Abstract
Circular RNAs (circRNAs) are covalently closed single-stranded RNAs with regulatory activity and regarded as new types of therapeutic targets in diseases such as cancers. By means of RNA-Seq technology, numerous cardiac circRNAs were discovered. Although some candidates were detected to involve in heart disease in murine model, relative low sequence conservation and expression level of their human homologs might result in an insignificant, even distinct effect in the human heart. Therefore, the therapeutic significance of circRNAs should be more strictly considered. It is also necessary to discuss which circRNA is suitable for being applied in heart disease treatment. Here, we are willing to introduce a ~ 1830 nt circular transcript generated from single exon of sodium/calcium exchanger 1 (ncx1) gene (also called solute carrier family 8 member A1, slc8a1), usually named circNCX1 or circSLC8A1, which is gradually coming into our view. circNCX1 is one of the most cardiac-enriched circRNAs. It is widely existent in vertebrate and relatively conserved, indicating its indispensability during the evolution of species. Indeed, circNCX1 was shown to involve in heart development by some expression analysis. It was further revealed that the dysregulation of circNCX1 is one of the key pathogeneses of heart diseases including ischemic cardiac injury and hypertrophic cardiomyopathy. To make the significance of circNCX1 in the heart clear, we comprehensively dissected circNCX1 in the aspects of its parental gene structure, conservation, biogenesis and expression profiles, function, molecular mechanisms, and clinical application in this review. New medicine or therapeutic schedules based on circNCX1 are expected in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heart disease has been the most lethal events of human beings for a long time. In cellular level, the dysregulation of cell differentiation, proliferation, metabolism, and death in the heart lead to a series of heart diseases. Essentially, aberrant gene expression results in the disruption of cellular homeostasis. Therefore, molecular mechanisms of cardiac developmental, physiological, and pathological processes are always being studied aiming to fully understand the pathogenesis and discover effective therapeutic schedules of heart diseases. It has been known that gene expression is regulated in various stages including chromatin activation (histone chemical modification and DNA methylation), transcription (transcription activation and inhibition), and post-transcription (RNA splicing, RNA degeneration, protein translation, and protein degradation). Evidence showed that defect in each step in cardiac cells could disorder cardiac physiology and cause heart disease [1,2,3,4,5].
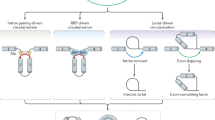
In 2013, a new type of RNA classified as circRNA was broadly identified in vertebrates [6, 7]. circRNAs are generated from back-spliced exons of precursor mRNAs (pre-mRNAs) or intronic lariats which escape from degradation [8]. Distinct covalently closed circular structure makes circRNAs stable and tolerates RNase digestion [9]. Hence, circRNAs could be active in a relatively longer term. Functionally, exonic circRNAs localized in the cytoplasm are able to interact with RNA-/DNA-binding proteins to modulate their subcellular localization, stability. or cofactor association [10,11,12]. Nevertheless, miRNA sponge is the more widely known function of circRNA. The degradation or translation repression of mRNA modulated by miRNA can be inhibited by the interaction of circRNA and miRNA [7, 13]. Intronic circRNAs can directly regulate parental gene transcription [14, 15]. In addition, a handful of circRNAs have coding ability and act via encoded proteins or peptides [16,17,18,19,20,21,22,23]. Versatile mechanisms make circRNAs defined as key regulator of gene expression.
The expression of circRNAs is spatio-temporal specific [6, 24, 25]. According to high-throughput RNA-Seq technology, thousands of circRNAs were detected in murine and human hearts [26, 27]. More and more candidates were demonstrated to function in the murine heart and affect gene expression in various levels during cardiac physiological and pathological processes [28,29,30]. However, whether these candidates involved in human heart disease need to be further determined. The back-splicing events are quite different among species [6, 7, 31, 32], suggesting some murine circRNAs might not be expressed in the human heart. In addition, some circRNAs with low absolute cellular concentration were just screened out due to their differential changed expression level in murine heart disease model. They might not show significant activities in the human heart without artificial intervention because of the low probability of interacting with target molecules [33].
In this review, we propose to introduce a circRNA which has therapeutic potency on heart disease treatment. This circRNA is usually named circNCX1 or circSLC8A1 and generated from the second ~ 1830 nt exon of ncx1 (also called slc8a1) gene. circNCX1 had been detected in vertebrates including primate, murine, pig, chicken, rabbit, amphibian, and teleost and rather conserved [26, 27, 34,35,36,37,38]. It is also a most abundant circRNA in murine and human hearts [26, 27]. Studies demonstrated that circNCX1 is involved in ischemic myocardial injury and cardiac hypertrophy [39, 40]. Evidence also suggested possible roles of circNCX1 in heart development [41,42,43,44]. We will comprehensively characterize circNCX1 in each aspect to reveal its therapeutic value in heart disease treatment.
circNCX1 Is an Evolutionary Conserved circRNA Transcribed from ncx1 Gene
Ncx1 gene, also named slc8a1, encodes a transmembrane protein which modulates cellular Ca2+ homeostasis and Ca2+ involved processes [45], which is predominantly expressed in the myocardium regulating heart excitation–contraction [46,47,48,49]. circNCX1 is just generated from the ~ 1830 nt single exon (normally the second exon of most NCX1 transcripts) of NCX1 pre-mRNA. Multiple alignment of ncx1 genomic locus by PhyloP reveals that this circularized region is remarkably conserved in mammals [39]. High conservation among the species indicates similar target molecules and functions. This is important for medical studies involving circRNAs which used non-primates as experimental animals. Nowadays, murine is the most frequently used animal model for cardiac circRNA research. However, if the murine circRNAs were not conserved or not spliced into the same isoform, even not existing in human, the therapeutic and clinical value of the results would be discounted. For instance, circRNA ACR (tagged as mm9_circ_006636 in database [7]) was demonstrated to protect the heart from I/R injury by attenuating cardiomyocyte autophagy in mice [29], but its homolog has not been detected in human so far. CDR1as, the first circRNA characterized as a miRNA sponge, was found to aggravate cardiomyocyte apoptosis in mice myocardial infarction (MI) [50]. Although CDR1as homologs had been identified in both human and mouse, their lengths are quite distinct (human CDR1as, hsa_circ_0001946, 1485nt. Mouse CDR1as, mmu_circ_0001878, 2927nt). Whether it causes functional difference is unknown. circNCX1 is not only expressed in human and rodent heart, but also detected by RNA-Seq or accurately identified in various classes of vertebrates including pig, chicken, toad, and zebrafish [26, 27, 34,35,36,37,38], suggesting its evolutional importance. Alignment of mammal circNCX1 locus sequence by MegAlign showed high sequence identity. It means that conserved miRNA- or RNA-binding protein (RBP)-interacting motifs are reserved during the process of animal evolution. Similar regional secondary structure or tertiary structure of circNCX1 can also be formed to interact with similar group of molecules due to conserved primary structure among the species. Importantly, miR-133a-3p-binding region in circNCX1 is conserved. It had been demonstrated in murine heart that circNCX1 primarily works as a miR-133a-3p sponge (it will be discussed in detail in the following section) (Fig. 1). Therefore, it could be inferred that in other species, circNCX1 would also function through a miR-133a-dependent way.
What Makes circNCX1 the Most Abundant circRNA in the Heart: the Expression Profile of circNCX1 and Its Upstream Regulators
circNCX1 is the most abundant circRNA in murine and human hearts as revealed by multiple RNA-Seq data [26, 27, 38]. To well understand the underlying mechanisms of cardiac enrichment of circNCX1, we should first talk about how circRNAs are generated. At present, the mainstream view believes that both circRNAs and mRNAs originate from pre-mRNAs (or heterogeneous nuclear RNAs, hnRNAs) [8]. Therefore, factors transcriptionally regulated the parental genes of circRNAs including cis-acting elements, trans-acting factors, DNA methylation, and histone modification, also affecting the expression pattern of circRNAs. ncx1 is the parental gene of circNCX1. The NCX1 mRNA transcripts are enriched in the heart, showing similar tissue-specific expression pattern with circNCX1 [47,48,49]. Moreover, the expression level of NCX1 mRNA is positively correlated with circNCX1 [43]. It could be inferred that the heart-specific expression pattern of circNCX1 might be mediated by some transcription regulatory factors.
Enhancer is a regulatory DNA sequence classified as one type of cis-acting element which can be recognized by specific transcription factors. These transcription factors would promote transcription of the enhancer-regulated genes and endow them with tissue- or cell-type-specific expression pattern [51, 52]. A fraction of enhancers are localized in cluster and extend for long range in genome. Generally, they are featured by high level of histone modification, particular sensitivity to Dnase I and intensive binding site of multiple transcription activators. This class of enhancers being able to strongly stimulate cell-type-specific gene expression and decide cell fate is defined as super enhancers (SEs) [53]. Transcription of ncx1 had been demonstrated to be regulated by histone deacetylase (HDAC) and homeobox protein Nkx-2.5 which is a core transcription co-activator in the differentiation of the myocardial lineage [54,55,56]. According to data of chromatin immunoprecipitation and sequencing (ChIP-seq) collected in the SE database, it could be found that heart-specific SEs with enrichment of H3K27ac modification are localized in human and mouse ncx1 locus (hg17, chr2:40,530,157–40,742,603, mm9, chr17:82,124,844–82,140,852) [57, 58], suggesting the heart-specific SEs might regulate transcription of ncx1 and further result in heart-specific abundance of circNCX1. The analysis by Huang et al. also inferred that circNCX1 was a predominant SE-regulated circRNA which play significant roles in the heart [59].
However, it was observed that some other circRNAs generated from ncx1 are not highly expressed in the heart (Table 1). It means that post-transcriptional mechanisms probably involve in the heart-specific regulation of circNCX1. We have known that exonic circRNAs originate from some special single exons or adjacent several exons (sometimes containing the introns in between) which can be circularized through so-called pre-mRNA back-splicing. The back-splicing is triggered by pairing of the introns flanking the circularized exons. Intron pairing makes the branchpoint in the upstream intron attacks the 5′ splicing site in the downstream intron and results in covalent ligand of 5′ and 3′ end of the exon. Specific elements such as short interspersed repeat segments (SINEs) distributed in the flanking introns could recruit RBPs and guide the bridging of upstream and downstream introns [6, 60, 61]. Therefore, the tissue-specific expression of circRNAs is also determined by RBPs during back-splicing. For example, the production of circRNAs from titin gene is regulated by heart-specific RBP-RBM20. In dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM), the expression of titin circRNAs was disturbed by the dysregulation of RBM20 [62]. Remarkably, circNCX1 has extremely long flanking introns in various species, especially in primate and murine (Table 2). The overlength introns harbor numerous SINEs and provide capacious platforms for RBPs binding. By means of RBPmap [63], it can be observed that binding motifs of RBPs which affect heart development or potentially involve in circRNA biogenesis including polypyrimidine tract-binding protein 1 (PTBP1), muscleblind-like protein 1 (MBNL1), CUGBP Elav-like family member 1 (CELF1), serine/arginine-rich splicing factor (SRSF) 3, and 5 are densely distributed in the flanking introns of human circNCX1 (Fig. 2). PTBP1 regulates alternative splicing of pre-mRNA and plays a role in exon exclusion during muscle cell differentiation [64]. It was further reported to correlate with circRNA expression. In glioma endothelial cells, PTBP1 positively affected the level of circRNA_001160 [65]. Knockdown of PTBP1 would downregulate circRNA_001160 expression. Both of MBNL1 and CELF1 can mediate the alternative splicing of cardiac-specific troponin-T (TNNT2) [66, 67]. It was notable that the homolog of MBNL1 in fruit fly named muscleblind was demonstrated to process the production of its isogeneic circRNA [68]. Whether MBNL1 involved in the biogenesis of circRNAs remains to be verified. Serine–arginine (SR) proteins were early discovered to control circRNA expression in fruit fly [69]. Recently, SR proteins were also found to be correlated with circRNA production in human cell line. Naqvi et al. reported in a preprint that SRSF3 not only regulated mRNA splicing, but also affected circRNA generating in P493-6 cell [70]. These evidence implied that the RBPs mentioned above might mediate the back-splicing process of NCX1 pre-mRNA in the heart. It is notable that several binding motifs of CELF1, MBNL1, PTBP1, and SRSF5 were also predicted to locate in circNCX1 locus. Cytoplasmic circRNA could retain the nucleic proteins bound with it in the cytoplasm and inhibit their function [11, 17]. circNCX1 predominately locates in the cytoplasm, if circNCX1 interacted with these RBPs and simultaneously was regulated by these RBPs at back-splicing stage; a feedback loop might be formed that circNCX1 would detain the excessed RBPs to keep circNCX1 expression level in a certain range, resembling circMBL in fruit fly [68]. In brief, transcriptional and post-transcriptional regulatory mechanisms both determine the expression profile of circNCX1 (Fig. 3).
The distribution of RBP binding motifs in flanking introns of human circNCX1. The conserved binding motifs of CUGBP, MBNL1, PTBP1, SRSF3, and SRSF5 were calculated by RBPmap using high stringency and conservation filter. One colorized line represents one RBP binding motif. Longer line means higher reliability
circNCX1 Is a Key Regulator Associated with miR-133a in Cardiomyopathies
Some circRNAs had been identified to affect the activity of factors associated with death, differentiation, proliferation, or stress by directly binding or indirectly regulation. The dysregulation of these circRNAs might trigger cell aberration and aggravate the pathological processes. Artificially modified cellular circRNA level would rescue the status [71]. The core circRNA related to heart disease remains to be screened out. circNCX1 had been detected as a highly abundant circRNA in the human heart. It also has a relatively large molecular size among the circRNA group. The length of circNCX1 is ~ 1830 nt, while the average length of circRNAs calculated by Jeck et al. is ~ 690 nt [6]. The supernormal expression level and size allow circNCX1 to serve as a broad platform for molecular interacting. An argonaute 2 (Ago2)-ribonucleoprotein immunoprecipitation (RIP)-Seq in rat cardiac myocytes performed by Werfel et al. suggested that circNCX1 has the highest Ago2 affinity and might act as a significant miRNA sponge [26].
Our previous work and Lim et al. predicted that there are abundant putative miR-133a binding sites in circNCX1 sequence. The interaction of circNCX1 and miR-133a was further solidly verified by RNA pull-down assays, luciferase assay, and fluorescence in situ hybridization [39, 40]. Moreover, Lim et al. uncovered that miR-133a is one of the miRNAs which were most highly enriched in the endogenous circNCX1-captured fraction in cardiomyocyte among the total 752 miRNAs [40]. miR-133a is a myocardium and skeletal muscle-specific miRNA. In skeletal myoblast and myocardium, miR-133a regulates cell proliferation and differentiation [72,73,74]. On the other hand, as a cardiac-enriched miRNA, miR-133a plays key roles in DCM, HCM, and ischemic cardiac injury [75]. For instance, in adult heart, miR-133a can protect cardiomyocyte from hypertrophy by suppressing stress-related genes. Knockdown of miR-133a would cause cardiac hypertrophy [76]. miR-133a also inhibits oxidative stress-induced cardiomyocyte apoptosis and attenuates ischemic myocardial injury [39]. Genetic depletion of miR-133a will also cause significant apoptosis in the heart [73].
As an effective sponge of miR-133a, circNCX1 is reasonable to involve in ischemic cardiac injury and cardiac hypertrophy by regulating miR-133a (Fig. 4). Oxidative stress will increase circNCX1 level in rat cardiomyocytes and mice myocardium. The excessive circNCX1 shows pro-apoptotic activity both in vitro and in vivo. Silencing of circNCX1 can inhibit cardiomyocyte apoptosis induced by H2O2 and hypoxia–reoxygenation (H/R) or decrease I/R caused by myocardial infarction size [39]. A lasted published data also showed that circNCX1 was markedly upregulated in ischemic myocardium tissues [38]. The upregulation of circNCX1 under oxidative stress is likely due to the mechanisms in the transcriptional level. Both hypoxia-inducible factor 1-alpha (HIF1α) and histone deacetylases (HDACs) had been demonstrated to be induced in I/R and regulate the transcription of ncx1 [77, 78]. Whether circNCX1 being affected by post-transcriptional ways under oxidative stress is uncertain. The expression of pro-apoptotic cell death-inducing protein (CDIP1) is elevated by overexpressed circNCX1, whereas miR-133a level is not affected because the sequence of miR-133a and circNCX1 is not perfectly complementary. Therefore, circNCX1 is only able to inhibit miR-133a targeting CDIP1 mRNA and not lead to miR-133a decay [39].
On the other hand, knockdown of circNCX1 in the heart could attenuate transverse aortic constriction (TAC)-induced cardiac hypertrophy and heart failure in vivo. The overexpression of circNCX1 will lead to rat neonatal cardiomyocyte hypertrophy. Noticeably, cardiac-specific overexpression of circNCX1 in day 1 postnatal (P1) rats causes mortality. Significant ventricle dilation and heart failure were observed in the rest alive rats. The miR-133a targets related with cardiac stress were upregulated without significant change of miR-133a level [40]. This phenotype is approximately coincident with miR-133a-KO mice. miR-133a-KO was also partially lethal. Dilated ventricles were developed in the survivals with falling cardiac function [73]. It can be concluded that circNCX1 affects process of cardiomyopathy and cardiac injury by mediating miR-133a predominately. However, postnatal overexpression of circNCX1 did not result in myocardial fibrosis which was observed in miR-133a-KO heart. Perhaps, genetic abolish of miR-133a disrupted the cardiac development and function in embryonic stage.
In addition, there is conflict about the expression of circNCX1 in DCM and HCM. Actually, NCX1 mRNA levels had been detected to be increased with pressure overload in feline and mouse model of cardiac hypertrophy [79, 80]. As a SE-regulated circRNA, the expression of circNCX1 should also be stimulated by pressure overload. qRT-PCR data from Lei et al. and Jakobi et al. both revealed that circNCX1 was remarkably increased in human HCM and DCM. However, Tan et al. reported that the average circNCX1 levels in DCM and HCM samples were not obviously higher than that in normal myocardial samples [27, 38, 42]. In fact, it can be observed that there was a sample with extremely high circNCX1 level in normal group of the data from Tan et al. which might disrupt the results of average expression folds and significance calculation [27]. Lim et al. found that circNCX1 abundance was unchanged in mice cardiomyocytes in closer detail across different time points post-TAC surgery [40]. However, it was also reported that the expression of circNCX1 was increased in the myocardial tissues of rats injected with ISO solution [81]. This controversial result could be explained by differences in the models and sampling methods used.
circNCX1 Is a Potential Modulator During Embryonic Heart Development
As early as 2014, circNCX1 was detected to approximately fourfold enriched in the neonatal rat heart samples as heart samples derived from adult rats [26]. Mature mammalian cardiomyocyte is a type of terminal differentiated cells which negligibly proliferate, while neonatal mammalian cardiomyocytes still preserve some proliferation ability. Therefore, the distinct amount of circNCX1 in neonatal heart and adult heart is very intriguing and implied a possible connection between circNCX1 and cardiomyocyte proliferation. If downregulation of circNCX1 was correlated with loss of proliferation ability of mature cardiomyocyte, the overexpression of circNCX1 would rescue the proliferation of cardiomyocyte. Unexpectedly, increased circNCX1 in cardiomyocytes would induce hypertrophy and apoptosis [39, 40].
So another question was raised: whether circNCX1 involved in earlier biological processes in fetal heart development? Actually, the production of circNCX1 in the heart was consistently increased during the stage of fetal cardiac development [41]. Analysis of circNCX1 level during the transformation from human embryonic stem cells (hESCs) to cardiomyocytes and transformation from induced pluripotent stem cells (iPSC) to cardiomyocytes also revealed that circNCX1 was gradually upregulated during cardiomyocyte differentiation in vitro [41,42,43,44]. Intriguingly, miR-133a was shown to be synchronously increased with circNCX1 during differentiation from stem cells to cardiomyocytes and regulates cardiac differentiation in vitro [74]. Possibly, circNCX1 was a potential regulator via miR-133a. However, it can’t be concluded from these data that circNCX1 was involved in cardiomyocyte differentiation because circNCX1 might only serve as a marker of cardiomyocyte which maintained the feature of cardiomyocyte (Fig. 5).
A Pendent Problem of circNCX1: Translatable or Non-coding?
Although most of circRNAs are defined as non-coding RNAs because they are lacking 5′ cap and 3′ poly-A tail, a few circRNAs are proved to have coding ability. circNCX1 was proposed to have protein-coding potency when it was initially identified at 1999 by Li and Lytton [34]. There is a putative ORF stretching across the junction site in human, monkey, and rabbit circNCX1 sequences. The ORF was able to encode a ~ 600aa protein which was almost completely overlapping the N-terminal of full-length NCX1. Here, we named this hypothetical protein as truncated NCX1. Li and Lytton transfected the plasmid containing linearized ORF of truncated NCX1 into HEK293 cells and successfully obtained a ~ 70 KDa protein, indicating that the truncated NCX1 could be folded into correct conformation and showed no cytotoxicity. A protein band corresponding to the molecular weight of the truncated NCX1 was recognized in rabbit heart by monoclonal antibodies C2C12 (epitope between amino acids 372 and 525 of NCX1) while not recognized by 6H2 antibody (epitope in the extracellular NH2-terminal 40 amino acids). No corresponding band was recognized in LLC-MK2 cells by these two antibodies. In fact, whether the truncated NCX1 could be translated from circNCX1 was not directly demonstrated in this research. It was actually due to technical and cognitive limitation on circRNAs at 20 years ago. What’s more, an antibody raised from truncated NCX1 should be applied to identify the existence of endogenous truncated NCX1. In recent years, technologies involving in transcriptomics, epigenomics, and proteomics are rapidly developing. The circRNAs with coding potency are characterized one after another [16,17,18,19,20,21,22,23]. The 5′ cap-independent translation mechanism in circRNAs had also been revealed. Besides ORF, circRNAs need special elements located in the UTR such as internal ribosomal entry sites (IRES) or N-6-methyladenosine (m6A), to recruit translation initiation factors [82, 83]. Significantly, circNCX1 was detected by ribosome sequencing of RNAs in human or mouse heart, indicating the association of circNCX1 and ribosome [43, 84]. However, the peptide fragments of putative truncated NCX1 were not identified in human heart by mass spectrum analysis [84]. We searched the human-truncated NCX1-specific peptide fragment “GGGEDFEDTCGELEFQNDEIVR” in published human proteomics data and did not find the matching candidate either [85, 86]. Murine circNCX1 sequences are lacking stop codon to form full ORFs. Lim et al. had further used a mouse circNCX1 expression vector containing 3 × HA tag and artificially C-terminal inserted stop codon TAA to definitely demonstrate that mouse circNCX1 could not be translated [40]. Maybe murine circNCX1 was just a non-coding circRNA. But the translation potency of human circNCX1 cannot be completely judged yet. Comparing with the detected fragment “GGGEDFEDTCGELEFQNDEIVK” in full-length human NCX1, only the last alkaline amino acid residue is unmatched. Additionally, the efficiency of cap-independent translation is much lower than the canonical translation [16]. Therefore, it was also possible that the high sequence coverage rate of human-truncated NCX1 with full-length NCX1 and relative weak signal strength made it hard to detect by mass spectrum. By means of Transcirc database, it can be observed that abundant IRES and m6A sites are spread in the sequence of human circNCX1, suggesting relative high translation possibility [87]. Genetic editing in vivo and in vitro cooperated with specific immunologic analysis might be helpful to identify the potential protein product of circNCX1. Moreover, if truncated NCX1 could be translated from human circNCX1, there should be some difference between the biological function of human circNCX1 and murine circNCX1.
Therapeutic Potency of circNCX1
Currently, it has been recognized that excessive cardiac circNCX1 is closely associated with ischemic cardiac injury and stress-induced cardiomyopathies. Decreasing of circNCX1 can be considered as a strategy of heart disease treatment because of highly conserved sequence and expression profile of circNCX1. Comparing to supplementing the protective circRNAs, inhibiting the pathogenic circRNAs is more feasible clinically. The main reason is that circRNAs can be targeted by small interfering RNA (siRNA) specifically and the technologies of siRNA synthesis, modification and delivery are developing rapidly [88, 89].
Indeed, shRNA carrying cardiac-specific circNCX1 silencing did not cause any obvious defects, at least in adult mice [39, 40]. It indicated that circNCX1-targeted nucleotide drugs is feasible and harmless enough to be brought into clinical experiments. Nevertheless, the potential extra-cardiac effect of circNCX1 should also be considered in case decreasing of circNCX1 affected other biological and pathological processes (Table 3). In addition to cardiac development and disease, circNCX1 was also suggested to be involved in Parkinson’s disease (PD). The substantia nigra (SN) is the brain region whose lesion is strongly associated with PD. Hanan et al. observed that SN samples of PD patients had increased circNCX1 expression level compared to healthy samples. To further mimic PD-related oxidative stress in vitro, oxidative agent Paraquat was used to treat human SH-SY neuronal cells. The expression of circNCX1 was increased by Paraquat treatment, while the protective agents Simvastatin and PF-06447475 could reduce circNCX1 level in neuronal cells. Although the exact function of circNCX1 in neuronal cells or in PD was not verified, a potential target of circNCX1- miR-128 suggested that circNCX1 might be correlated with neurodegeneration and neuronal aging. All the miR-128-targeted neurodegeneration and neuronal aging-related genes were co-upregulated with circNCX1 in PD in vivo and under oxidative stress in vitro [90]. According to these results, silencing of circNCX1 seemed not to be harmful to the brain. However, the downregulation of circNCX1 was associated with the process of bladder cancer. Decreased circNCX1 level was detected in both of bladder tumor and bladder cancer cell lines. The overexpression of circNCX1 could inhibit the migration, invasion, and proliferation of bladder cancer by regulating the miR-130b/miR-494-PTEN axis [91]. It implied that bladder cancer patients should avoid using the RNAi drugs of circNCX1 to treat cardiomyopathy. Moreover, in view of circNCX1 might involve in embryonic cardiac development, therapy of circNCX1-silencing was not suitable for pregnant woman. On the other hand, the RNAi drugs should be safely and specifically delivered to the heart. Adeno-associated virus (AAV) is a type of ideal delivery vehicle. However, a recently published long-term study suggested that AAV had potential genotoxicity and oncogenicity [92]. The appropriate delivery material of RNAi of circNCX1 needs to be cautiously selected.
Covalent closed structure makes circRNAs tolerable from nuclease digestion and relatively stable, even sensitively detectable in body fluid. Aberrant cells usually showed dysregulation of certain circRNAs. The level of circRNAs released into extracellular fluid from the cells was also altered with it. Therefore, circRNAs are regarded as new types of biomarker in the diagnosis of diseases such as cancers, diabetes, and neuropathy. The level of some specific circRNA in patients’ peripheral blood was significantly changed [93,94,95]. circRNAs could also be applied to the diagnosis of cardiac injury. Under ischemia, toxicity, or myocardial remodeling, the processes of expression and transportation of circRNAs would both be affected, resulting in the alteration of circRNA amount in the peripheral blood. For example, a circular RNA of znf-609 gene named myocardial infarction-associated circular RNA (MICRA) was referred as a biomarker of acute myocardial infarction (MI) and predictor of left ventricular dysfunction after acute MI. The level of MICRA in blood samples of MI patients was decreased comparing with healthy people. MI patients with low level of MICRA in the blood showed more serious left ventricular dysfunction [96]. circNCX1 was not only demonstrated to aggravate ischemic myocardial injury in murine, but also detected to be upregulated in human ventricle samples of MI patients [38, 81], indicating its close relationship with MI. This cardiac-enriched circRNA was also reported to be increased in the peripheral blood of MI patients. Li et al. observed that circNCX1 level in MI samples was approximately twice as that in healthy samples. Receiver operating characteristic (ROC) curve analysis revealed that the area under the curve (AUC) value reached 0.8 [97]. These results suggested that blood circNCX1 level could be referred in MI diagnosis.
Conclusions and Research Perspective
CircNCX1 is the most abundant and relatively conserved circRNAs in vertebrate hearts, implying its evolutional indispensability. The cardiac enrichment of circNCX1 is attributed to both transcriptional and post-transcriptional regulations. Transcriptionally, circNCX1 is regulated by super-enhancer. Histone acetylation and Nkx-2.5 affect the transcription of circNCX1 pre-mRNA. The extremely long flanking introns of circNCX1 harbor numerous binding motifs of multiple RBPs which mediate RNA splicing in the heart. Post-transcriptionally, these RBPs might be involved in back-splicing of circNCX1. circNCX1 is demonstrated as a miR-133a sponge. The development of heart disease including ischemic cardiac injury, HCM, and DCM is closely associated with circNCX1 dysregulation. Under pathological conditions such as oxidative stress and pressure overload, the expression of circNCX1 is induced by HIF1a or HDAC in cardiomyocytes and subsequently upregulate miR-133a-targeted stress-related or cell death-related genes. circNCX1 is potentially a key regulator of cardiac development. Probably, it also modulates the differentiation and maturation of myocardium by miR-133a-dependent way. In some species, although circNCX1 has the most characters of protein-coding circRNA, circNCX1 is very likely a non-coding RNA. It could be considered to apply AAV with siRNA of circNCX1 in heart disease treatment. It might not be suitable for cancer patients and pregnant women.
Other miRNAs were also reported to interact with circNCX1 in extra-cardiac tissues [90, 91, 98,99,100]. Some of these miRNAs had been proven to involve in cardiomyopathies. For instance, miR-21, miR-128, and miR-494 could protect the myocardium from ischemic injury [101,102,103,104,105]. miR-21 and miR-128 also had inhibitory effect on cardiac hypertrophy [106]. However, they did not highly enrich in circNCX1 pull-down assay in cardiomyocytes [40]. It cannot be estimated whether the pro-apoptosis and pro-hypertrophy activities of circNCX1 acted through these miRNAs. Perhaps, forced expression of circNCX1 with disrupted miRNA binding sites was necessary to solve this problem. What’s more, the RBP sponge potency of circNCX1 remains to be explored. Besides the self-restriction mentioned above, binding with RBPs was also likely to affect the expression of other genes [107, 108], even circRNAs. It needs further investigation by some omics analysis.
Change history
22 November 2021
A Correction to this paper has been published: https://doi.org/10.1007/s12265-021-10183-z
Abbreviations
- Ago2:
-
Argonaute 2
- CELF1:
-
CUGBP Elav-like family member 1
- ChIP-seq:
-
Chromatin immunoprecipitation and sequencing
- circRNAs:
-
Circular RNAs
- HIF1α:
-
Hypoxia-inducible factor 1α
- HDAC:
-
Histone deacetylase
- hnRNAs:
-
Heterogeneous nuclear RNAs
- hESCs:
-
Human embryonic stem cells
- iPSC:
-
Induced pluripotent stem cells
- I/R:
-
Myocardial ischemia–reperfusion
- MI:
-
Myocardial infarction
- MBNL1:
-
Muscleblind-like protein 1
- ncx1:
-
Sodium/calcium exchanger 1
- ORF:
-
Open reading frame
- PTBP1:
-
Polypyrimidine tract-binding protein 1
- PD:
-
Parkinson’s disease
- RBP:
-
RNA-binding protein
- slc8a1:
-
Solute carrier family 8 member A1
- SEs:
-
Super enhancers
- SINEs:
-
Short interspersed repeat segments
- SRSF:
-
Serine-/arginine-rich splicing factor
References
Akazawa, H., & Komuro, I. (2003). Roles of cardiac transcription factors in cardiac hypertrophy. Circulation Research, 92, 1079–1088. https://doi.org/10.1161/01.RES.0000072977.86706.23.
Hannan, R., Jenkins, A., Jenkins, A., & Brandenburger, Y. (2003). Cardiac hypertrophy: A matter of translation. Clinical and Experimental Pharmacology and Physiology, 30, 517–527. https://doi.org/10.1046/j.1440-1681.2003.03873.x
Preissl, S., Schwaderer, M., Raulf, A., Hesse, M., Gruning, B. A., Kobele, C., et al. (2015). Deciphering the epigenetic code of cardiac myocyte transcription. Circulation Research, 117, 413–423. https://doi.org/10.1161/CIRCRESAHA.115.306337.
Papait, R., Serio, S., Pagiatakis, C., Rusconi, F., Carullo, P., Mazzola, M., Salvarani, N., Miragoli, M., & Condorelli, G. (2017). Histone methyltransferase G9a is required for cardiomyocyte homeostasis and hypertrophy. Circulation, 136, 1233–1246. https://doi.org/10.1161/circulationaha.117.028561
Poller, W., Dimmeler, S., Heymans, S., Zeller, T., Haas, J., Karakas, M., Leistner, D. M., Jakob, P., Nakagawa, S., Blankenberg, S., Engelhardt, S., Thum, T., Weber, C., Meder, B., Hajjar, R., & Landmesser, U. (2018). Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. European Heart Journal, 39, 2704–2716. https://doi.org/10.1093/eurheartj/ehx165
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., Marzluff, W. F., & Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19, 141–157. https://doi.org/10.1261/rna.035667.112
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., Maier, L., Mackowiak, S. D., Gregersen, L. H., Munschauer, M., Loewer, A., Ziebold, U., Landthaler, M., Kocks, C., le Noble, F., & Rajewsky, N. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495, 333–338. https://doi.org/10.1038/nature11928
Chen, L. L. (2016). The biogenesis and emerging roles of circular RNAs. Nature Reviews Molecular Cell Biology, 17, 205–211. https://doi.org/10.1038/nrm.2015.32
Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., & Mayeda, A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Research, 34, e63. https://doi.org/10.1093/nar/gkl151.
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nature Communications, 7, 12429. https://doi.org/10.1038/ncomms12429.
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., & Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research, 44, 2846–2858. https://doi.org/10.1093/nar/gkw027
Du, W., Fang, L., Yang, W., Wu, N., Awan, F., Yang, Z., & Yang, B. B. (2017). Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death and Differentiation, 24, 357–370. https://doi.org/10.1038/cdd.2016.133
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., & Kjems, J. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. https://doi.org/10.1038/nature11993
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., Zhu, S., Yang, L., & Chen, L. L. (2013). Circular intronic long noncoding RNAs. Molecular Cell, 51, 792–806. https://doi.org/10.1016/j.molcel.2013.08.017
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., Zhong, G., Yu, B., Hu, W., Dai, L., Zhu, P., Chang, Z., Wu, Q., Zhao, Y., Jia, Y., Xu, P., Liu, H., & Shan, G. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structure and Molecular Biology, 22, 256–264. https://doi.org/10.1038/nsmb.2959
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., Hanan, M., Wyler, E., Perez-Hernandez, D., Ramberger, E., Shenzis, S., Samson, M., Dittmar, G., Landthaler, M., Chekulaeva, M., & Rajewsky N,&Kadener S, . (2017). Translation of CircRNAs. Molecular Cell, 66(9–21), e27. https://doi.org/10.1016/j.molcel.2017.02.021
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., Fatica, A., Santini, T., Andronache, A., Wade, M., Laneve, P., Rajewsky, N., & Bozzoni, I. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular Cell, 66(22–37), e29. https://doi.org/10.1016/j.molcel.2017.02.017
Liang, W., Wong, C., Liang, P., Shi, M., Cao, Y., Rao, S., Tsui, S., Waye, M., Zhang, Q., Fu, W., & Zhang, J. (2019). Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biology, 20, 84. https://doi.org/10.1186/s13059-019-1685-4
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, & Zhang N (2018) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. Journal of the National Cancer Institute 110
Zhang, M., Zhao, K., Xu, X., Yang, Y., Yan, S., Wei, P., Liu, H., Xu, J., Xiao, F., Zhou, H., Yang, X., Huang, N., Liu, J., He, K., Xie, K., Zhang, G., Huang, S., & Zhang, N. (2018). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature Communications, 9, 4475. https://doi.org/10.1038/s41467-018-06862-2
Xia, X., Li, X., Li, F., Wu, X., Zhang, M., Zhou, H., Huang, N., Yang, X., Xiao, F., Liu, D., Yang, L., & Zhang, N. (2019). A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Molecular Cancer, 18, 131. https://doi.org/10.1186/s12943-019-1056-5
Zhang, M., Huang, N., Yang, X., Luo, J., Yan, S., Xiao, F., Chen, W., Gao, X., Zhao, K., Zhou, H., Li, Z., Ming, L., Xie, B., & Zhang, N. (2018). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene, 37, 1805–1814. https://doi.org/10.1038/s41388-017-0019-9
Li, J., Ma, M., Yang, X., Zhang, M., Luo, J., Zhou, H., Huang, N., Xiao, F., Lai, B., Lv, W., & Zhang, N. (2020). Circular HER2 RNA positive triple negative breast cancer is sensitive to pertuzumab. Molecular Cancer, 19, 142. https://doi.org/10.1186/s12943-020-01259-6
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., & Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE, 7, e30733. https://doi.org/10.1371/journal.pone.0030733
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L., & Brown, P. O. (2013). Cell-type specific features of circular RNA expression. PLoS Genetics, 9, e1003777. https://doi.org/10.1371/journal.pgen.1003777
Werfel, S., Nothjunge, S., Schwarzmayr, T., Strom, T. M., Meitinger, T., & Engelhardt, S. (2016). Characterization of circular RNAs in human, mouse and rat hearts. Journal of Molecular and Cellular Cardiology, 98, 103–107. https://doi.org/10.1016/j.yjmcc.2016.07.007
Tan, W. L., Lim, B. T., Anene-Nzelu, C. G., Ackers-Johnson, M., Dashi, A., See, K., Tiang, Z., Lee, D. P., Chua, W. W., Luu, T. D., Li, P. Y., Richards, A. M., & Foo, R. S. (2017). A landscape of circular RNA expression in the human heart. Cardiovascular Research, 113, 298–309. https://doi.org/10.1093/cvr/cvw250
Wang, K., Long, B., Liu, F., Wang, J. X., Liu, C. Y., Zhao, B., Zhou, L. Y., Sun, T., Wang, M., Yu, T., Gong, Y., Liu, J., Dong, Y. H., Li, N., & Li, P. F. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal, 37, 2602–2611. https://doi.org/10.1093/eurheartj/ehv713
Zhou, L. Y., Zhai, M., Huang, Y., Xu, S., An, T., Wang, Y. H., Zhang, R. C., Liu, C. Y., Dong, Y. H., Wang, M., Qian, L. L., Ponnusamy, M., Zhang, Y. H., Zhang, J., & Wang, K. (2019). The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell death and differentiation, 26, 1299–1315. https://doi.org/10.1038/s41418-018-0206-4
Du, W. W., Yang, W., Chen, Y., Wu, Z. K., Foster, F. S., Yang, Z., Li, X., & Yang, B. B. (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European Heart Journal, 38, 1402–1412. https://doi.org/10.1093/eurheartj/ehw001
Rybak-Wolf, A., Stottmeister, C., Glazar, P., Jens, M., Pino, N., Giusti, S., Hanan, M., Behm, M., Bartok, O., Ashwal-Fluss, R., Herzog, M., Schreyer, L., Papavasileiou, P., Ivanov, A., Ohman, M., Refojo, D., Kadener, S., & Rajewsky, N. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell, 58, 870–885. https://doi.org/10.1016/j.molcel.2015.03.027
Maass, P. G., Glazar, P., Memczak, S., Dittmar, G., Hollfinger, I., Schreyer, L., Sauer, A. V., Toka, O., Aiuti, A., Luft, F. C., & Rajewsky, N. (2017). A map of human circular RNAs in clinically relevant tissues. Journal of Molecular Medicine (Berlin), 95, 1179–1189. https://doi.org/10.1007/s00109-017-1582-9
Thomson DW,&Dinger ME, . (2016). Endogenous microRNA sponges: Evidence and controversy. Nature Reviews Genetics, 17, 272–283. https://doi.org/10.1038/nrg.2016.20
Li XF,&Lytton J, . (1999). A circularized sodium-calcium exchanger exon 2 transcript. Journal of Biological Chemistry, 274, 8153–8160. https://doi.org/10.1074/jbc.274.12.8153
Ouyang, H., Chen, X., Wang, Z., Yu, J., Jia, X., Li, Z., Luo, W., Abdalla, B. A., Jebessa, E., Nie, Q., & Zhang, X. (2018). Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Research, 25, 71–86. https://doi.org/10.1093/dnares/dsx039
Sai, L., Li, L., Hu, C., Qu, B., Guo, Q., Jia, Q., Zhang, Y., Bo, C., Li, X., Shao, H., Ng, J. C., & Peng, C. (2018). Identification of circular RNAs and their alterations involved in developing male Xenopus laevis chronically exposed to atrazine. Chemosphere, 200, 295–301. https://doi.org/10.1016/j.chemosphere.2018.02.140
Sharma, D., Sehgal, P., Mathew, S., Vellarikkal, S. K., Singh, A. R., Kapoor, S., Jayarajan, R., & ScariaSivasubbu, V. S. (2019). A genome-wide map of circular RNAs in adult zebrafish. Scientific Reports, 9, 3432. https://doi.org/10.1038/s41598-019-39977-7
Jakobi T, Siede D, Eschenbach J, Heumuller AW, Busch M, Nietsch R, Meder B, Most P, Dimmeler S, Backs J, Katus HA, & Dieterich C (2020) Deep characterization of circular RNAs from human cardiovascular cell models and cardiac tissue. Cells 9https://doi.org/10.3390/cells9071616
Li, M., Ding, W., Tariq, M. A., Chang, W., Zhang, X., Xu, W., Hou, L., Wang, Y., & Wang, J. (2018). A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics, 8, 5855–5869. https://doi.org/10.7150/thno.27285
Lim, T. B., Aliwarga, E., Luu, T. D. A., Li, Y. P., Ng, S. L., Annadoray, L., Sian, S., Ackers-Johnson, M. A., & Foo, R. S. (2019). Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovascular Research, 115, 1998–2007. https://doi.org/10.1093/cvr/cvz130
Szabo, L., Morey, R., Palpant, N. J., Wang, P. L., Afari, N., Jiang, C., Parast, M. M., Murry, C. E., Laurent, L. C., & Salzman, J. (2015). Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biology, 16, 126. https://doi.org/10.1186/s13059-015-0690-5
Lei, W., Feng, T., Fang, X., Yu, Y., Yang, J., Zhao, Z. A., Liu, J., Shen, Z., Deng, W., & Hu, S. (2018). Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Research and Therapy, 9, 56. https://doi.org/10.1186/s13287-018-0793-5
Siede, D., Rapti, K., Gorska, A. A., Katus, H. A., Altmuller, J., Boeckel, J. N., Meder, B., Maack, C., Volkers, M., Muller, O. J., Backs, J., & Dieterich, C. (2017). Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. Journal of Molecular and Cellular Cardiology, 109, 48–56. https://doi.org/10.1016/j.yjmcc.2017.06.015
Humphreys, D. T., Fossat, N., Demuth, M., Tam, P. P. L., & Ho, J. W. K. (2019). Ularcirc: Visualization and enhanced analysis of circular RNAs via back and canonical forward splicing. Nucleic Acids Research, 47, e123. https://doi.org/10.1093/nar/gkz718
Dong, H., Dunn, J., & Lytton, J. (2002). Electrophysiological studies of the cloned rat cardiac NCX1.1 in transfected HEK cells: A focus on the stoichiometry. Annals of the New York Academy of Sciences, 976, 159–165. https://doi.org/10.1111/j.1749-6632.2002.tb04737.x
Wakimoto, K., Kobayashi, K., Kuro, O. M., Yao, A., Iwamoto, T., Yanaka, N., Kita, S., Nishida, A., Azuma, S., Toyoda, Y., Omori, K., Imahie, H., Oka, T., Kudoh, S., Kohmoto, O., Yazaki, Y., Shigekawa, M., Imai, Y., Nabeshima, Y., & Komuro, I. (2000). Targeted disruption of Na+/Ca2+ exchanger gene leads to cardiomyocyte apoptosis and defects in heartbeat. Journal of Biological Chemistry, 275, 36991–36998. https://doi.org/10.1074/jbc.M004035200
Komuro, I., Wenninger, K. E., Philipson, K. D., & Izumo, S. (1992). Molecular cloning and characterization of the human cardiac Na+/Ca2+ exchanger cDNA. Proc Natl Acad Sci U S A, 89, 4769–4773. https://doi.org/10.1073/pnas.89.10.4769
Kofuji, P., Hadley, R. W., Kieval, R. S., Lederer, W. J., & Schulze, D. H. (1992). Expression of the Na-Ca exchanger in diverse tissues: A study using the cloned human cardiac Na-Ca exchanger. American Journal of Physiology, 263, C1241-1249. https://doi.org/10.1152/ajpcell.1992.263.6.C1241
Fagerberg, L., Hallstrom, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., Habuka, M., Tahmasebpoor, S., Danielsson, A., Edlund, K., Asplund, A., Sjostedt, E., Lundberg, E., Szigyarto, C. A., Skogs, M., Takanen, J. O., Berling, H., Tegel, H., Mulder, J., … Uhlen, M. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular and Cellular Proteomics, 13, 397–406. https://doi.org/10.1074/mcp.M113.035600
Geng, H. H., Li, R., Su, Y. M., Xiao, J., Pan, M., Cai, X. X., & Ji, X. P. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE, 11, e0151753. https://doi.org/10.1371/journal.pone.0151753
Spitz, F., & Furlong, E. E. (2012). Transcription factors: From enhancer binding to developmental control. Nature Reviews Genetics, 13, 613–626. https://doi.org/10.1038/nrg3207
Bulger, M., & Groudine, M. (2011). Functional and mechanistic diversity of distal transcription enhancers. Cell, 144, 327–339. https://doi.org/10.1016/j.cell.2011.01.024
Pott, S., & Lieb, J. D. (2015). What are super-enhancers? Nature genetics, 47, 8–12. https://doi.org/10.1038/ng.3167
Lee, Y., Shioi, T., Kasahara, H., Jobe, S. M., Wiese, R. J., Markham, B. E., & Izumo, S. (1998). The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Molecular and cellular biology, 18, 3120–3129. https://doi.org/10.1128/mcb.18.6.3120
Chandrasekaran, S., Peterson, R. E., Mani, S. K., Addy, B., Buchholz, A. L., Xu, L., Thiyagarajan, T., Kasiganesan, H., Kern, C. B., & Menick, D. R. (2009). Histone deacetylases facilitate sodium/calcium exchanger up-regulation in adult cardiomyocytes. FASEB journal : Official publication of the Federation of American Societies for Experimental Biology, 23, 3851–3864. https://doi.org/10.1096/fj.09-132415
Harris, L. G., Wang, S. H., Mani, S. K., Kasiganesan, H., Chou, C. J., & Menick, D. R. (2016). Evidence for a non-canonical role of HDAC5 in regulation of the cardiac Ncx1 and Bnp genes. Nucleic acids research, 44, 3610–3617. https://doi.org/10.1093/nar/gkv1496
Hnisz, D., Abraham, B. J., Lee, T. I., Lau, A., Saint-Andre, V., Sigova, A. A., Hoke, H. A., & Young, R. A. (2013). Super-enhancers in the control of cell identity and disease. Cell, 155, 934–947. https://doi.org/10.1016/j.cell.2013.09.053
Khan, A., & Zhang, X. (2016). dbSUPER: A database of super-enhancers in mouse and human genome. Nucleic acids research, 44, D164-171. https://doi.org/10.1093/nar/gkv1002
Huang, S., Li, X., Zheng, H., Si, X., Li, B., Wei, G., Li, C., Chen, Y., Chen, Y., Liao, W., & Liao Y,&Bin J, . (2019). Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation, 139, 2857–2876. https://doi.org/10.1161/CIRCULATIONAHA.118.038361
Zhang, X., Wang, H., Zhang, Y., Lu, X., Chen, L., & Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell, 159, 134–147. https://doi.org/10.1016/j.cell.2014.09.001
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., Roslan, S., Schreiber, A. W., Gregory, P. A., & Goodall, G. J. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell, 160, 1125–1134. https://doi.org/10.1016/j.cell.2015.02.014
Khan, M. A., Reckman, Y. J., Aufiero, S., van den Hoogenhof, M. M., van der Made, I., Beqqali, A., Koolbergen, D. R., Rasmussen, T. B., van der Velden, J., Creemers, E. E., & Pinto, Y. M. (2016). RBM20 regulates circular RNA production from the titin gene. Circulation research, 119, 996–1003. https://doi.org/10.1161/CIRCRESAHA.116.309568
Paz, I., Kosti, I., Ares, M., Jr., Cline, M., & Mandel-Gutfreund, Y. (2014). RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic acids research, 42, W361-367. https://doi.org/10.1093/nar/gku406
Kafasla, P., Lin, H., Curry, S., & Jackson, R. J. (2011). Activation of picornaviral IRESs by PTB shows differential dependence on each PTB RNA-binding domain. RNA, 17, 1120–1131. https://doi.org/10.1261/rna.2549411
Li, H., Shen, S., Ruan, X., Liu, X., Zheng, J., Liu, Y., Yang, C., Wang, D., Liu, L., Ma, J., Ma, T., Wang, P., Cai, H., Li, Z., Zhao, L., & Xue, Y. (2019). Biosynthetic CircRNA_001160 induced by PTBP1 regulates the permeability of BTB via the CircRNA_001160/miR-195-5p/ETV1 axis. Cell Death & Disease, 10, 960. https://doi.org/10.1038/s41419-019-2191-z
Warf, M. B., Diegel, J. V., von Hippel, P. H., & Berglund, J. A. (2009). The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci U S A, 106, 9203–9208. https://doi.org/10.1073/pnas.0900342106
Kalsotra, A., Xiao, X., Ward, A. J., Castle, J. C., Johnson, J. M., Burge, C. B., & Cooper, T. A. (2008). A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A, 105, 20333–20338. https://doi.org/10.1073/pnas.0809045105
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., Evantal, N., Memczak, S., Rajewsky, N., & Kadener, S. (2014). circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell, 56, 55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S., & Wilusz, J. E. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes & Development, 29, 2168–2182. https://doi.org/10.1101/gad.270421.115
Ammar S. Naqvi, Mukta Asnani, Kathryn L. Black, Katharina E. Hayer, Deanne Taylor, Andrei Thomas-Tikhonenko. The role of SRSF3 splicing factor in generating circular RNAs. 2019. BioRxiv.
Li, M., Ding, W., Sun, T., Tariq, M. A., Xu, T., Li, P., & Wang, J. (2018). Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. The FEBS journal, 285, 220–232. https://doi.org/10.1111/febs.14191
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., Conlon, F. L., & Wang, D. Z. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature genetics, 38, 228–233. https://doi.org/10.1038/ng1725
Liu, N., Bezprozvannaya, S., Williams, A. H., Qi, X., Richardson, J. A., Bassel-Duby, R., & Olson, E. N. (2008). microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes & Development, 22, 3242–3254. https://doi.org/10.1101/gad.1738708
Izarra, A., Moscoso, I., Canon, S., Carreiro, C., Fondevila, D., Martin-Caballero, J., Blanca, V., Valiente, I., Diez-Juan, A., & Bernad, A. (2017). miRNA-1 and miRNA-133a are involved in early commitment of pluripotent stem cells and demonstrate antagonistic roles in the regulation of cardiac differentiation. Journal of Tissue Engineering and Regenerative Medicine, 11, 787–799. https://doi.org/10.1002/term.1977
Meder, B., Katus, H. A., & Rottbauer, W. (2008). Right into the heart of microRNA-133a. Genes & Development, 22, 3227–3231. https://doi.org/10.1101/gad.1753508
Care, A., Catalucci, D., Felicetti, F., Bonci, D., Addario, A., Gallo, P., Bang, M. L., Segnalini, P., Gu, Y., Dalton, N. D., Elia, L., Latronico, M. V., Hoydal, M., Autore, C., Russo, M. A., Dorn, G. W., 2nd., Ellingsen, O., Ruiz-Lozano, P., Peterson, K. L., … Condorelli, G. (2007). MicroRNA-133 controls cardiac hypertrophy. Nature Medicine, 13, 613–618. https://doi.org/10.1038/nm1582
Valsecchi, V., Pignataro, G., Del Prete, A., Sirabella, R., Matrone, C., Boscia, F., Scorziello, A., Sisalli, M. J., Esposito, E., Zambrano, N., Di Renzo, G., & Annunziato, L. (2011). NCX1 is a novel target gene for hypoxia-inducible factor-1 in ischemic brain preconditioning. Stroke, 42, 754–763. https://doi.org/10.1161/strokeaha.110.597583
Formisano, L., Guida, N., Valsecchi, V., Cantile, M., Cuomo, O., Vinciguerra, A., Laudati, G., Pignataro, G., Sirabella, R., Di Renzo, G., & Annunziato, L. (2015). Sp3/REST/HDAC1/HDAC2 complex represses and Sp1/HIF-1/p300 complex activates ncx1 gene transcription, in brain ischemia and in ischemic brain preconditioning, by epigenetic mechanism. Journal of Neuroscience, 35, 7332–7348. https://doi.org/10.1523/JNEUROSCI.2174-14.2015
Kent, R. L., Rozich, J. D., McCollam, P. L., McDermott, D. E., Thacker, U. F., Menick, D. R., McDermott, P. J., & Gt, C. (1993). Rapid expression of the Na(+)-Ca2+ exchanger in response to cardiac pressure overload. American Journal of Physiology, 265, H1024-1029. https://doi.org/10.1152/ajpheart.1993.265.3.H1024
Muller, J. G., Isomatsu, Y., Koushik, S. V., O’Quinn, M., Xu, L., Kappler, C. S., Hapke, E., Zile, M. R., Conway, S. J., & Menick, D. R. (2002). Cardiac-specific expression and hypertrophic upregulation of the feline Na(+)-Ca(2+) exchanger gene H1-promoter in a transgenic mouse model. Circulation research, 90, 158–164. https://doi.org/10.1161/hh0202.103231
Tian, M., Xue, J., Dai, C., Jiang, E., Zhu, B., & Pang, H. (2021). CircSLC8A1 and circNFIX can be used as auxiliary diagnostic markers for sudden cardiac death caused by acute ischemic heart disease. Science and Reports, 11, 4695. https://doi.org/10.1038/s41598-021-84056-5
Chen, X., Han, P., Zhou, T., Guo, X., Song, X., & Li, Y. (2016). circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Science and Reports, 6, 34985. https://doi.org/10.1038/srep34985
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., Jin, Y., Yang, Y., Chen, L. L., Wang, Y., Wong, C. C., Xiao, X., & Wang, Z. (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Research, 27, 626–641. https://doi.org/10.1038/cr.2017.31
van Heesch, S., Witte, F., Schneider-Lunitz, V., Schulz, J. F., Adami, E., Faber, A. B., Kirchner, M., Maatz, H., Blachut, S., Sandmann, C. L., Kanda, M., Worth, C. L., Schafer, S., Calviello, L., Merriott, R., Patone, G., Hummel, O., Wyler, E., Obermayer, B., … Hubner, N. (2019). The translational landscape of the human heart. Cell, 178(242–260), e229. https://doi.org/10.1016/j.cell.2019.05.010
Craig, R., Cortens, J. P., & Beavis, R. C. (2004). Open source system for analyzing, validating, and storing protein identification data. Journal of Proteome Research, 3, 1234–1242. https://doi.org/10.1021/pr049882h
Kim, M. S., Pinto, S. M., Getnet, D., Nirujogi, R. S., Manda, S. S., Chaerkady, R., Madugundu, A. K., Kelkar, D. S., Isserlin, R., Jain, S., Thomas, J. K., Muthusamy, B., Leal-Rojas, P., Kumar, P., Sahasrabuddhe, N. A., Balakrishnan, L., Advani, J., George, B., Renuse, S., … Pandey, A. (2014). A draft map of the human proteome. Nature, 509, 575–581. https://doi.org/10.1038/nature13302
Huang, W., Ling, Y., Zhang, S., Xia, Q., Cao, R., Fan, X., Fang, Z., & Wang Z,&Zhang G, . (2021). TransCirc: An interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic acids research, 49, D236–D242. https://doi.org/10.1093/nar/gkaa823
Saw PE,&Song EW, . (2020). siRNA therapeutics: A clinical reality. Sci China Life Sci, 63, 485–500. https://doi.org/10.1007/s11427-018-9438-y
Kanasty, R., Dorkin, J. R., Vegas, A., & Anderson, D. (2013). Delivery materials for siRNA therapeutics. Nature Materials, 12, 967–977. https://doi.org/10.1038/nmat3765
Hanan, M., Simchovitz, A., Yayon, N., Vaknine, S., Cohen-Fultheim, R., Karmon, M., Madrer, N., Rohrlich, T. M., Maman, M., Bennett, E. R., Greenberg, D. S., Meshorer, E., Levanon, E. Y., Soreq, H., & Kadener, S. (2020). A Parkinson’s disease CircRNAs resource reveals a link between circSLC8A1 and oxidative stress. EMBO Molecular Medecine, 12, e13551. https://doi.org/10.15252/emmm.202013551
Lu, Q., Liu, T., Feng, H., Yang, R., Zhao, X., Chen, W., Jiang, B., Qin, H., Guo, X., Liu, M., Li, L., & Guo, H. (2019). Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Molecular Cancer, 18, 111. https://doi.org/10.1186/s12943-019-1040-0
Nguyen, G. N., Everett, J. K., Kafle, S., Roche, A. M., Raymond, H. E., Leiby, J., Wood, C., Assenmacher, C. A., Merricks, E. P., Long, C. T., Kazazian, H. H., Nichols, T. C., Bushman, F. D., & Sabatino, D. E. (2021). A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nature Biotechnology, 39, 47–55. https://doi.org/10.1038/s41587-020-0741-7
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., Chen, D., Gu, J., He, X., & Huang, S. (2015). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Research, 25, 981–984. https://doi.org/10.1038/cr.2015.82
Zhao, Z., Li, X., Jian, D., Hao, P., Rao, L., & Li, M. (2017). Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetologica, 54, 237–245. https://doi.org/10.1007/s00592-016-0943-0
Cui, X., Niu, W., Kong, L., He, M., Jiang, K., Chen, S., Zhong, A., Li, W., Lu, J., & Zhang, L. (2016). hsa_circRNA_103636: Potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomarkers in Medicine, 10, 943–952. https://doi.org/10.2217/bmm-2016-0130
Vausort, M., Salgado-Somoza, A., Zhang, L., Leszek, P., Scholz, M., Teren, A., Burkhardt, R., Thiery, J., Wagner, D. R., & Devaux, Y. (2016). Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. Journal of the American College of Cardiology, 68, 1247–1248. https://doi.org/10.1016/j.jacc.2016.06.040
Qi Li, Zhongjie Yu, Mengyang Li et al. The expression profiles and role of circular RNA in peripheral blood of myocardial infarction patients, 09 June 2020, Preprint (Version 1) available at Research Square.
Zhu, Q., Zhang, X., Zai, H. Y., Jiang, W., Zhang, K. J., He, Y. Q., & Hu, Y. (2021). circSLC8A1 sponges miR-671 to regulate breast cancer tumorigenesis via PTEN/PI3k/Akt pathway. Genomics, 113, 398–410. https://doi.org/10.1016/j.ygeno.2020.12.006
Lin, C., Zhong, W., Yan, W., Yang, J., Zheng, W., & Wu, Q. (2020). Circ-SLC8A1 regulates osteoporosis through blocking the inhibitory effect of miR-516b-5p on AKAP2 expression. The Journal of Gene Medicine, 22, e3263. https://doi.org/10.1002/jgm.3263
Wang, D., Yan, S., Wang, L., Li, Y., & Qiao, B. (2021). circSLC8A1 acts as a tumor suppressor in prostate cancer via sponging miR-21. BioMed Research International, 2021, 6614591. https://doi.org/10.1155/2021/6614591
Qiao, L., Hu, S., Liu, S., Zhang, H., Ma, H., Huang, K., Li, Z., Su, T., Vandergriff, A., Tang, J., Allen, T., Dinh, P. U., Cores, J., Yin, Q., Li, Y., & Cheng, K. (2019). microRNA-21-5p dysregulation in exosomes derived from heart failure patients impairs regenerative potential. The Journal of Clinical Investigation, 129, 2237–2250. https://doi.org/10.1172/JCI123135
Cheng, Y., Zhu, P., Yang, J., Liu, X., Dong, S., Wang, X., Chun, B., Zhuang, J., & Zhang, C. (2010). Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovascular research, 87, 431–439. https://doi.org/10.1093/cvr/cvq082
Wang, X., Zhang, X., Ren, X. P., Chen, J., Liu, H., Yang, J., Medvedovic, M., Hu, Z., & Fan, G. C. (2010). MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation, 122, 1308–1318. https://doi.org/10.1161/CIRCULATIONAHA.110.964684
Zhao, D., Shun, E., Ling, F., Liu, Q., Warsi, A., Wang, B., Zhou, Q., Zhu, C., Zheng, H., Liu, K., & Zheng, X. (2020). Plk2 regulated by miR-128 induces ischemia-reperfusion injury in cardiac cells. Molecular therapy. Nucleic acids, 19, 458–467. https://doi.org/10.1016/j.omtn.2019.11.029
Li, H., Zhang, X., Wang, F., Zhou, L., Yin, Z., Fan, J., Nie, X., Wang, P., Fu, X. D., Chen, C., & Wang, D. W. (2016). MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation, 134, 734–751. https://doi.org/10.1161/CIRCULATIONAHA.116.023926
Qi H, Ren J, Ba L, Song C, Zhang Q, Cao Y, Shi P, Fu B, Liu Y, & Sun HJMtNa (2020) MSTN attenuates cardiac hypertrophy through inhibition of excessive cardiac autophagy by blocking AMPK /mTOR and miR-128/PPARγ/NF-κB. 19:507–522. https://doi.org/10.1016/j.omtn.2019.12.003
Tsitsipatis, D., Grammatikakis, I., Driscoll, R. K., Yang, X., Abdelmohsen, K., Harris, S. C., Yang, J. H., Herman, A. B., Chang, M. W., Munk, R., Martindale, J. L., Mazan-Mamczarz, K., De, S., Lal, A., & Gorospe, M. (2021). AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic acids research, 49, 1631–1646. https://doi.org/10.1093/nar/gkaa1246
Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., Kim, J., Noh, J. H., Kim, K. M., Martindale, J. L., & Gorospe, M. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biology, 14, 361–369. https://doi.org/10.1080/15476286.2017.1279788
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81900259) and the Natural Science Foundation of Shandong Province (grant number JQ201815).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Research Involving Human Participants and/or Animals
No human or animal studies were carried out by the authors for this review article.
Conflict of Interest
The authors declare no competing interests.
Additional information
Communicated by Associate Editor Junjie Xiao oversaw the review of this article.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: A note has been added to indicate that Lin Ding and Mengyang Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ding, L., Li, M., Yang, F. et al. CircNCX1: the “Lord of the Ring” in the Heart — Insight into Its Sequence Characteristic, Expression, Molecular Mechanisms, and Clinical Application. J. of Cardiovasc. Trans. Res. 15, 571–586 (2022). https://doi.org/10.1007/s12265-021-10176-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-021-10176-y