Abstract
Oral squamous cell carcinoma (OSCC) has a dismal 50% five-year survival rate, emphasizing the need to develop reliable and sensitive tools for early diagnosis. In this study we evaluated the performance of 7 previously identified, potential mRNA biomarkers of OSCC in saliva samples of Hungarian patients. Expression of the putative OSCC biomarkers (DUSP1, OAZ1, H3F3A, IL1B, IL8, SAT and S100P), 2 biomarkers of inflammation (IL6 and TNFα) and 8 putative normalizing genes was quantified from each sample using real-time quantitative PCR. In contrast with previous studies, the expression pattern of the 7 mRNA biomarkers was similar between OSCC patients and age-matched control patients in the Hungarian patient population. On the other hand, 5 of the 7 mRNA biomarkers were present at significantly higher levels in saliva samples of OSCC patients when compared to young control patients. The best biomarker combination could distinguish only the OSCC vs. young control patients, but not the OSCC vs. age-matched control patients. In conclusion, the significant differences between our results and previous studies, and the clinical characteristics of the patients suggest that inflammatory processes in the oral cavity may affect the performance of the 7 putative salivary mRNA biomarkers. Lastly, since IL6 mRNA was quantifiable in the majority of OSCC cases, but only in a few control samples, salivary IL6 mRNA may be utilized as part of a biomarker combination to detect OSCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the past decade, saliva has emerged as a promising diagnostic material in oral squamous cell carcinoma (OSCC), and the number of studies investigating the diagnostic value of salivary proteins, nucleic acids or metabolites is increasing (recently reviewed by Guerra EN et al.) [1]. In contrast to pessimistic expectations that RNA molecules are quickly degraded by salivary ribonucleases, mRNAs and microRNAs proved to be readily detectable and potential analytes for salivary diagnostics. More than 3000 mRNA species were identified in the saliva of healthy individuals, and over 1600 mRNAs were expressed differently in the saliva of patients with OSCC and healthy controls [2, 3]. A persisting problem of the salivary diagnostics of OSCC is that the specificity and sensitivity of individual biomarkers are less than optimal. Thus far, the combination of proteomic and transcriptomic salivary biomarkers yielded the highest predictive value for OSCC, but the area under the curve (AUC) values of the receiver operating characteristic (ROC) curves for the multiple marker model still did not reach the ideal >0.90 level in the majority of different cohorts [4,5,6].
The Wong group has repeatedly confirmed the predictive value of seven RNA biomarkers, human H3 histone family 3A (H3F3A), interleukin-8 (IL-8), interleukin-1-beta (IL-1B), dual specificity phosphatase 1 (DUSP1), ornithine decarboxylase antizyme 1 (OAZ1), spermidine/spermine N1-acetyltransferase 1 (SAT1), and S100 calcium binding protein P (S100P) [5], but other groups identified different salivary mRNA species as most distinctive biomarkers for OSCC [7, 8]. It should be noted that the salivary gene expression profile is not just a passive mirror image of the tumor cells, as differentially expressed mRNAs exhibit only a partial overlap in OSCC tissue samples and saliva samples from OSCC patients [3, 4, 9,10,11,12]. Some of these mRNAs may be originated from the cancer cells, or they may be produced by tumor-infiltrating lymphocytes and macrophages, or even by immune cells from coexisting oral inflammatory lesions of the patients, which might explain some of the differences in the published studies. In this regard, the overlap between the biomarker profile of OSCC and periodontal inflammation has been recently reviewed [13]. Therefore, oral inflammatory status of patients, which show characteristic differences according to nationality and socio-economic situation, may influence the results of studies executed in different populations.
The heterogeneity and the suboptimal performance of salivary biomarkers in OSCC warrants further studies. The investigation of Hungarian OSCC patients is particularly important, since Hungary is characterized by very unfavorable epidemiologic data regarding OSCC. The average annual crude incidence and mortality rates are about 15.3/100,000 inhabitants and 7.2/100,000 inhabitants, respectively. The same US figures are approximately 8.3/100,000 inhabitants and 1.5/100,000 inhabitants, respectively [8, 14]. Even more disturbing is the fact that Hungary successfully reduced the mortality rates for several cancer types, but mortality from OSCC showed a fourfold elevation from the 1970-ies to the new millennium [15]. Among the causative factors we consider the smoking and ethanol consumption habits, and the poor oral hygiene of the Hungarian population [16,17,18]. In this pilot study our goal was to validate the applicability of salivary RNA-based diagnostics among Hungarian patients with OSCC, using the same putative mRNA biomarkers and the same approach as Wong et al. [2,3,4].
Materials and Methods
Patient Enrollment and Characterization
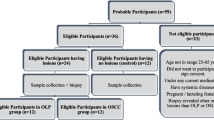
Within the frames of a case-control study, three independent patient groups were recruited: the cancer patients group included 31 consecutive patients with recently diagnosed and untreated OSCC, recruited from the Department of Oral Surgery (Faculty of Dentistry, University of Debrecen, Hungary), between 06.2011–05.2012. The two control groups included 30 elderly (age- and gender-matched) subjects, and 29 young healthy subjects consecutively admitted for dental check-up at the Department of Restorative Dentistry and Department of Periodontology (Faculty of Dentistry, University of Debrecen, Hungary) (recruited between 11.2012–02.2013, and 02.2014–04.2014, respectively). Exclusion criteria were: a prior history of cancer, coexisting diabetes, autoimmune disorder or contagious diseases. The study was approved by the Ethics Committee of the University of Debrecen (Approval number: 3244–8/2011), and was carried out in accordance with the Code of Ethics of the World Medical Association, and the ethical standards of the Helsinki Declaration (Revision 2000). The study participants were all invited, and all gave their written, informed consent. Detailed characteristics of OSCC patients’ and controls’ are summarized in Table 1. Socioeconomic status, previous and coexisting diseases, smoking habits and history, as well as alcohol consumption habits were reported in questionnaires filled in at the time of recruitment. Cumulative tobacco consumption was quantified as pack years. Pack years were calculated by multiplying the number of packs (defined as 20 manufactured cigarettes) of cigarettes smoked per day by the number of years the person had smoked. Calibrated oral surgeons and dentists carried out the clinical examinations of all study subjects patients according to the standard methods and criteria of the World Health Organization (World Health organization: Oral Health Surveys: Basic Methods. Geneva, WHO; 1997): for the recognition of the dental condition DMFT and DMFS indices were recorded. DMFT and DMFS scores provide a numerical value for caries prevalence and are obtained by calculating the number of decayed (D), missing (M) and filled (F) teeth (T; DMFT) or surfaces (S; DMFS). Periodontal status was characterized by measuring gingival and plaque indices (GI and PI). GI is a widely used measure of impaired (or undisturbed) periodontal health, indicating the severity and location of gingival lesions. PI characterizes the status of oral hygiene by measuring dental plaques located in areas bordering the gingival margin [19]. Plaque is a biofilm strongly associated with the etiology of periodontitis, dental caries and systemic microinflammation of odontogenic origin [20]. OSCC diagnosis was confirmed by the gold (reference) standard method uniformly in each case, i.e. histopathologic examination of the biopsy specimen. Briefly, formalin-fixed biopsy samples were paraffin embedded, then tissue sections were prepared and stained with hematoxylin and eosin according to standard methods. The histological differentiation grade was defined according to the classification of the World Health Organization (WHO) [21]. Staging was achieved according to the seventh edition of TNM classification of the International Union Against Cancer (UICC) [22]. The histopathologic results were interpreted by an experienced pathologist within 14 days of sampling of the biopsy material, i.e. well before conducting RNA extraction and analysis. Location, histological staging and grading of the OSCC tumors and patient survival are summarized in Table 2.
Saliva Collection and Processing
Unstimulated saliva samples were collected between 9 a.m. and 11 a.m. Patients were asked to refrain from eating, drinking, smoking, or using oral hygiene products for at least 1 h before sample collection. Saliva samples were kept on ice throughout the collection and processing - no more than 60 min elapsed from sample collection to freezing as RLT-lysate. Yeast tRNA and RNase inhibitor was added to the saliva samples before centrifugation: 5 μL RiboLock (Thermo Scientific, #ER00382), and 5 μg yeast tRNA (Sigma-Aldrich #R5636)/ 200 μL whole saliva sample. Samples were centrifuged at 4100 x g for 15 min at 4 °C. The supernatants were transferred to fresh tubes and 3X volume of RLT buffer (Qiagen, #74104) was added to them. RLT-lysed samples (from 600 μL whole saliva sample) were stored at −70 °C until RNA isolation. RNA was isolated from the RLT-lysed samples with the RNeasy Mini Kit (Qiagen, #74104) according to the manufacturer’s instructions, with on-column DNase I treatment, with the following modification: after elution, 0.5 μg yeast tRNA was added to the samples to prevent sample loss due to adsorption to plastic surfaces.
Reverse Transcription and Real-Time Quantitative PCR
5 μL of each saliva total RNA sample was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystem® Cat.No:#43,688,414) according to the manufacturer’s instructions, in a total volume of 20 μL. Expression of 7 previously identified, potential OSCC biomarkers (DUSP1, OAZ1, H3F3A, IL1B, IL8, SAT and S100P mRNAs), two biomarkers of inflammation (TNFα and IL6) and 8 potential normalizing genes (ANXA2, RPL37, B2M, GAPDH, ACTB, GNB2L1, HPRT1 and YWHAZ mRNAs) was quantified from each sample using real-time quantitative PCR (R-qPCR), with FAM-MGB labelled probes. Gene symbols and TaqMan® qPCR assay IDs are summarized in Supplementary Table 1. qPCR quantification of each mRNA was carried out in a 10 μL reaction mix containing 2 μL cDNA (diluted 4.5-fold from the RT reaction), 0.3 μM TaqMan® gene expression assay (Applied Biosystem®, final concentration), and 5 μL 2× TaqMan® Gene Expression Master Mix (Applied Biosystem®, #4369016). PCR cycle parameters were as follows: 10 min at 95 °C, then 40 or 50 cycles of 15 s at 95 °C and 60 s at 60 °C. Each gene was measured in duplicates. Samples from cancer patients and age-matched control patients were measured on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystem®). Samples from young control patients were measured on an QuantStudio™ 12 K Flex Real Time PCR System (Applied Biosystem®). Platform comparison (ABI PRISM 7900 HT vs. QuantStudio™ 12 K Flex) was carried out with samples of the OSCC and age-matched control group with IL8 – as expected, the measurements were in excellent correlation between the two platforms (data not shown). Raw data analysis was performed using the ABI PRISM SDS 2.1 software or ExpressionSuite Software v1.0.3 (Applied Biosystem®). The parameters (threshold and baseline settings) were the same in both analyses. PCR efficiency values were determined for each TaqMan® gene expression assay using the serial cDNA dilution method (data not shown). Cutoff Ct values (limit of quantitation) for each TaqMan® assay were also determined in these experiments, and were set at Ct = 38. Cts above this value were not used in the calculations. IL8 and IL1B were analyzed using total RNA isolated from immature dendritic cells, whereas the other assays were analysed using total RNA isolated from HeLa cells. For these analyses, 100 ng total RNA from each cell type was reverse transcribed and quantified with qPCR on a QuantStudio™ 12 K Flex Real Time PCR System as described above, with 4 replicates for each dilution.
Data Processing
To identify the best reference genes, raw Ct values were analyzed using the geNorm and NormFinder algorithms in the GenEx software (TATAA Biocenter), in addition to another ANOVA-based analysis [23]. Based on these analyses, the GAPDH/ACTB was identified as the best reference gene combination for our study. For PCR efficiency correction, raw Ct values of the cDNA serial dilutions were used to determine the slope, using the linear regression function of GraphPad Prism software (GraphPad Software, Inc., CA, USA). The PCR efficiency values were calculated with the following equation:
Ct values of each gene were corrected with efficiency values, and data was normalized using the following equations:
“Q” is the normalized expression of the gene of interest (GOI), “NF” is the normalizing factor calculated from the two reference genes, “ref1” and “ref2” are the reference genes used for normalizing (in the study, GAPDH and ACTB, respectively), whereas “E” is the PCR efficiency for each gene.
Statistics
Differential expression of each potential biomarker mRNA was assessed between the patient cohorts with GraphPad Prism, using the Mann-Whitney U-test on the efficiency-corrected, normalized expression values. Classification of the patients as “cancer” or “control” (based on the pathologist’s and the calibrated oral surgeons’ and dentists’ evaluation) was known to the investigators before data analysis. XLSTAT (Addinsoft) was used to construct receiver operating characteristic (ROC) curves, and to calculate threshold cutoff values and sensitivity/specificity values. Logistic regression analysis was also performed with XLSTAT, using the logit method and the best model/likelihood selection, with CI 95%, and stop conditions of 100 iterations/10−6 convergence.
Results
Normalizing Gene Selection
Since previous works on saliva mRNA biomarkers utilized different combinations of reference genes for normalizing [5, 24, 25], we analyzed the expression of 8 candidate reference genes (CRG) in our samples: ANXA2, RPL37, B2M, GAPDH, ACTB, GNB2L1, HPRT1 and YWHAZ. HPRT1 and YWHAZ mRNAs were not detectable in the saliva samples. To identify the best reference gene(s), the dataset was analyzed with three algorithms: geNorm [26], NormFinder [27] and the ANOVA-based analysis of Khanlou et al. [23]. All three analyses agreed in that B2M is the least stable CRG, but otherwise the results were different. GeNorm suggested the RPL37 and ANXA2 combination, NormFinder suggested the GAPDH and ACTB combination, whereas the method of Khanlou et al. suggested GAPDH and ANXA2 as the best reference genes (Supplementary Fig. 1) Both NormFinder and the ANOVA-based method of Khanlou et al. takes into consideration the inter-group, as well as the intra-group variability of CRGs; however, since ANXA2 (and RPL37) mRNA levels were below the quantification limit in close to 50% of the young control samples, we used the GAPDH/ACTB combination to calculate the normalizing factor in our study.
Relative Expression of the Putative mRNA Biomarkers between the Patient Cohorts
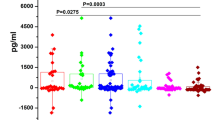
In contrast to previously published results [3, 5, 25], none of the 7 mRNAs were present in significantly higher quantities in the saliva samples of OSCC patients, when compared to samples of age-matched control patients (Fig. 1). In fact, the levels of 3 mRNAs (especially for DUSP1 and OAZ1) tended to be higher in the age-matched control group than in OSCC patients, although the high intra-group variability characteristic of this control group may have been partially responsible for the difference. Since the 7 mRNAs can be associated with pro-inflammatory and pro-proliferative pathways, and the elderly OSCC patients and age-matched control patients all suffered from periodontal disease (Table 1), we decided to extend the analysis to young control patients, who had significantly better periodontal status (Fig. 1 and Table 1). Comparative analysis of samples derived from young control patients vs. OSCC patients gave the expected results: with the exception of H3F3A and IL1B, the other 5 mRNAs were present in significantly higher quantities in the saliva samples of OSCC patients. These differences were reflected in the AUC values of the receiver operator characteristic (ROC) curves as well (Table 3 and Supplementary Fig. 2). Interestingly, although IL6 mRNA was originally tested as a potential marker of inflammation, it was quantifiable in 20 of the 31 OSCC patients, but only in 5 of 30 age-matched control patients, and in 1 of 29 young control patients (Fig. 1h). The logistic regression model analysis identified a 4-biomarker combination from the original 7 mRNAs (DUSP1/IL8/OAZ1/H3F3A) that could differentiate between the age-matched control and OSCC patients with a combined AUC of only 0.770, whereas the 4-marker combination identified in a similar analysis for OSCC vs. young control patients (DUSP1/OAZ1/H3F3A/SAT) had a combined AUC = 0.925 (Fig. 2a–b). If IL6 was also included in the logistic regression analysis from a 8-gene panel, the new 4-marker combinations both selected IL6, and the combined AUC values increased to 0.870 and 0.982, respectively (Fig. 2c–d).
Normalized expression of the mRNA biomarkers in saliva samples of the three patient groups. Normalized expression values were calculated after qPCR efficiency correction and normalization with the normalizing factor (from GAPDH and ACTB). Means are indicated, and p values are derived from Mann-Whitney U test (p<0.05 is significant). LOQ = limit of quantification. For IL6, samples with non-quantifiable qPCR are also shown, as having a normalized mRNA value = 0.00. These samples had Ct > 38, or gave no signal in the qPCR. The number of quantifiable/total samples are shown for each patient group
Discussion
Selecting the appropriate reference gene(s) for normalizing is a key step in real-time qPCR studies. Several earlier publications used different combinations of reference genes for saliva analysis of OSCC patients: ANXA2, RPL37 and S100AB [24], GAPDH, ACTB and RPS9 [5], or RPS9 and ACTB [25]. The preprocessing of saliva samples presumably removed intact cells and large apoptotic bodies, but retained exosomes, microvesicles and potentially small apoptotic bodies, all of which could contribute to the RNA content of the samples. Unlike total RNA isolated from intact human cells, preprocessed saliva contains small RNAs and larger RNA species in variable ratios, derived from both the oral microbiota and human cells; hence, the same quantity of saliva RNA may hide widely different human mRNA content in the different samples. Therefore, reverse transcription reactions were equalized by adding the same amount of yeast tRNA to each sample, and we analysed a 8-gene candidate reference gene panel across all samples to identify the best reference gene(s). We selected the GAPDH-ACTB combination from the partially overlapping recommendations of geNorm, NormFinder and Kahlou et al. ANOVA analyses, because these genes reflected best the intra- and inter-group variability of the samples’ human mRNA content, and because both mRNAs could be quantified reliably in all samples.
In contrast to previously published results of other groups, we could not detect higher expression of the 7 mRNAs in the saliva samples of OSCC patients vs. age-matched control patients. Instead, 3 mRNAs (DUSP1, OAZ1 and H3F3A) had even higher expression in the samples of age-matched control vs. OSCC patients. On the other hand, when samples of young control patients and OSCC patients were compared, results of the previous studies [3, 5, 25] were better replicated: 5 out of the 7 mRNAs were present in higher amounts in the samples of OSCC patients. The age-matched control patients were examined twice by the clinicians to exclude the presence of precancerous lesions or cancer in the oral cavity: first, at the time of recruitment (2013), and the second time, 2 years after that (2015). Importantly, all age-matched control patients still alive in 2015 (28) were free of pre-cancerous oral lesions or cancer, and those who died (2) perished from non-cancer related causes. Therefore, we concluded that the overlapping expression patterns of the 7 mRNAs in the saliva of age-matched control and OSCC patients were not the result of undetected, but active precancerous processes in the oral cavity of the control patients. Our results also suggest that the optimal cutpoint values in the ROC analysis for DUSP1 and OAZ1 (which are among the best biomarkers identified in previous studies) should be applied reciprocally in the Hungarian at-risk population – namely, expression below rather than above the cutpoint being associated with the presence of OSCC. If that difference is accepted, the best biomarker combination to identify OSCC in the elderly, at-risk population (DUSP1/IL8/OAZ1/H3F3A) still has mediocre performance: sensitivity 63%, specificity 70% and combined AUC = 0.770 - although the performance of a 4-biomarker set can be improved slightly with the inclusion of IL6 (sensitivity 81.5%, specificity 76.7%, combined AUC = 0.870) (Supplementary Table 2).
Such differences in the results of biomarker studies are not uncommon, and might be caused by a combination of different factors - the first being technical differences in sample collection and processing. However, we followed closely the protocols described in previous studies [3], with small differences that should not have a major impact on the measurements. In addition, differences in 5 of the 7 mRNAs were validated succesfully in the young control vs. OSCC comparison. Therefore, we hypothesize that the lack of difference in 4 mRNAs, and higher expression for 3 mRNAs in the age-matched controls vs. OSCC patients may have biological, rather than technical explanation - most likely, the inflammatory processes in the oral cavity of both patient groups affecting the performance of the putative biomarkers. The high salivary TNFα mRNA levels (data not shown), the DMFT and DMFS values, as well as the plaque and gingival indices (PI and GI) support that the elderly patients (OSCC and control both) have similarly poor oral health, and that the oral health of the young control patients is markedly better. It should be noted that although these indices were not reported in previous studies for the American or Serbian patient cohorts, the average age of the OSCC patients was significantly higher than that of the control patients in the Serbian study: 61 ± 12.3 years vs. 38 ± 12.3 years [5]. Most importantly, a recent study of Cheng et al. [28] found that the presence of periodontitis increased the salivary levels of the same mRNA biomarkers (with the exception of S100P), rendering them unreliable as OSCC-detecting biomarkers. It should also be noted that elevated levels of proteins encoded by some of the genes in our study were also detected in other diseases, which may affect the salivary mRNA levels even in the absence of oral cancer. For instance, salivary IL1B protein levels can be indicative of active psoriasis (29), and salivary IL8 levels are elevated in cystic fibrosis patients (30) or in the presence of chronic inflammatory diseases (31).
In summary, further analyses are required to determine the biological factors that may affect the performance of salivary biomarkers for OSCC in certain patient populations, in the context of oral inflammatory conditions, as well as in the context of ailments commonly found in the elderly, at-risk patient populations. In addition, salivary IL6 mRNA and protein levels should be analyzed in a larger OSCC/age-matched control patient cohort, to validate its suitability as an OSCC-specific biomarker (work currently in progress).
References
Guerra EN, Acevedo AC, Leite AF, Gozal D, Chardin H, De Luca CG (2015) Diagnostic capability of salivary biomarkers in the assessment of head and neck cancer: a systematic review and meta-analysis. Oral Oncol 51(9):805–818. https://doi.org/10.1016/j.oraloncology.2015.06.010
Li Y, Zhou X, St John MA, Wong DT (2004) RNA profiling of cell-free saliva using microarray technology. J Dent Res 83(3):199–203. https://doi.org/10.1177/154405910408300303
Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH, Wong DT (2004) Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res 10(24):8442–8450. https://doi.org/10.1158/1078-0432.CCR-04-1167
Yakob M, Fuentes L, Wang MB, Abemayor E, Wong DT (2014) Salivary biomarkers for detection of oral squamous cell carcinoma - current state and recent advances. Curr Oral Health Rep 1(2):133–141. https://doi.org/10.1007/s40496-014-0014-y
Brinkmann O, Kastratovic DA, Dimitrijevic MV, Konstantinovic VS, Jelovac DB, Antic J, Nesic VS, Markovic SZ, Martinovic ZR, Akin D, Spielmann N, Zhou H, Wong DT (2011) Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol 47(1):51–55. https://doi.org/10.1016/j.oraloncology.2010.10.009
Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, Henson B, Hu S, Arellano M, Sinha U, Le A, Messadi D, Wang M, Nabili V, Lingen M, Morris D, Randolph T, Feng Z, Akin D, Kastratovic DA, Chia D, Abemayor E, Wong DT (2012) Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomark Prev 21(4):664–672. https://doi.org/10.1158/1055-9965.EPI-11-1093
Lallemant B, Evrard A, Combescure C, Chapuis H, Chambon G, Raynal C, Reynaud C, Sabra O, Joubert D, Hollande F, Lallemant J-G, Lumbroso S, Brouillet J-P (2009) Clinical relevance of nine transcriptional molecular markers for the diagnosis of head and neck squamous cell carcinoma in tissue and saliva rinse. BMC Cancer 9:370. https://doi.org/10.1186/1471-2407-9-370
Ervik M, Lam F, Ferlay J, Mery L, Soerjomataram I, Bray F (2016) Cancer today. Lyon, France: International Agency for Research on Cancer. Cancer today. Available from: http://gco.iarc.fr/today, accessed [01/05/2017]
Alevizos I, Mahadevappa M, Zhang X, Ohyama H, Kohno Y, Posner M, Gallagher GT, Varvares M, Cohen D, Kim D, Kent R, Donoff RB, Todd R, Yung CM, Warrington JA, Wong DT (2001) Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene 20(43):6196–6204. https://doi.org/10.1038/sj.onc.1204685
Chen C, Méndez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, Schwartz SM, Zhao LP (2008) Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomark Prev 17(8):2152–2162. https://doi.org/10.1158/1055-9965.EPI-07-2893
Sajnani MR, Patel AK, Bhatt VD, Tripathi AK, Ahir VB, Shankar V, Shah S, Shah TM, Koringa PG, Jakhesara SJ, Joshi CG (2012) Identification of novel transcripts deregulated in buccal cancer by RNA-seq. Gene 507(2):152–158
Jiang Q, Yu YC, Ding XJ, Luo Y, Ruan H (2014) Bioinformatics analysis reveals significant genes and pathways to target for oral squamous cell carcinoma. Asian Pac J Cancer Prev 15(5):2273–2278. https://doi.org/10.1016/j.gene.2012.07.036
Liu J, Duan Y (2012) Saliva: A potential media for disease diagnostics and monitoring. Oral Oncol 48(7):569–577. https://doi.org/10.1016/j.oraloncology.2012.01.021
Ottó S, Kásler M (2005) Trends in cancer mortality and morbidity in Hungarian and international statistics. Characteristics and potential outcome of public health screening programmes. Magy Onkol 49(2):99–101 103–7
Suba Z, Mihályi S, Takács D, Gyulai-Gaál S (2009) Oral cancer: morbus Hungaricus in the 21st century. Fogorv Sz 102(2):63–68 PMID:19514245
Hermann P, Gera I, Borbély J, Fejérdy P, Madléna M (2009) Periodontal health of an adult population in Hungary: findings of a national survey. J Clin Periodontol 36(6):449–457. https://doi.org/10.1111/j.1600-051X.2009.01395.x
Nemes JA, Nemes Z, Márton IJ (2005) p21WAF1/CIP1 expression is a marker of poor prognosis in oral squamous cell carcinoma. J Oral Pathol Med 34(5):274–279. https://doi.org/10.1111/j.1600-0714.2005.00310.x
Nemes JA, Deli L, Nemes Z, Márton IJ (2006) Expression of p16(INK4A), p53, and Rb proteins are independent from the presence of human papillomavirus genes in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 102(3):344–352. https://doi.org/10.1016/j.tripleo.2005.10.069
Lang NP, Corbet EF (1995) Periodontal diagnosis in daily practice. Int Dent J 45(1):3–15 PMID:7607742
Beikler T, Flemmig TF (2011) Oral biofilm-associated diseases: trends and implications for quality of life, systemic health and expenditures. Periodontol 2000 55(1):87–103. https://doi.org/10.1111/j.1600-0757.2010.00360.x
Pindborg JJ, Reichart PA, Smith CJ, Van der Waal I (1997) Histological typing of cancer and precancer of the oral mucosa. WHO international histological classification of tumours, 2nd edn. Springer, Berlin
Head and neck tumors: Lip and oral cavity (2009) In: Sobin LH, Gospodarowitz MK, Wittekind CH, editors. TNM classification of malignant tumors. 7th ed. Wiley-Blackwell
Mehdi Khanlou K, Van Bockstaele E (2012) A critique of widely used normalization software tools and an alternative method to identify reliable reference genes in red clover (Trifolium Pratense L.) Planta 236(5):1381–1393. https://doi.org/10.1007/s00425-012-1682-2
Hu Z, Zimmermann BG, Zhou H, Wang J, Henson BS, Yu W, Elashoff D, Krupp G, Wong DT (2008) Exon-level expression profiling: a comprehensive transcriptome analysis of oral fluids. Clin Chem 54(5):824–832. https://doi.org/10.1373/clinchem.2007.096164
Cheng YS, Jordan L, Rees T, Chen HS, Oxford L, Brinkmann O, Wong D (2014) Levels of potential oral cancer salivary mRNA biomarkers in oral cancer patients in remission and oral lichen planus patients. Clin Oral Investig 18(3):985–993. https://doi.org/10.1007/s00784-013-1041-0
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034 PMCID: PMC126239
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Cheng YL, Jordan L, Chen HS, Kang D, Oxford L, Plemons J, Parks H, Rees T (2017) Chronic periodontitis can affect the levels of potential oral cancer salivary mRNA biomarkers. J Periodontal Res 52(3):428–437. https://doi.org/10.1111/jre.12407
Ganzetti G, Campanati A, Santarelli A, Sartini D, Molinelli E, Brisigotti V, Di Ruscio G, Bobyr I, Emanuelli M, Offidani A (2016) Salivary interleukin-1β: oral inflammatory biomarker in patients with psoriasis. J Int Med Res 44(1 suppl):10–14. https://doi.org/10.1177/0300060515598902
Nie S, Zhang H, Mayer KM, Oppenheim FG, Little FF, Greenberg J, Uluer AZ, Walt DR (2015) Correlations of salivary biomarkers with clinical assessments in patients with cystic fibrosis. PLoS One 10(8):e0135237. https://doi.org/10.1371/journal.pone.0135237
Rathnayake N, Akerman S, Klinge B, Lundegren N, Jansson H, Tryselius Y, Sorsa T, Gustafsson A (2013) Salivary biomarkers for detection of systemic diseases. PLoS One 8(4):e61356. https://doi.org/10.1371/journal.pone.0061356
Acknowledgements
M. Fera, T. Kerekes and E. Mátyás are thanked for their excellent technical assistance.
Funding
This work was supported by the Hungarian Research Fund OTKA (http://www.otka.hu/), grant ID K 105034 (principal investigator: I. Márton). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Horváth, J., Szabó, A., Tar, I. et al. Oral Health May Affect the Performance of mRNA-Based Saliva Biomarkers for Oral Squamous Cell Cancer. Pathol. Oncol. Res. 24, 833–842 (2018). https://doi.org/10.1007/s12253-017-0296-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0296-1