Abstract
Background
The lncRNA HOTAIR is frequently overexpressed in breast cancer tissues and plays an important role in the development of breast cancer. Here, we investigated the effect of the lncRNA HOTAIR on the biological behaviour of breast cancer cells and its molecular mechanism.
Methods
We investigated the level of HOTAIR in breast cancer and its clinical pathological characteristics by bioinformatic methods. Then, we evaluated the effects of HOTAIR and miRNA-1 expression on the biological behaviour of breast cancer cells by qPCR, Cell Counting Kit-8 (CCK-8) assay, clonogenic assays, Transwell assay and flow cytometry for cell proliferation, invasion migration and apoptosis, and cell cycle analysis. Finally, the target genes of the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis were validated by luciferase reports.
Results
The expression of HOTAIR in breast cancer tissues was significantly higher than that in normal breast tissues (P < 0.05). Silencing of HOTAIR suppressed cell proliferation, invasion and migration, promoted apoptosis and induced G1 phase block in breast cancer (P < 0.0001). We also verified that miR-1 is a target of HOTAIR and that GOLPH3 is a target of miR-1 by luciferase reporter assays (P < 0.001).
Conclusions
The expression of HOTAIR was significantly elevated in breast cancer tissues. Reducing the expression of HOTAIR inhibited the proliferation, invasion and migration of breast cancer cells and promoted apoptosis, and the mechanism was mainly the effect of the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis on the biological behaviour of breast cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the latest data, breast cancer has surpassed lung cancer as the most prevalent cancer worldwide [1]. The number of new cases of breast cancer worldwide is 2.26 million, accounting for 24.5% of all new cancers in women worldwide [1]. The main treatments for breast cancer patients include surgery, radiotherapy, endocrine therapy, chemotherapy and targeted therapy, but 40% of breast cancer patients still experience tumour recurrence or distant metastasis [2]. Therefore, it is imperative to explore the molecular mechanism of breast cancer and to find new potential therapeutic targets.
In 1990, Brannan et al. identified the first long-stranded noncoding RNA (lncRNA)—H19—in mammalian cells [3]. lncRNAs are a class of RNA molecules greater than 200 nucleotides in length that cannot encode proteins but have biologically relevant regulatory functions [4, 5]. lncRNAs have been linked to a variety of diseases, particularly tumours, by examining differences in transcript expression levels in disease-associated and non-disease-associated states and directed experiments in model organisms, such as mice [6]. The role of long noncoding RNAs in cancer has been extensively described, highlighting their ability to influence cell cycle regulation, cell proliferation, transdifferentiation, survival, immune response, metastatic progression and therapeutic response [7]. Ramnarine et al. [8] found that lncRNAs play an important role in the development and progression of prostate cancer and can be used as biomarkers for the diagnosis and prediction of the prognosis of prostate cancer. Deng et al. [9] demonstrated that lncRNA plays an oncogenic role in colorectal cancer, is an independent prognostic factor, and is a potential therapeutic target. Zhao et al. [10] found that lncRNAs may be potential biomarkers for the diagnosis and prognosis of gastric cancer. HOTAIR (HOX transcript antisense intergenic RNA) is a type of lncRNA, and HOTAIR and other lncRNAs are aberrantly expressed in breast cancer, affecting breast cancer progression by influencing the biological behaviour of breast cancer cells [11]; moreover, the expression level of HOTAIR in primary tumours could be a strong predictor of eventual metastasis and death of patients [12]. However, the molecular mechanism of HOTAIR in BC remains unknown.
In recent years, noncoding RNAs such as lncRNAs have been shown to act as competitive endogenous RNAs (ceRNAs) to sequester microRNAs to regulate mRNA transcripts containing common microRNA recognition elements (MREs) [13,14,15]. They act as sponges for common miRNAs and eliminate the endogenous repressive effects of these miRNAs on their true target transcripts [16]. MicroRNA (miRNA) is a small noncoding RNA of approximately 22 nucleotides (nt) in length [17]. Many studies have shown that miRNAs can act as tumour suppressors and oncogenes, which in turn affect tumour progression [18]. miR-1 is a type of miRNA that is usually expressed at low levels in malignant tumours. High expression of miR-1 inhibits tumour recurrence and metastasis [19]. miRNAs regulate the expression of downstream genes by binding to miRNA recognition elements (MERs) located in the 30-untranslated region (30-UTR) of the target messenger RNA (mRNA), leading to their translational repression or degradation [20]. Therefore, the function of lncRNA HOTAIR and miRNA in breast cancer and the molecular mechanism of its regulation deserve further in-depth study.
Materials and methods
Materials
RNASEqV2 data from the breast cancer dataset were downloaded from the TCGA database via the Bioconductor/TCGA biolinks function package, along with pre-processing, for a specific analysis of HOTAIR clinicopathological correlation, relative expression and prognosis.
Methods
Cell culture, siRNA for HOTAIR and miRNA-1 and transfection
The human breast cancer cells MDA-MB-231 and MCF7 and normal breast epithelial cells MCF10A used in this experiment were obtained from Guangzhou Saiku Biotechnology Company. The cells were cultured in Dulbecco’s Modified Eagle Medium with 15% foetal bovine serum at 37 °C and 5% CO2. The lentivirus targeting knockdown of HOTAIR and miRNA-1 and the negative control lentivirus (sh-HOTAIR; Lv-miRNA-1; NC) designed by Shanghai Jima Pharmaceutical Technology Ltd. were transfected into MDA-MB-231 and MCF7 cells, respectively. After 72 h, qRT-PCR was performed to examine the levels of HOTAIR and miRNA-1 in the breast cancer cell lines after cell transfection.
Cell counting kit-8 (CCK-8) assay
The successfully transfected cells were counted under a microscope using a cell counting plate, and 100 μl of cell suspension was added to each well of a 96-well plate. Then, 100 μl of CCK8 solution was added on days 1–6 and incubated for 2.5 h at 37 °C. The absorbance values were measured by an enzyme marker set at 450 nm.
Clonogenic assays
Cells were added to the cell counting plate at 500 cells/well, 2 ml of complete medium was added to each well, and the culture was continued at 37 °C and 5% CO2 for 2 weeks. Images were collected under the microscope after staining.
Flow cytometry analysis
Flow cytometry was used for cell apoptosis cell cycle analysis. The transfected cells were washed with PBS and centrifuged to obtain cell precipitates. Then, 500 µl of 70% ethanol was added and fixed overnight at − 4 °C. Then, 500 μl of PI/RNase A staining solution was added and incubated for 30 min at room temperature and protected from light. Cell cycle assays were performed in a flow cytometer after incubation.
Transwell assay
The transfected MDA-MB-231 and MCF7 cells were suspended in serum-free medium and removed from the 24-well plate, after 700 μl of complete medium containing 20% FBS was added to the lower chamber and 300 μl of cell suspension was spread in the upper chamber (for invasion experiments, Matrigel was also spread in the upper chamber), and they were placed in a 37 °C 5% CO2 cell incubator for 24 h. The cells and Matrigel were removed from the upper chamber. Formaldehyde was added to fix the cells in the lower chamber, which were then stained with Giemsa stain. Then, the chambers were placed under a microscope for photographing and counting.
Luciferase reporter assay
X3′-UTR wild-type and mutant luciferase reporter gene vectors were designed; pGL3-HOTAIR-Mut 3′-UTR, pGL3-HOTAIR-Wt 3′-UTR, pGL3-GOLPH3-Mut 3′-UTR and pGL3-GOLPH3-Wt 3′-UTR plasmids were constructed; and subsequently, the plasmids were transfected into cells. The luciferase activity was measured using a dual-luciferase assay system.
Western blot
The cells were taken from the cell culture incubator and lysed by a mixture of RIPA and PMSF, and the supernatant was obtained by centrifugation. After electrophoresis, the separated proteins were transferred to PVDF membranes, incubated with antibodies, washed and colour developed. Finally, the results of the protein bands were measured in greyscale using ImageJ.
Statistical analysis
SPSS 25.0 software was used to statistically analyse the data obtained from the experiment, and all results are expressed as the mean ± standard deviation (mean ± SD). The difference between two groups of data was compared using Student’s t test (two-tailed method) for comparisons between two samples conforming to the normal distribution, and ANOVA was used for two comparisons between multiple groups, with P < 0.05 indicating that the two groups of data were statistically significant.
Results
HOTAIR is highly expressed in breast cancer tissues and cell lines
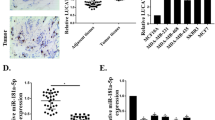
We analysed the relative expression differences of HOTAIR in breast cancer tissues and normal tissues and the relationship between HOTAIR and clinical indicators, such as ER, PR and HER2, through the TCGA database. The results showed that the level of HOTAIR was significantly higher in breast cancer tissues than in normal breast tissues (Fig. 1A). The AUC was also calculated by plotting the ROC curve of HOTAIR, and the results suggested that HOTAIR has the potential to predict normal and cancerous tissues in breast cancer (Fig. 1B). The HOTAIR expression levels were significantly associated with ER status (P < 0.001), PR status (P = 0.002), HER2 status (P = 0.005), T stage (P = 0.03), N stage (P < 0.001), molecular staging (P < 0.001) and pathological type (P < 0.001) of breast cancer patients (Fig. 1C), whilst they were not significantly associated with patient age (P = 0.117), M stage (P = 0.108) or clinical stage (P = 0.358) (Fig. 1D). Meanwhile, we classified HOTAIR expression into high and low expression by median and analysed its relationship with clinical baseline characteristics in breast cancer patients (Table 1).
Overexpression of HOTAIR in breast cancer tissues. A HOTAIR expression in normal and cancerous tissues. B The ROC curve of HOTAIR: HOTAIR can predict normal and cancerous tissues in breast cancer. C HOTAIR expression in breast cancers with different oestrogen status, progesterone status, HER2 status, tumour size, lymph node status, molecular staging and pathological histological type. D HOTAIR expression at different ages, distant metastasis status and clinical staging periods. E Kaplan‒Meier survival analysis of HOTAIR levels
We concluded by Kaplan‒Meier survival analysis that HOTAIR expression was not associated with OS or DFS in breast cancer patients (Fig. 1E, P> 0.05). According to the univariate analysis, age, TNM stage, T stage, N stage, M stage, HER2 status, molecular typing and pathological type were factors affecting the prognosis of breast cancer patients (Table 2, P value < 0.05); according to the multivariate analysis, age, T stage, N stage and M stage were all independent factors affecting the prognosis of breast cancer patients (Table 2, P value < 0.05). The expression of HOTAIR was not related to the prognosis of breast cancer patients and could not be used as one of the risk factors to evaluate the prognosis of breast cancer patients (Table 2, P > 0.05).
In vitro assay validates the effect of HOTAIR and miRNA-1 on the biological behaviour of breast cancer
Expression levels of miRNA-1 and GOLPH3 after silencing of HOTAIR
We examined the expression of HOTAIR in the breast cancer cell lines MDA-MB-231 and MCF7 and the normal breast epithelial cell line MCF10A by real-time fluorescence quantitative PCR assay (Fig. 2A). Then, sh-HOTAIR was transfected into MDA-MB-231 and MCF7 cells, and the transfection efficiency was observed under inverted phase contrast fluorescence microscopy after 72 h (Fig. 2B). It was concluded by RT-PCR experiment that the level of HOTAIR was significantly decreased and the level of miRNA-1 was significantly increased after transfection of cells with sh-HOTAIR, and the expression level of GOLPH3 was not significantly changed (Fig. 2C).
Expression levels of miRNA-1 and GOLPH3 after silencing of HOTAIR. A Relative expression levels of HOTAIR in different cell lines (**P < 0.01, ***P < 0.001). B Effect of sh-HOTAIR lentivirus transfection (100 ×). C RT-PCR evaluation of the expression levels of HOTAIR in MDA-MB-231 and MCF7 cells after transfection with sh-HOTAIR as well as miRNA-1 and GOLPH3
Effect of silencing HOTAIR and miRNA-1 on the biological behaviour of breast cancer cells
In further evaluating the effects of HOTAIR and miRNA-1 on the biological behaviour of breast cancer cells, we found that silencing of HOTAIR could inhibit cell proliferation and silencing of miRNA-1 could promote cell proliferation by CCK8 assay (Fig. 3A). The results of the clone formation assay showed that the number of clones was significantly reduced when silencing HOTAIR cells compared with NC cells, and the number of clones was significantly increased when silencing miRNA-1 cells compared with NC cells, which led to the conclusion that silencing HOTAIR inhibited the proliferation ability of breast cancer cells and silencing miRNA-1 promoted the proliferation ability of breast cancer cells (Fig. 3B). Transwell assays showed that the number of cells crossing the ventricular wall and Matrigel ventricular wall was significantly reduced after silencing of HOTAIR compared with NC cells, indicating that the migration and invasion ability of breast cancer cells were reduced after silencing of HOTAIR; the opposite result was obtained after silencing of miRNA-1, indicating that the migration and invasion ability of breast cancer cells were enhanced after silencing of miRNA-1 (Fig. 3C).
Effect of silencing HOTAIR and miRNA-1 on the biological behaviour of MCF-7 and MDA-MB-231 breast cancer cells transfected with si-NC, sh-HOTAIR or Lv-miRNA-1. A CCK8 assay: silencing of HOTAIR inhibited cell proliferation and silencing of miRNA-1 promoted cell proliferation (*P < 0.05, **P < 0.01). B Clonogenic assays: silencing of HOTAIR inhibited cell proliferation and silencing of miRNA-1 promoted cell proliferation. C Transwell invasion and migration assay: silencing of HOTAIR decreased cell migration and invasion and silencing of miRNA-1 increased cell migration and invasion. D Flow cytometry analysis: silencing of HOTAIR increased the percentage of G0/G1 phase cells of MCF7 and MDA-MB-231. E Flow cytometry analysis: Representative images of flow cytometry using Annexin V-FITC and PI staining. Upper right quadrant: cells producing late apoptosis; lower right quadrant: cells producing early apoptosis; lower left quadrant: surviving cells. Bar graph showing the increased proportion of early and late apoptotic cells after knockdown of HOTAIR in MCF-7 cells
We assessed the effects of HOTAIR and miRNA-1 on cell apoptosis and the cell cycle of breast cancer cells by flow cytometer analysis, and the results showed that the proportion of G0/G1 phase cells increased in MCF7 and MDA-MB-231 cells after silencing of HOTAIR, and the difference was statistically significant (Fig. 3D, P < 0.05). After silencing of miRNA-1 in breast cancer cells, there was no significant change in the cell cycle. The number of apoptotic breast cancer cells was significantly increased after silencing HOTAIR in MCF7 cells, and the difference was statistically significant. The apoptotic cells of breast cancer cells were not significantly changed after silencing of miRNA-1, and the difference was not statistically significant; the apoptotic cells of breast cancer cells were not significantly changed after silencing of both HOTAIR and miRNA-1 in MDA-MB-231 cells. The differences were not statistically significant (Fig. 3E, P< 0.001).
Validating lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis target genes
In addition, we analysed the sequences of HOTAIR and miR-1 using the starBase 2.0 prediction program and found that they have binding sites. To further confirm this, we designed luciferase reporter assays to verify whether paired sequences can exist between miR-1 and HOTAIR, and the results showed that transfection of the miR-1 mimic resulted in significant downregulation of the luciferase activity of the HOTAIR-WT reporter but had little effect on the luciferase activity of the HOTAIR-MUT reporter (Fig. 4A). Similarly, we applied the TargetScan Human 7.1 target prediction system to predict the potential targets of miR-1 and found that GOLPH3 was the target of miR-1. Luciferase reports were constructed again to verify the existence of paired sequences between GOLPH3 and miR-1, and the results showed that the transfection of miR-1 mimic inhibited the luciferase activity of the GOLPH3-WT reporter, whilst it had almost no effect on the luciferase activity of the GOLPH3-MUT reporter. It can be concluded that there is a regulatory relationship between HOTAIR and miR-1 and between miR-1 and GOLPH3 (Fig. 4A). Furthermore, lv-miRNA-1 was transfected into MDA-MB-231 and MCF7 cells, and the transfection efficiency was observed under inverted phase contrast fluorescence microscopy after 72 h (Fig. 4B). After transfection, the levels of miRNA-1 in MDA-MB-231 and MCF7 cells and the level of GOLPH3 after silencing of miRNA-1 were detected by RT-PCR, and the results showed that the level of miRNA-1 decreased significantly after transfection of cells with Lv-miRNA-1, and there was no significant difference in the change in GOLPH3 level (Fig. 4C). Western blotting was performed to detect the level of GOLPH3 after transfection, and the results showed that no significant changes in the downstream target protein GOLPH3 were observed after silencing HOTAIR and miRNA-1 (Fig. 4D). Kaplan‒Meier survival analysis was performed on GOLPH3, and the results showed that the OS of patients with high GOLPH3-expressing breast cancer was lower than that of patients with low GOLPH3-expressing breast cancer, and the difference was statistically significant (Fig. 4E, P < 0.05).
Luciferase reports validate lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis target genes. A Luciferase reports showed reduced miR-1 expression in the HOTAIR-WT group but not in the HOTAIR-MUT group. miR-1 could reduce the expression of the GOLPH3-WT group but not the GOLPH3-MUT group. B Effect of Lv-miRNA-1 lentivirus transfection (100 ×). C RT-PCR to evaluate the knockdown effect of miRNA-1 in MDA-MB-231 and MCF7 cells after transfection with Lv-miRNA-1 and the level of GOLPH3 after silencing miRNA-1. D Kaplan‒Meier survival analysis of GOLPH3 levels and OS in non-triple-negative breast cancer patients
Discussion
In this paper, we analysed the role of the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis in the proliferation, invasion and migration of breast cancer. We used a bioinformatic approach to determine that the level of HOTAIR was significantly higher in breast cancer tissues than in normal breast tissues and to verify the relationship between the expression level of HOTAIR and relevant clinical indicators. Knockdown of HOTAIR was found to inhibit proliferation, invasion and migration and promote apoptosis of breast cancer cells by qPCR, CCK-8, Transwell invasion and migration assays and flow cytometer analysis, whilst knockdown of miR-1 showed the opposite result. Furthermore, bioinformatics analysis was used to predict the targets of HOTAIR and miR-1, and luciferase reporter assays were established to further verify that HOTAIR can target miR-1 and that miR-1 can target GOLPH3.
The aberrant expression of lncRNAs in malignant tumours has been well documented both domestically and internationally, highlighting their ability to influence cell cycle regulation, cell proliferation, differentiation, survival, immune response, metastatic progression and therapeutic response, and their relationship with breast cancer has been verified by multiple sources [21, 22]. The lncRNA HOTAIR has been shown to be a key regulator of chromatin state and mediator of transcriptional silencing that reprograms chromatin state and induces cancer metastasis [12]. Consistent with the above study, we found that HOTAIR expression levels were significantly higher in breast cancer tissues than in normal breast tissues and that HOTAIR expression was associated with ER status, PR status, HER2 status, T stage, N stage, molecular staging and pathological type. Loss-of-function experiments revealed that silencing HOTAIR inhibited the proliferation, invasion and migration of breast cancer cells and promoted apoptosis. This demonstrates that HOTAIR can have an impact on the biological behaviour of breast cancer cells and, to some extent, illustrate the potential of HOTAIR as a diagnostic, prognostic and predictive marker and a potential therapeutic target for BC.
Regarding the mechanism, micro(mi)RNAs are small noncoding RNAs that negatively regulate the expression of most mRNAs [23]. LncRNAs can act as microRNA decoys, with the sequestration of microRNAs favouring the expression of repressed target mRNAs [24]. Sorensen et al. [25] found that the miR-148a levels in breast cancer patients were negatively correlated with the HOTAIR levels. Zhao et al. [26] reported that HOTAIR regulates BC cell growth, migration, invasion and apoptosis through the miR-20a-5p/HMGA2 axis. Wang et al. [27] found that HOTAIR promotes BC proliferation, migration and invasion by regulating the miR-601/ZEB1 axis. Zhang et al. [28] showed that the ESR1/miR-130b-3p/HOTAIR regulatory axis promotes breast cancer progression and is significantly associated with endocrine therapy resistance. In our study, we predicted that HOTAIR has binding sites with miR-1 by bioinformatics and constructed luciferase reporter assays to verify this conclusion, which suggested a possible regulatory relationship between HOTAIR and miR-1. In addition, further experiments showed that downregulation of HOTAIR expression was followed by upregulation of miR-1 expression, which also verified again that HOTAIR has a regulatory effect on miR-1. In summary, it can be inferred that there is a regulatory relationship between lncRNA HOTAIR and miR-1 in breast cancer. This conclusion is similar to that obtained by the abovementioned scholars.
GOLPH3 (Golgi phosphorylated protein 3), also known as GMx33 or GPP34, is located in the Golgi apparatus and is a highly conserved transmembrane protein [29]. GOLPH3 has been shown to be a pro-oncogenic factor in many cancers [30]. Zhao et al. [31] found that in breast cancer, GOLPH3 is highly expressed and promotes the metastasis and proliferation of cancer cells. In our study, we applied bioinformatics to predict the potential targets of miR-1 and found that GOLPH3 is the target of miR-1. We again constructed luciferase reporter assays to verify that the two have binding sites, indicating a regulatory relationship between miR-1 and GOLPH3. Meanwhile, we further knocked down the expression of miR-1, and no significant difference was seen in the expression of downstream GOLPH3, indicating that the regulatory relationship between miR-1 and GOLPH3 needs to be further verified. Kaplan‒Meier survival analysis of GOLPH3 suggested a lower OS in patients with high GOLPH3-expressing non-triple-negative breast cancer than in patients with low GOLPH3-expressing breast cancer. In summary, we confirmed the existence of the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis, which provides new ideas for the molecular mechanism of breast cancer development.
In conclusion, the lncRNA HOTAIR/miR-1/GOLPH3 regulatory axis has a regulatory role in breast cancer proliferation, invasion and migration. This is likely to be a novel regulatory mechanism for breast cancer cell metastasis. This topic is important to further reveal the molecular mechanism of noncoding RNA in breast cancer metastasis, discover new potential breast cancer therapeutic targets, and solve the problem of breast cancer recurrence and metastasis. However, there are some shortcomings in this study: the regulatory relationship between the lncRNA HOTAIR and miR-1 on the downstream target protein GOLPH3 could not be verified, and further verification of the regulatory relationship is needed.
Data availability
Data availability statement is available for this paper at https://doi.org/10.6084/m9.figshare.21506364.
Change history
02 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s12094-023-03224-3
References
World Health Organization. WHO mortality database. http://www.who.int/healthinfo/statistics/mortality_rawdata/en/. Published online 2020. Accessed 2022 Oct 01.
Sharifian A, Pourhoseingholi MA, Emadedin M, Nejad MR, Ashtari S, Hajizadeh N, et al. Burden of breast cancer in Iranian women is increasing. Asian Pac J Cancer Prev. 2015;16:5049–52.
Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36.
Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5:379.
Reiche K, Kasack K, Schreiber S, Luders T, Due E, Naume B, et al. Long non-coding RNAs differentially expressed between normal versus primary breast tumor tissues disclose converse changes to breast cancer-related protein-coding genes. PLoS ONE. 2014;9:e106076.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7.
Ramnarine VR, Kobelev M, Gibb EA, Nouri M, Lin D, Wang Y, et al. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur Urol. 2019;76:546–59.
Deng X, Li S, Kong F, Ruan H, Xu X, Zhang X, et al. Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics. 2020;10:265–80.
Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68.
Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146:353–8.
Sumazin P, Yang X, Chiu H-S, Chung W-J, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81.
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–83.
Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18.
Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Alves C, Fonseca A, Muys B, Bueno R, Burger M, de Souza J, et al. Brief report: the lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31:2827–32.
Guo J, McKenna SL, O’Dwyer ME, Cahill MR, O’Driscoll CM. RNA interference for multiple myeloma therapy: targeting signal transduction pathways. Expert Opin Ther Targets. 2016;20:107–21.
Bayram S, Sumbul A, Batmaci C, Genc A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumor Biol. 2015;36:3863–70.
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–100.
Hua Y, Duan S, Murmann AE, Larsen N, Kjems J, Lund AH, et al. miRConnect: identifying effector genes of miRNAs and miRNA families in cancer cells. PLoS ONE. 2011;6: e26521.
Yoon J-H, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14.
Sorensen K, Thomassen M, Tan Q, Bak M, Cold S, Burton M, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142:529–36.
Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–55.
Wang Y, Gong G, Xu J, Zhang Y, Wu S, Wang S. Long noncoding RNA HOTAIR promotes breast cancer development by targeting ZEB1 via sponging miR-601. Cancer Cell Int. 2020;20:320.
Zhang M, Wu K, Zhang P, Qiu Y, Bai F, Chen H. HOTAIR facilitates endocrine resistance in breast cancer through ESR1/miR-130b-3p axis: comprehensive analysis of mRNA-miRNA-lncRNA network. Int J Gen Med. 2021;14:4653–63.
Wu C, Taylor R, Lane D, Ladinsky M, Weisz J, Howell K. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1:963–75.
Buschman M, Rahajeng J, Field S. GOLPH3 links the Golgi, DNA damage, and cancer. Can Res. 2015;75:624–7.
Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R, et al. Overexpression of GOLPH3 Promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res. 2012;18:4059–69.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81960542 and 81960517), Science and Technology Project of Yunnan Provincial Science and Technology Department (grant no. 202001AU070053, 202001AU070093 and 202201AY070001-169), Yunnan Health Training Project of High Level Talents (grant no. H-2019075) and Beijing Science and Technology Innovation Medical Development Foundation (grant no. KC2021-JK-0044-6).
Author information
Authors and Affiliations
Contributions
LZ and JWZ contributed to the experiments and clinical data interpretation. JWZ and SCT discussed the results and analysed the data and wrote the manuscript conceived and designed the experiments. YZZ, NW and XZ contributed to the statistical analysis. RG, HML and CXL contributed to polishing the English to improve the quality of this manuscript. SCT, KZ and DQL supervised and directed the overall project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised to include the co-corresponding authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Zhang, L., Zhao, Y. et al. Long noncoding RNA HOTAIR promotes breast cancer development through the lncRNA HOTAIR/miR-1/GOLPH3 axis. Clin Transl Oncol 25, 3420–3430 (2023). https://doi.org/10.1007/s12094-023-03197-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03197-3