Abstract
The genetically modified rice G6H1 expressing a fused protein of Cry1Ab/Vip3H and a 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) G6 is a genetically modified event that has been approved for preproduction field testing in China. The purpose of this study was to establish an event-specific qualitative and quantitative detection method that could provide a stable, reliable system for monitoring this new transgenic event. In this study, event-specific qualitative and quantitative detection methods based on the 3′ integration junction sequence between host plant DNA and the integrated gene were developed. The limit of detection (LOD) of qualitative PCR was assessed to be 0.1%. The LOD of quantitative PCR was estimated to be ten haploid genome copies. The quantitative PCR detection method was verified with three mixed rice samples with known G6H1 contents, and the results agreed with the expected values. Evaluation of specificity and sensitivity indicated that the developed qualitative and quantitative PCR methods are reliable and can be used for the detection and quantification of G6H1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To meet the increasing food demand of the growing human population, genetically modified crops were developed to boost agricultural productivity despite the lack of arable land. Since 1989, with the government’s support, numerous transgenic rice lines were developed and tested at various stages based on the regulatory process for agricultural genetically modified organisms (GMOs) in China (Mäde et al. 2006). Most of these transgenic rice lines were genetically modified to express insecticidal genes with lepidopteran activity from bacterium Bacillus thuringiensis (Bt), with single or dual insecticidal genes or stacked with other types of transgenes (Li et al. 2015). In 2009, China’s Ministry of Agriculture issued biosafety certificates for the commercial production of two transgenic Bt rice lines, Huahui 1 and Bt Shanyou 63, in restricted areas, and the biosafety certificates were renewed in 2014. At the same time, several other transgenic rice lines have been extensively tested and are now waiting for approval (Chen et al. 2011). However, in the face of public opposition to GM rice, the adoption of Bt rice has been stalled by the Chinese government.
To ensure that consumers have the freedom to choose new products, several countries have established a mandatory or voluntary labeling of products containing genetically modified (GM) material, including specifications of thresholds for labeling. For example, in the European Union (EU), the threshold is set at 0.9%. In Japan, the value is 5%, and in Korea, the value is 3% (Regulation (EC) No. 1829/ 2003; Akiyama et al. 2008; Ministry of Agriculture and Forestry of South Korea 2000). All published labeling laws are directed against approved GMOs. As for unapproved GMOs, they cannot be present in food or feed at any level. To meet the legislation requirements, there is consequently a need for effective and accurate analytical methods for detection, identification, and quantification of GMOs and other derived products. However, the lack of information inevitably makes it challenging to detect and identify GMOs, especially the unauthorized GMOs (Fraiture et al. 2013, 2014; Jensen et al. 2012).
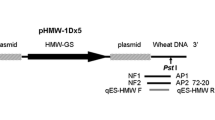
Transgenic rice G6H1 was genetically modified by Zhejiang University to express a fused protein consisting of Cry1Ab/Vip3H and an EPSPS protein G6. The Cry1Ab and Vip3H were stacked to facilitate a delay in the development of Bt resistance and to enhance the insecticidal activity of the transgenic rice. The G6 gene is a glyphosate tolerance gene, which is a synthetic 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene from Pseudomonas putida (Zhao et al. 2011). Previous studies have shown that G6H1 is highly resistant to the Asiatic rice borer, Chilo suppressalis (Walker) and the stem borer Sesamia inferens (Walker) (Chen et al. 2010). In addition, it shows higher resistance to Asiatic rice borer in the field than transgenic rice lines containing a single gene of Cry1Aa, Cry1Ac, Cry1C, and a fused gene of Cry1Ab/Cry1Ac and Cry1Ac/CpTI (Chen et al. 2010). Because of its good performance, G6H1 is one of the several transgenic rice lines that has been approved for preproduction field trials and has thus been included in the pipeline for commercialization. The integration flanking sequence of the G6H1 rice was characterized and provided by the developers. In this study, event-specific qualitative and quantitative PCR detection methods were established based on the 3′ flanking sequence of transgenic rice G6H1. We refer to the Right Border and the Left Border sequences as 5′ and 3′ regions of the construct, respectively (Fig. 1).
Materials and Methods
Plant Materials
GM rice G6H1 and non-transgenic rice Xiushui 110 seeds were kindly supplied by Zhejiang University. G6H1 and non-GM rice seeds were sown and cultivated in a greenhouse. The fresh seedlings leaves were collected for DNA extraction.
Five groups of genetically modified crop seed powder combinations were selected for determining the specificity of established method, including six transgenic maize events (maize lines MON810, MON863, Bt176, Bt11, MIR604, and TC1507), seven transgenic soybean events (soybean lines GTS 40-3-2, A2704-12, MON89788, DP356043, A5547-127, CV127, and DP305423), eight transgenic rapeseed events (oilseed rape lines MS1, Topas 19/2, OXY-235, MS8, RF1, RF2, RF3, and T45), five transgenic rice events (rice line TT51-1, Kefeng6, G281, Keming rice, and M12), and five transgenic cotton events (cotton lines MON531, MON88913, MON1445, MON15985, and LLCotton25). In each transgenic crop seed powder combination, every transgenic crop line accounts for 1% of the total weight. All transgenic crop line materials were kindly provided by the corresponding developers. The GM samples with 100%, 5%, 1%, and 0.5% mass ratio of G6H1 event and the same genetic background of non-transgenic Xiushui 110 seeds powders were provided by the Chinese Ministry of Agriculture-certificated GM test lab. The preparation procedure was conducted according to the standard material candidate process (Chinese Ministry of Agriculture Bulletin No. 1782-Technical specification of preparation of the GMO standard materials).
DNA Extraction and Purification
Genomic DNA was extracted using a QIAGEN DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. DNA samples were quantified by measuring absorbance at a wavelength of 260 nm using an ND-2000C spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). Rice genome copy numbers were calculated by the DNA quantity and a rice haploid genome size of 0.45 pg (Arumuganathan and Earle 1991).
Oligonucleotide Primers and Probes
The genomic structure of the locus in transgenic rice G6H1 was identified by Zhejiang University and shown in Fig. 2. The qualitative PCR primers were designed based on the obtained 3′ integration flanking sequence of G6H1, with a resulting amplification fragment of 247 bp. The rice gene sucrose phosphate synthase (SPS) was used as the endogenous reference gene for GM rice detection, and the qualitative PCR detection primers for the SPS genes were synthesized as previously reported (Jiang et al. 2009). The PCR primers and a corresponding TaqMan probe for quantitative real-time PCR targeting event G6H1 3′ flanking sequence were designed using the Primer Premier 5.0 software (Premier Biosoft, USA). The primers were searched against Oryza sativa Nucleotide collection database (nt) and no target templates were found. For the relative quantification of G6H1, the rice PLD gene was used as the rice endogenous reference gene (Mazzara et al. 2006). All oligonucleotide probes were labeled with FAM as reporter dye at its 5′ end and Black Hole Quencher-1 (BHQ1) at its 3′ end. The primers and probes used in this study were synthesized by Sangon (Sangon, Shanghai, China). All PCR primers and probes used in this study are listed in Table 1.
Primers design for G6H1 event. The nucleotide sequence of the 3′ junction sequence between rice genome and integrated event was used for designing the primers. Primers and probes used for conventional and quantitative PCR amplification are indicated by arrows. Arrow head defines 3′ end. Lowercase letters are the genome flanking sequence, and uppercase letters are the sequence of exogenous vector
Qualitative PCR Assay for the Presence of the Transgene
All PCR reactions were set up in 25 μl volume, using 50 ng of genomic DNA, 1× PCR buffer (Takara, Japan), 1.5 mM MgCl2, 200 mM of each dNTP, 0.4 μM each primer, and 0.6 U of Taq DNA Polymerase (Takara, Japan). All reactions were performed in the T3000 Thermocycler (Biometra), with the following program: 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 45 s, and 72 °C for 60 s, and then a final elongation step of 72 °C for 7 min. Amplification products were electrophoresed in 2% agarose gels for approximately 30 min at 120 V and stained with ethidium bromide for visualization.
Quantitative Real-time PCR
In the development of the method, quantitative real-time PCR was carried out on a 7500 real-time PCR cycler (Life Technologies AB) using the FastStart Universal Probe Master (Roche) with ROX reference dye according to the manufacturer’s instructions. Reactions contained the following: 12.5 μL of the FastStart universal probe master mix (ROX), 0.25 μL of the probe (25 μM), 0.25 μL of the forward primer (90 μM), 0.25 μL of the reverse primer (90 μM), 9.25 μL of water, and 2.5 μL of the genomic DNA. The samples were amplified in semi-skirted 96-well plates sealing with adhesive optical film. The following temperature profile was used for real-time PCR: 50 °C for 120 s (UNG incubation), 95 °C for 120 s, and 50 cycles of 95 °C for 15 s, and 60 °C for 30 s. Data were analyzed with 7500 System SDS Software version 1.4.0.25 (Applied Biosystems). For robustness tests, CFX 96 (Bio-Rad, Munich, Germany) and Fast Advanced Master Mix (Applied Biosystems, Waltham, MA, USA) were used with the PCR cycling conditions as mentioned above.
Inter-lab Applicability of the Developed Methods
The developed qualitative PCR method was validated by six GMO detection laboratories (from the Ministry of Agriculture). Each laboratory received ten genomic samples: one sample labeled as PC was used as positive control, one sample labeled NC was used as negative control, other samples labeled with number from S1–S8 were used as blind samples, samples labeled with S1–S5 representing five groups of genetically modified crop seed powder combinations which were selected for determining the specificity of the developed method as mentioned above, samples labeled with S6–S8 representing G6H1 content levels of 1% and 0.1% and autoclaved G6H1 flour (1%) respectively.
The inter-lab applicability of the developed qPCR method was tested by two other GMO detection laboratories (Oil Crops Research Institute, Chinese Academy of Agricultural Sciences and National Center for the Molecular Characterization of Genetically Modified Organisms, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University), where the CFX96 Real-Time PCR Detection System (Bio-Rad, USA) and the ABI 7900 (Applied Biosystems, Carlsbad, CA, USA) were used respectively. In their assays, the genomic DNA samples were diluted to different concentrations (100.0, 10.0, 1.0, 0.1, and 0.01 ng/μL) and used as calibrators for constructing the standard curve and evaluating the sensitivity in G6H1 event real-time PCR analysis. The specificity of G6H1 event real-time PCR assay was tested employing other GM events.
Results and Discussion
Qualitative Detection of the G6H1 Rice by Conventional PCR
The event-specific primers G6H1F/R were designed based on the G6H1 3′ junction site and used for developing conventional PCR detection assay for the G6H1 rice. The primer G6H1-R targets the flanking rice genome, and the primer G6H1-F targets the inserted DNA sequence of the event G6H1, yielding a 247-bp amplicon (Fig. 2). The rice SPS gene was selected as endogenous reference gene, and the primers SPS F/R were used for rice characterization (PCR primers are shown in Table 1). As expected, a specific 247-bp amplicon was obtained from G6H1 rice, and no fragment was detected with non-transgenic rice and water as templates (Fig. 3). To study the specificity of established conventional PCR method, the primer pair G6H1F/G6H1R were tested by amplifying the genomic DNA samples of five groups of genetically modified crop seed powder combinations, including six transgenic maize events (maize lines MON810, MON863, Bt176, Bt11, MIR604, and TC1507), seven transgenic soybean events (soybean lines GTS 40-3-2, A2704-12, MON89788, DP356043, A5547-127, CV127, and DP305423), eight transgenic rapeseed events (oilseed rape lines MS1, Topas 19/2, OXY-235, MS8, RF1, RF2, RF3, and T45), five transgenic rice events (rice lines TT51-1, Kefeng6, G281, Keming rice, and M12), and five transgenic cotton events (Cotton Line MON531, MON88913, MON1445, MON15985, and LLCotton25). As expected, no amplification fragment was observed from any of those samples except for G6H1 rice (Fig. 3). These data confirmed the fragment obtained was unique to G6H1 rice and from the G6H1 event-specific region between 3′ end of exogenous integration and rice genomic DNA.
Validation of the event-specific detection system of G6H1 with primer pair G6H1F/R. Lane 1, 100 bp DNA ladder; Lane 2, blank control; Lane 3, negative control; Lane 4, 1% G6H1; Lane5, 0.1% G6H1; Lane 6, other transgenic rice combination; Lane 7, transgenic maize combination; Lane 8, transgenic soybean combination; Lane 9, transgenic oilseed combination; Lane 10, transgenic cotton combination
Sensitivity of the Qualitative PCR Method
The sensitivity of the established G6H1 event-specific PCR assay was measured by the limit of detection (LOD). The G6H1 rice powder was mixed with non-transgenic rice powder with 10%, 5%, 1%, 0.5%, 0.1%, and 0.05% of GMO content. Genomic DNA was extracted from these mixed rice powder sample respectively and used as template. The amplification result showed that the target fragment was detected in all mixtures (Fig. 4). Twenty rice powder samples with 0.1% G6H1 content were used for the assay LOD determination. Each sample was amplified with three replicates. The result showed that the expected fragment could be detected from all 60 reactions, i.e., the LOD was 0.1% (data was not shown).
Sensitivity of the event-specific detection system of G6H1 with different amounts of G6H1. Lane 1, 100 bp DNA ladder; Lane 2, blank control; Lane 3, negative control; Lane 4, relative amounts of G6H1 with 10%; Lane 5, relative amounts of G6H1 with 5%; Lane 6, relative amounts of G6H1 with 1%; Lane 7, relative amounts of G6H1 with 0.5%; Lane 8, relative amounts of G6H1 with 0.1%; Lane 9, relative amounts of G6H1 with 0.05%
Study of Potential Food Processing Impact
As food processing is well-known to cause DNA damage, the developed qualification PCR assay was carried out on processed rice flour. Rice powder containing 1% G6H1 was autoclaved at 121 °C for 30 min. DNA was extracted from the processed rice powder sample and used as PCR template. PCR results indicated that the expected 247-bp fragment could be amplified from the processed rice powder using the developed qualitative PCR method (Fig. 5). The assay indicated that the developed qualitative PCR method can be applied for processed food GMO detection.
Inter-lab Applicability of the Qualitative PCR Method
Six GMO detection laboratories were invited to validate the developed qualitative PCR method. The results were shown in Table S1. All test samples are the same as samples used in our in-house validation experiments. The amplification results were consistent with our in-house experiment, and the limit of detection of 0.1% was confirmed. This collaborative trial proved that our developed qualitative PCR method produces reliable and reproducible results for a given sample set and can be adopted for testing laboratories.
G6H1 Rice Event-Specific Quantitative Real-time PCR
The event-specific primers and probe for quantitative detecting G6H1 were designed based on the 3′ flanking sequence in G6H1 event using the software Primer Premier 5. The primers and probe locations in the junction sequence are shown in Fig. 2, and the sequences were shown in Table 1. The primer F98 targets the inserted DNA sequence of event G6H1. The primer R187 and the probe were located at the G6H1 genomic DNA region. The rice PLD gene was selected as endogenous reference gene in quantitative real-time PCR detection assay, and the primers PLD-F/R and the probe PLD-P were used for rice characterization. The specificity of the developed quantitative assay was evaluated on non-transgenic rice and other transgenic crops. The primer pair and probe G6H1-F98/G6H1-R187/G6H1-P were tested by amplifying the genomic DNA samples of five groups of genetically modified crop seed powder combinations and non-transgenic rice used in qualitative PCR specificity investigation. The test results showed that no signals were detected in other GM plants, which confirmed that our quantitative PCR assay is specific for characterizing G6H1 event (Table 2).
Construction of Standard Curves
To evaluate the G6H1 event-specific real time PCR assay, standard curves were established. Genomic DNA of the G6H1 was serially four-fold diluted with ddH2O to a final concentration equivalent to 6.8 ng/μL, 1.7 ng/μL, 0.425 ng/μL, 0.106 ng/μL, 0.027 ng/μL, and 0.007 ng/μL in a solution. Five microliters was used for each reaction, so each amplification reaction contained roughly 75,556, 18,889, 4722, 1181, 295, and 74 copies of the transgenic haploid genome, considering the Japonica rice genomic size to be 0.45 pg per haploid genome (Arumuganathan and Earle 1991). The assays were repeated three times, each time with triple parallels. The threshold of cycle (Ct) value for each standard sample was listed in Table 3, and standard curves for the three replicates were generated by plotting the Ct values against the common logarithm of the copy number as illustrated in Fig. S1. Two parameters, R2 coefficient and the amplification efficiency, which were calculated from the slopes of the standard curve, were employed to evaluate the standard curves. The R2 values were all > 0.99 and ranged from 0.997 to 0.998, which showed good linearity between the amount of Ct values. Based on the slope of the standard curves, the event-specific quantitative amplification efficiencies in all experiments were close to 100% (Fig. S1). The acceptable linearity and the high efficiency of the standard curves indicated that the established real-time PCR method could be used for ingredient identification and quantification of the G6H1 rice.
In-house and Inter-lab Validation of the Quantitative PCR Method
The robustness of the developed quantitative assay was tested on CFX 96 (Bio-Rad, Munich, Germany) with Fast Advanced Master Mix (Applied Biosystems, Waltham, MA, USA). The genomic DNA samples were diluted to approximately 20,000, 4000, 800, 160, 32, and 6.4 copies per reaction and used as calibrators for constructing the standard curve. The assays were repeated three times, each time with five parallels. The results were showed in Supplementary Fig. S2. In three repeats, the R2 values were all > 0.99 and the PCR amplification efficiencies ranging from 96.1 to 101.1%, In consequence, the method performance is not influenced by the choice of the qPCR instrument, or the master mix product used. Assays at two other GMO detection laboratories also demonstrated the high specificity and sensitivity of the G6H1 event real-time PCR assay. And this quantitative method has been used for the development of certified matrix-based reference material of G6H1 (Yang et al. 2018).
Sensitivity of the Quantitative PCR Method
The sensitivity of the G6H1 quantification method was evaluated by determining the limit of detection (LOD). The G6H1 genomic DNA dilution series containing approximately 20, 10, 5, and 1 haploid genomic copies (copy) of G6H1 genomic DNA per reaction were analyzed with 20 repeats. With the template copy number decreasing, the developed quantitative method’s ability to detect G6H1 decreased. Amplification signals were detected in all reactions when the template decreased down to approximately 20 or 10 haploid copies (Table 4). When approximately five haploid copy of G6H1 DNA sample was used, amplification was detected with the mean Ct value > 38 and the test positive signals appeared 18 out of 20 reactions (Table 4). Based on the results, the LOD of the G6H1 event-specific quantification detection was estimated to be 10 haploid copies.
Accuracy and Precision
The accuracy of the G6H1 quantification method was assessed using the prepared test samples with 0.5%, 1.0%, and 5.0% GM levels. The PCR assay was repeated three times for precision estimation. The samples were prepared with seed powder of G6H1 and its non-GM control by mass/mass. The relative content of G6H1 was calculated as (mean copy number of G6H1 of three replicates)/(mean copy number of rice DNA of three replicates) × 100. The results were shown in Table 5. The calculated GM contents corresponding to 5.0%, 1.0%, and 0.5% were 4.48%, 1.05%, and 0.48%, respectively, with the bias ranging from − 10.40 to 5.00%, and the RSDr ranged from 4.68 to 14.58%. These results showed that the trueness and precision of the quantification were within the ENGL method acceptance criteria that the bias% should be within 25% of the target value over the entire dynamic range and the relative standard deviation of repeatability (RSDr) should be ≤ 25% over the entire dynamic range (ENGL 2015).
Conclusions
In this study, we established sensitive and reliable qualitative and quantitative PCR detection methods for event-specific detection of the G6H1 rice line. For the event-specific qualitative method, a 247-bp fragment was specified amplified from the G6H1 event. The detection limit of the developed qualitative PCR method was 0.1%. The event-specific quantitative PCR method was evaluated with construction of standard curves. The assay results indicated amplification efficiencies in all experiments were close to 100% and acceptable linearity between copy numbers and fluorescent values. The absolute LOD of the event quantitative PCR method for the G6H1 was estimated to be about ten haploid copies. The accuracy of the quantitative method was evaluated as the % bias of the experimental mean value from the theoretical value, and the calculated average G6H1 event was close to the expected value. All results demonstrate the developed event-specific qualitative and quantitative PCR method is creditable and suitable for the detection and quantification of G6H1 and its derivatives.
References
Akiyama H, Sakata K, Kondo K, Tanaka A, Liu MS, Oguchi T, et al (2008) Individual detection of genetically modified maize varieties in non-identity-preserved maize samples. J Agric Food Chem 56:1977–1983
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Report 9:208–218
Chen Y, Tian J, Shen ZC, Peng YF, Hu C, Guo YY, Ye GY (2010) Transgenic rice plants expressing a fused protein of Cry1Ab/Vip3H has resistance to rice stem borders under laboratory and field conditions. J Econ Entomol 103:1444–1453
Chen M, Shelton A, Ye GY (2011) Insect-resistant genetically modified rice in China: from research to commercialization. Annu Rev Entomol 56:81–101
ENGL (2015) Definition of minimum performance requirements for analytical methods of GMO testing. Available at: http://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm
Fraiture MA, Herman P, Taverniers I, Loose MD, Deforce D, Roosens NH (2013) An innovative and integrated approach based on DNA walking to identify unauthorized GMOs. Food Chem 147:60–69
Fraiture MA, Herman P, Taverniers I, Loose MD, Nieuwerburgh FV, Deforce D, Roosens NH (2014) Validation of a sensitive DNA walking strategy to characterize unauthorized GMOs using model food matrices mimicking common rice products. Food Chem 173:1259–1265
Hernandez M, Esteve T, Pla M (2005) Real-time polymerase chain reaction based assays for quantitative detection of barley, rice, sunflower, and wheat. J Agric Food Chem 53:7003–7009
Jensen AH, Bertheau Y, Loose MD, Grohmann L, Hamels S, Hougs L et al (2012) Detecting un-authorized genetically modified organisms (GMOs) and derived materials. Biotechnol Adv 30:1318–1335
Jiang L, Yang L, Zhang H, Guo J, Mazzara M, Van den Eede G et al (2009) International collaborative study of the endogenous reference gene, sucrose phosphate synthase (SPS), used for qualitative and quantitative analysis of genetically modified rice. J Agric Food Chem 57:3525–3532
Li Y, Hallerman EM, Liu Q, Wu K, Peng Y (2016) The development and status of Bt rice in China. Plant Biotechnol J 14:839–848
Mäde D, Degner C, Grohmann L (2006) Detection of genetically modified rice: a construct-specific real-time PCR method based on DNA sequences from transgenic Bt rice. Eur Food Res Technol 224:271–278
Mazzara M, Grazioli E, Savini C, Van Den Eede G (2006) Event-specific method for the quantitation of rice line LLRICE62 using real-time PCR - validation report and protocol - sampling and DNA extraction of rice. EUR 22490 EN. JRC34091
Ministry of Agriculture and Forestry of South Korea (2000) Guidelines for labeling of genetically modified agricultural products. MAF Notification 31
Commission Regulation (EC) No. 1829 (2003) Of the European parliament and of the council of 22 September 2003 on geneticallymodified food and feed. Off J Eur Union L 268:1–23
Yang Y, Li L, Yang H, Li X, Zhang X, Xu J, Zhang D, Jin W, Yang L (2018) Development of certified matrix-based reference material as calibrator for genetically modified rice G6H1 analysis. J Agric Food Chem 66:3708–3715
Zhao T, Lin CY, Shen ZC (2011) Development of transgenic glyphosate-resistant rice with G6 gene encoding 5-Enolpyruvylshikimate-3-Phophate synthase. Agric Sci China 10:1307–1312
Acknowledgements
We greatly thank Shen Zhicheng of Zhejiang University for providing the seeds of the G6H1 event and non-transgenic rice Xiushui 110.
Funding
This work was financially supported by the National Transgenic Plant Special Fund (2014ZX08012-001) and a grant from Public Technology Application Research of Zhejiang Province (LGN18C200029).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Xiaoli Xu declares that she has no conflict of interest. Xiaoyun Chen declares that she has no conflict of interest. Yongmin Lai declares that he has no conflict of interest. Qing Zhang declares that he has no conflict of interest. Yuhua Wu declares that she has no conflict of interest. Jun Li declares that he has no conflict of interest. Cheng Peng declares that he has no conflict of interest. Xiaofu Wang declares that he has no conflict of interest. Qingmei Miao declares that she has no conflict of interest. Hui Liu declares that she has no conflict of interest. Junfeng Xu declares that he has no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Not applicable
Rights and permissions
About this article
Cite this article
Xu, X., Chen, X., Lai, Y. et al. Event-Specific Qualitative and Quantitative Detection of Genetically Modified Rice G6H1. Food Anal. Methods 12, 440–447 (2019). https://doi.org/10.1007/s12161-018-1375-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-018-1375-y