD2/c-JUN pathway. The initial finding in our research was that CAV1 was downregulated in myocardial tissues with MI. A corroborating earlier report elaborated that CAV1 expression was lower in AMI mice than in sham-operated mice [42]. Furthermore, we also observed that mBMSC-derived EVs delivered CAV1 into hypoxia-exposed CFs to repress fibrosis and apoptosis of CFs, accompanied by downregulated α-SMA, Collagen I, TNF-α, and IL-1β. Besides, such results were reproduced in MI mice. α-SMA and Collagen I have been widely recognized as fibrosis-related biomarkers [43]. Additionally, the etiology of fibrosis is primarily featured with inflammatory and fibro-proliferative changes by secreting TNF-α and IL-1β [44]. Of note, α-SMA, Collagen I, TNF-α, and IL-1β expression were previously documented to be upregulated in rats with isoproterenol hydrochloride-induced MI [45, 46]. Concordant with our finding, prior research illustrated that BMSCs diminished fibrosis and apoptotic cardiomyocytes and preserved the function of infarcted hearts [47]. Another research unveiled that myocardial injection of BMSC-released EVs improved cardiac function and restrained MF in MI rats [48]. It has generally been accepted that BMSC-secreted EVs are protected against MI by carrying proteins [49]. Our research showed that BMSC-derived EVs containing CAV1 restricted MF in MI. More importantly, it was observed that CAV1 knockout facilitated cardiac dysfunction in mice with MI, which was concurrent with the results in our animal experiments [50]. Also, CAV1 downregulation triggered fibrosis of CFs following MI [51].
The functionality of BMSC-EVs was directly validated through the inhibition of GW4869. This is consistent with previous research that indicates no separate GW4869 control group had an impact [52]. However, we acknowledge that the absence of a separate GW4869 control group in our experiment prevents us from directly determining the effect of GW4869 residues in the culture medium and isolated exosomes on CF cells. This limitation is also one of the shortcomings of our study. And in previous studies on exosomes, Calnexin has often been used as an endoplasmic reticulum marker, but its presence in exosomes is considered as contamination [53, 54]. This coincides with the latest MISEV guidelines [55], which classify calnexin as a contaminant. In our study, Calnexin was also used as one of the indicators of contamination, as its presence in exosomes indicates minimal cytoplasmic contamination, which is often observed alongside exosomes.
Another critical result in our research was that CAV1 delivered by mBMSC-EVs disrupted the TGF-β1/SMAD2 pathway to downregulate c-JUN, thus attenuating MF and cell apoptosis after MI. Similarly, a previous report manifested that CAV1 downregulation contributed to atrial fibrosis by activating the TGF-β1/SMAD2 pathway [56]. Notably, CAV1 inactivates the TGF-β1/SMAD2 signaling pathway [57]. Specifically, blocking the expression of TGF-β1/SMAD2 signaling pathway and its related molecules can inhibit cardiomyocyte fibrosis [58]. Also, another work documented the high TGF-β1 and SMAD2 expression in the infarction border zone of MI rats [59]. Furthermore, the inactivation of the TGF-β1/SMAD2 pathway alleviated MF and cell apoptosis in MI mice [60]. Of note, a study reports that the TGF-β1/SMAD2 pathway accelerates MF in rats by upregulating c-JUN [15]. The apoptotic signaling system JNK/c-Jun could indicate ischemic damage, while the repression of JNK/c-Jun exerted cardioprotective effects in MI [61]. Specifically, our findings further revealed that under TGF-β1 stimulation, the enrichment of SMAD2 protein was augmented in the promoters of α-SMA and Collagen I, which was nullified by c-JUN silencing. Consistently, TGF-β1 enhanced α-SMA and Collagen I expression in equine endometrial fibroblasts [62]. Similarly, TGF-β1/SMAD2 pathway activation caused elevation of α-SMA and Collagen I during fibroblast-myofibroblast transition [63]. Besides, c-JUN inhibition noticeably lowered TGF-β1-induced α-SMA and Collagen I expression in hepatic stellate cells [60], which consented to our finding.
Conclusions
In conclusion, mBMSC-EV-packaged CAV1 protein assumes a critical role in the treatment of MI due to its protective effects against MF following MI (Fig. 8). Investigation of mBMSC-EVs carrying CAV1 protein yields a better understanding of the mechanisms of MI and the potential of serving as a target for MI treatments in the future.
Molecular mechanism of mBMSC-EVs-CAV1-mediated TGF-β1/SMAD2/c-JUN axis in inhibiting cardiac fibroblast differentiation to improve MF after MI. mBMSC-EVs deliver CAV1 protein to CFs where the protein expression of CAV1 is upregulated upon hypoxia conditions. The TGF-β1/SMAD2 signaling pathway downstream of CAV1 is consequently inactivated, the transcription of c-JUN is inhibited, and transcription of SMAD2/c-JUN transcription complex target genes α-SMA and Collagen I is reduced. This mechanism suppresses CF fibrosis and apoptosis in vitro, and MF is ameliorated in MI mice
Data Availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
References
Chiang MH, Liang CJ, Lin LC, Yang YF, Huang CC, Chen YH, et al. miR-26a attenuates cardiac apoptosis and fibrosis by targeting ataxia-telangiectasia mutated in myocardial infarction. J Cell Physiol. 2020;235:6085–102. https://doi.org/10.1002/jcp.29537.
Zhao Z, Du S, Shen S, Wang L. microRNA-132 inhibits cardiomyocyte apoptosis and myocardial remodeling in myocardial infarction by targeting IL-1beta. J Cell Physiol. 2020;235:2710–21. https://doi.org/10.1002/jcp.29175.
Golforoush P, Yellon DM, Davidson SM. Mouse models of atherosclerosis and their suitability for the study of myocardial infarction. Basic Res Cardiol. 2020;115:73. https://doi.org/10.1007/s00395-020-00829-5.
Daseke MJ 2nd, Tenkorang MAA, Chalise U, Konfrst SR, Lindsey ML. Cardiac fibroblast activation during myocardial infarction wound healing: fibroblast polarization after MI. Matrix Biol. 2020;91-92:109–16. https://doi.org/10.1016/j.matbio.2020.03.010.
Kurose H. Cardiac Fibrosis and Fibroblasts. Cells. 2021;10 https://doi.org/10.3390/cells10071716.
Prabhu SD, Frangogiannis NG. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res. 2016;119:91–112. https://doi.org/10.1161/CIRCRESAHA.116.303577.
Miao C, Lei M, Hu W, Han S, Wang Q. A brief review: the therapeutic potential of bone marrow mesenchymal stem cells in myocardial infarction. Stem Cell Res Ther. 2017;8:242. https://doi.org/10.1186/s13287-017-0697-9.
Woith E, Fuhrmann G, Melzig MF. Extracellular vesicles-connecting kingdoms. Int J Mol Sci. 2019;20 https://doi.org/10.3390/ijms20225695.
Bian S, Zhang L, Duan L, Wang X, Min Y, Yu H. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J Mol Med (Berl). 2014;92:387–97. https://doi.org/10.1007/s00109-013-1110-5.
Keshtkar S, Azarpira N, Ghahremani MH. Mesenchymal stem cell-derived extracellular vesicles: novel frontiers in regenerative medicine. Stem Cell Res Ther. 2018;9:63. https://doi.org/10.1186/s13287-018-0791-7.
Lin CJ, Yun EJ, Lo UG, Tai YL, Deng S, Hernandez E, et al. The paracrine induction of prostate cancer progression by caveolin-1. Cell Death Dis. 2019;10:834. https://doi.org/10.1038/s41419-019-2066-3.
Gvaramia D, Blaauboer ME, Hanemaaijer R, Everts V. Role of caveolin-1 in fibrotic diseases. Matrix Biol. 2013;32:307–15. https://doi.org/10.1016/j.matbio.2013.03.005.
Shivshankar P, Halade GV, Calhoun C, Escobar GP, Mehr AJ, Jimenez F, et al. Caveolin-1 deletion exacerbates cardiac interstitial fibrosis by promoting M2 macrophage activation in mice after myocardial infarction. J Mol Cell Cardiol. 2014;76:84–93. https://doi.org/10.1016/j.yjmcc.2014.07.020.
Lu J, Zhang J, Wang Y, Sun Q. Caveolin-1 scaffolding domain peptides alleviate liver fibrosis by inhibiting TGF-beta1/Smad signaling in mice. Int J Mol Sci. 2018;19 https://doi.org/10.3390/ijms19061729.
Lu M, Qin Q, Yao J, Sun L, Qin X. Induction of LOX by TGF-beta1/Smad/AP-1 signaling aggravates rat myocardial fibrosis and heart failure. IUBMB Life. 2019;71:1729–39. https://doi.org/10.1002/iub.2112.
Qiu H, Liu W, Lan T, Pan W, Chen X, Wu H, et al. Salvianolate reduces atrial fibrillation through suppressing atrial interstitial fibrosis by inhibiting TGF-beta1/Smad2/3 and TXNIP/NLRP3 inflammasome signaling pathways in post-MI rats. Phytomedicine. 2018;51:255–65. https://doi.org/10.1016/j.phymed.2018.09.238.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. https://doi.org/10.1093/nar/gks1193.
Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, et al. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428:688–92. https://doi.org/10.1016/j.jmb.2015.09.019.
Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–3. https://doi.org/10.1038/nmeth.3484.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D13. https://doi.org/10.1093/nar/gky1131.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. https://doi.org/10.1101/gr.1239303.
Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W41. https://doi.org/10.1093/nar/gkz240.
Chen B, Luo L, Wei X, Gong D, Li Z, Li S, et al. M1 Bone marrow-derived macrophage-derived extracellular vesicles inhibit angiogenesis and myocardial regeneration following myocardial infarction via the MALAT1/MicroRNA-25-3p/CDC42 Axis. Oxid Med Cell Longev. 2021;2021:9959746. https://doi.org/10.1155/2021/9959746.
Zhu LP, Tian T, Wang JY, He JN, Chen T, Pan M, et al. Hypoxia-elicited mesenchymal stem cell-derived exosomes facilitates cardiac repair through miR-125b-mediated prevention of cell death in myocardial infarction. Theranostics. 2018;8:6163–77. https://doi.org/10.7150/thno.28021.
Wu Y, Peng W, Fang M, Wu M, Wu M. MSCs-derived extracellular vesicles carrying miR-212-5p alleviate myocardial infarction-induced cardiac fibrosis via NLRC5/VEGF/TGF-beta1/SMAD Axis. J Cardiovasc Transl Res. 2022;15:302–16. https://doi.org/10.1007/s12265-021-10156-2.
Gao W, Shao R, Zhang X, Liu D, Liu Y, Fa X. Up-regulation of caveolin-1 by DJ-1 attenuates rat pulmonary arterial hypertension by inhibiting TGFbeta/Smad signaling pathway. Exp Cell Res. 2017;361:192–8. https://doi.org/10.1016/j.yexcr.2017.10.019.
Han M, Gu Y, Lu P, Li J, Cao H, Li X, et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19:26. https://doi.org/10.1186/s12943-020-1145-5.
Hinkelbein J, Bohm L, Braunecker S, Adler C, De Robertis E, Cirillo F. Decreased tissue COX5B expression and mitochondrial dysfunction during sepsis-induced kidney injury in rats. Oxid Med Cell Longev. 2017;2017:8498510. https://doi.org/10.1155/2017/8498510.
Yu SY, Dong B, Fang ZF, Hu XQ, Tang L, Zhou SH. Knockdown of lncRNA AK139328 alleviates myocardial ischaemia/reperfusion injury in diabetic mice via modulating miR-204-3p and inhibiting autophagy. J Cell Mol Med. 2018;22:4886–98. https://doi.org/10.1111/jcmm.13754.
Borger V, Bremer M, Ferrer-Tur R, Gockeln L, Stambouli O, Becic A, et al. Mesenchymal stem/stromal cell-derived extracellular vesicles and their potential as novel immunomodulatory therapeutic agents. Int J Mol Sci. 2017;18 https://doi.org/10.3390/ijms18071450.
Han C, Zhou J, Liang C, Liu B, Pan X, Zhang Y, et al. Human umbilical cord mesenchymal stem cell derived exosomes encapsulated in functional peptide hydrogels promote cardiac repair. Biomater Sci. 2019;7:2920–33. https://doi.org/10.1039/c9bm00101h.
Zhao Y, Sun X, Cao W, Ma J, Sun L, Qian H, et al. Exosomes derived from human umbilical cord mesenchymal stem cells relieve acute myocardial ischemic injury. Stem Cells Int. 2015;2015:761643. https://doi.org/10.1155/2015/761643.
Sundqvist A, Vasilaki E, Voytyuk O, Bai Y, Morikawa M, Moustakas A, et al. TGFbeta and EGF signaling orchestrates the AP-1- and p63 transcriptional regulation of breast cancer invasiveness. Oncogene. 2020;39:4436–49. https://doi.org/10.1038/s41388-020-1299-z.
Sposito AC, de Lima-Junior JC, Moura FA, Barreto J, Bonilha I, Santana M, et al. Reciprocal multifaceted interaction between HDL (high-density lipoprotein) and myocardial infarction. Arterioscler Thromb Vasc Biol. 2019;39:1550–64. https://doi.org/10.1161/ATVBAHA.119.312880.
Wang N, Chen C, Yang D, Liao Q, Luo H, Wang X, et al. Mesenchymal stem cells-derived extracellular vesicles, via miR-210, improve infarcted cardiac function by promotion of angiogenesis. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2085–92. https://doi.org/10.1016/j.bbadis.2017.02.023.
Riaud M, Martinez MC, Montero-Menei CN. Scaffolds and extracellular vesicles as a promising approach for cardiac regeneration after myocardial infarction. Pharmaceutics. 2020;12 https://doi.org/10.3390/pharmaceutics12121195.
Viola M, de Jager SCA, Sluijter JPG. Targeting inflammation after myocardial infarction: a therapeutic opportunity for extracellular vesicles? Int J Mol Sci. 2021;22 https://doi.org/10.3390/ijms22157831.
van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28. https://doi.org/10.1038/nrm.2017.125.
Silva AM, Lazaro-Ibanez E, Gunnarsson A, Dhande A, Daaboul G, Peacock B, et al. Quantification of protein cargo loading into engineered extracellular vesicles at single-vesicle and single-molecule resolution. J Extracell Vesicles. 2021;10:e12130. https://doi.org/10.1002/jev2.12130.
Cohen AW, Park DS, Woodman SE, Williams TM, Chandra M, Shirani J, et al. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am J Physiol Cell Physiol. 2003;284:C457–74. https://doi.org/10.1152/ajpcell.00380.2002.
Quintas LE, Pierre SV, Liu L, Bai Y, Liu X, Xie ZJ. Alterations of Na+/K+-ATPase function in caveolin-1 knockout cardiac fibroblasts. J Mol Cell Cardiol. 2010;49:525–31. https://doi.org/10.1016/j.yjmcc.2010.04.015.
Zhu JZ, Bao XY, Zheng Q, Tong Q, Zhu PC, Zhuang Z, et al. Buyang Huanwu decoction exerts cardioprotective effects through targeting angiogenesis via caveolin-1/VEGF signaling pathway in mice with acute myocardial infarction. Oxid Med Cell Longev. 2019;2019:4275984. https://doi.org/10.1155/2019/4275984.
Bai L, Li A, Gong C, Ning X, Wang Z. Protective effect of rutin against bleomycin induced lung fibrosis: involvement of TGF-beta1/alpha-SMA/Col I and III pathway. Biofactors. 2020;46:637–44. https://doi.org/10.1002/biof.1629.
Kandhare AD, Bodhankar SL, Mohan V, Thakurdesai PA. Effect of glycosides based standardized fenugreek seed extract in bleomycin-induced pulmonary fibrosis in rats: decisive role of Bax, Nrf2, NF-kappaB, Muc5ac, TNF-alpha and IL-1beta. Chem Biol Interact. 2015;237:151–65. https://doi.org/10.1016/j.cbi.2015.06.019.
Radhiga T, Senthil S, Sundaresan A, Pugalendi KV. Ursolic acid modulates MMPs, collagen-I, alpha-SMA, and TGF-beta expression in isoproterenol-induced myocardial infarction in rats. Hum Exp Toxicol. 2019;38:785–93. https://doi.org/10.1177/0960327119842620.
Emran T, Chowdhury NI, Sarker M, Bepari AK, Hossain M, Rahman GMS, et al. L-carnitine protects cardiac damage by reducing oxidative stress and inflammatory response via inhibition of tumor necrosis factor-alpha and interleukin-1beta against isoproterenol-induced myocardial infarction. Biomed Pharmacother. 2021;143:112139. https://doi.org/10.1016/j.biopha.2021.112139.
Zhang Z, Yang C, Shen M, Yang M, Jin Z, Ding L, et al. Autophagy mediates the beneficial effect of hypoxic preconditioning on bone marrow mesenchymal stem cells for the therapy of myocardial infarction. Stem Cell Res Ther. 2017;8:89. https://doi.org/10.1186/s13287-017-0543-0.
Firoozi S, Pahlavan S, Ghanian MH, Rabbani S, Barekat M, Nazari A, et al. Mesenchymal stem cell-derived extracellular vesicles alone or in conjunction with a SDKP-conjugated self-assembling peptide improve a rat model of myocardial infarction. Biochem Biophys Res Commun. 2020;524:903–9. https://doi.org/10.1016/j.bbrc.2020.02.009.
Ning H, Chen H, Deng J, Xiao C, Xu M, Shan L, et al. Exosomes secreted by FNDC5-BMMSCs protect myocardial infarction by anti-inflammation and macrophage polarization via NF-kappaB signaling pathway and Nrf2/HO-1 axis. Stem Cell Res Ther. 2021;12:519. https://doi.org/10.1186/s13287-021-02591-4.
Jasmin JF, Rengo G, Lymperopoulos A, Gupta R, Eaton GJ, Quann K, et al. Caveolin-1 deficiency exacerbates cardiac dysfunction and reduces survival in mice with myocardial infarction. Am J Physiol Heart Circ Physiol. 2011;300:H1274–81. https://doi.org/10.1152/ajpheart.01173.2010.
Gao Y, Chu M, Hong J, Shang J, Xu D. Hypoxia induces cardiac fibroblast proliferation and phenotypic switch: a role for caveolae and caveolin-1/PTEN mediated pathway. J Thorac Dis. 2014;6:1458–68. https://doi.org/10.3978/j.issn.2072-1439.2014.08.31.
Zhao S, Li W, Yu W, Rao T, Li H, Ruan Y, et al. Exosomal miR-21 from tubular cells contributes to renal fibrosis by activating fibroblasts via targeting PTEN in obstructed kidneys. Theranostics. 2021;11:8660–73. https://doi.org/10.7150/thno.62820.
Samaeekia R, Rabiee B, Putra I, Shen X, Park YJ, Hematti P, et al. Effect of human corneal mesenchymal stromal cell-derived exosomes on corneal epithelial wound healing. Invest Ophthalmol Vis Sci. 2018;59:5194–200. https://doi.org/10.1167/iovs.18-24803.
Huang K, Garimella S, Clay-Gilmour A, Vojtech L, Armstrong B, Bessonny M, et al. Comparison of human urinary exosomes isolated via ultracentrifugation alone versus ultracentrifugation followed by SEC column-purification. J Pers Med. 2022;12 https://doi.org/10.3390/jpm12030340.
Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. https://doi.org/10.1080/20013078.2018.1535750.
Zhang M, Wang H, Bie M, Wang X, Lu K, Xiao H. Caveolin-1 deficiency induces atrial fibrosis and increases susceptibility to atrial fibrillation by the STAT3 signaling pathway. J Cardiovasc Pharmacol. 2021;78:175–83. https://doi.org/10.1097/FJC.0000000000001066.
Zhang M, Wang H, Wang X, Bie M, Lu K, Xiao H. MG53/CAV1 regulates transforming growth factor-beta1 signaling-induced atrial fibrosis in atrial fibrillation. Cell Cycle. 2020;19:2734–44. https://doi.org/10.1080/15384101.2020.1827183.
Tu JH, Xu Y, Dai Y, Dang L. Effect of alprostadil on myocardial fibrosis in rats with diabetes mellitus via TGF-beta1/Smad signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:9633–41. https://doi.org/10.26355/eurrev_201911_19457.
Chen Y, Li T, Gao Q, Wang LY, Cui LQ. MiR-1908 improves cardiac fibrosis after myocardial infarction by targeting TGF-beta1. Eur Rev Med Pharmacol Sci. 2018;22:2061–9. https://doi.org/10.26355/eurrev_201804_14736.
Shen Z, Shen A, Chen X, Wu X, Chu J, Cheng Y, et al. Huoxin pill attenuates myocardial infarction-induced apoptosis and fibrosis via suppression of p53 and TGF-beta1/Smad2/3 pathways. Biomed Pharmacother. 2020;130:110618. https://doi.org/10.1016/j.biopha.2020.110618.
Wang J, Ma Y, Sachs F, Li J, Suchyna TM. GsMTx4-D is a cardioprotectant against myocardial infarction during ischemia and reperfusion. J Mol Cell Cardiol. 2016;98:83–94. https://doi.org/10.1016/j.yjmcc.2016.07.005.
Szostek-Mioduchowska AZ, Lukasik K, Skarzynski DJ, Okuda K. Effect of transforming growth factor -beta1 on alpha-smooth muscle actin and collagen expression in equine endometrial fibroblasts. Theriogenology. 2019;124:9–17. https://doi.org/10.1016/j.theriogenology.2018.10.005.
Zhou JP, Tang W, Feng Y, Li N, Gu CJ, Li QY, et al. Angiotensin-(1-7) decreases the expression of collagen I via TGF-beta1/Smad2/3 and subsequently inhibits fibroblast-myofibroblast transition. Clin Sci (Lond). 2016;130:1983–91. https://doi.org/10.1042/CS20160193.
Funding
This study was supported by the Basic and Applied Basic Research Project of Guangzhou Basic Research Plan (202201011721).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Statement
Animal experimentations were authorized by the Animal Ethics Committee of Guangdong Provincial People's Hospital (Guangdong Academy of Medical Sciences; Guangdong Cardiovascular Institute) (No. KY-7.-2022-2317-01). Appropriate measures had been taken to avoid any unnecessary distress to the animals. This study was approved by the Ethics Committee of Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences; Guangdong Cardiovascular Institute), and the methods were carried out following the approved guidelines. All the patients have been informed and signed informed consent before the experiments.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Associate Editor Nicola Smart oversaw the review of this article
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yijin Wu and Wenying Peng are regarded as co-first authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Peng, W., Chen, S. et al. CAV1 Protein Encapsulated in Mouse BMSC-Derived Extracellular Vesicles Alleviates Myocardial Fibrosis Following Myocardial Infarction by Blocking the TGF-β1/SMAD2/c-JUN Axis. J. of Cardiovasc. Trans. Res. 17, 523–539 (2024). https://doi.org/10.1007/s12265-023-10472-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-023-10472-9