Abstract
Purpose
Long noncoding RNAs (lncRNAs) are outstanding as novel cancer biomarkers with great prospects. Herein, we focused on summarizing the overall diagnostic evaluation of lncRNAs for hepatocellular carcinoma (HCC).
Methods
Relevant literature was collected from the online databases. The Quality Assessment for Studies of Diagnostic Accuracy checklist was used to assess the quality of included studies. The pooled sensitivity, specificity, and diagnostic odds ratio (DOR) were plotted using random-effects models. Summary receiver operating characteristic curve and the area under the curve (AUC) were used to estimate the overall test performance. Statistical analysis was performed by STATA 14.0 and Meta-DiSc 1.4 software.
Results
Ten studies with a total of 820 HCC patients and 785 healthy controls were included. For overall lncRNAs, the pooled sensitivity, specificity, and DOR to predict HCC patients were 80% [95% confidence interval (CI) 77–82%], 79% (95% CI 76–81%), and 27.66 (95% CI 14.26–53.63), respectively, corresponding to an AUC of 0.91.
Conclusions
LncRNAs were a high diagnostic value for HCC and its expression could potentially be used as auxiliary biomarker in confirming HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors; its incidence ranks fifth and fatality rate ranks third in the world. It is a highly invasive and highly fatal tumor, easy to relapse and metastasize, and the prognosis is very poor [1]. The development of HCC is complex, involving multiple genes and pathways. Thus, the treatment of HCC is a global problem. Surgical resection is generally used to treat liver cancer, but it can be a difficult problem for patients who have lost the option of surgery [2]. With the continuous understanding of genes and the research of long noncoding RNA (lncRNA) in recent years, increasing evidence shows that the level of lncRNA expression in liver cancer has changed and has become important [3].
Long noncoding RNA is a noncoding RNA that is more than 200 nucleotides in length and has important biological function in the regulation of gene expression [4]. In recent years, a study showed that lncRNAs played a role in promoting cancer or tumor suppressor in the occurrence and development of cancer; they are involved in the process of cell apoptosis, tumor invasion, and metastasis [5]. LncRNA differentially expressed in the normal tissue and tumor tissue can be used as indicators of prevention and treatment, and prognosis of tumor [6]. Some literature has reported the relationship between lncRNAs and tumor prognosis by the method of meta-analysis [7, 8].
At present, although there are many studies in which lncRNAs can be used as biomarkers for tumor diagnosis, only a few studies have used meta-analysis to assess the overall value of lncRNAs in tumor diagnosis [9]. lncRNAs can be detected in the serum of HCC patients, which provides a new way for the diagnosis of HCC. In this study, on the basis of a comprehensive search of the relevant literature, the meta-analysis method is used to evaluate the overall diagnostic value of lncRNAs in HCC.
Materials and methods
Data sources and search strategy
We searched the PubMed and Embase with search terms of lncRNA or long non-coding RNA, liver or hepatocellular, cancer or carcinoma or tumor or neoplasm or cancers, serum or sera or serums or blood or plasma, diagnosis or sensitivity or specificity or ROC or AUC in October 30, 2016. Moreover, references in the literature and related literature reviews were also searched to prevent missed search results. Languages of the researched literature were limited to English, but there was no limit for the year of publication.
Inclusion and exclusion criteria
All the studies were carefully decided by three investigators independently on the basis of titles and abstracts, and the full text was then found for any potential eligibility. Any disagreement was resolved by full discussion to consensus. Furthermore, if necessary, we contacted the original authors for missing data. Inclusion standards for literature were as follows: (1) the diagnosis of HCC was based on histological examination; (2) the matched control individuals were included with a recent negative result of examination and without a personal history of any types of cancer; (3) all blood samples were collected prior to pathologic examination and without any treatment; (4) the researchers assessed the lncRNA in blood sample alone; (5) the studies should contain the data of sensitivity, specificity, or the possibility of deriving such values from the data; (6) only studies with more than 20 cases were included; and (7) all of the studies were published in English. Exclusion standards were as follows: (1) duplicate publications; (2) letters, editorials, meeting abstracts, case reports, and reviews; (3) unqualified patients and control subjects, as well as their blood samples; and (4) insufficient data. If the same authors reported their results acquired from the overlapping population or multiple published data in the different works, only the nearest or the most complete report was included.
Data extraction and quality assessment
Data extracted from each study included the first author, publication date, country, control sources, mean or median age, sample size or types, detection method, lncRNA expression patterns, and the diagnostic results. In case the study contained both training and validating tests, data from validating tests were extracted. Any disagreements were resolved by discussion. The quality of each study was assessed independently by three investigators according to Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) in Review Manager 5.3 (The Nordic Cochrane Center, Rigshospitalet, Denmark) [10]. The QUADAS-2 is recognized as an improved, redesigned tool that comprises four key domains (patient selection, index test, reference standard, and flow and timing) supported by signaling questions to aid judgment on risk of bias, rating risk of bias, and concerns about applicability as “high,” “unclear,” and “low,” and handling studies in which the reference standard consists of follow-up.
Statistical analysis
The STATA 14.0 (Stata Corporation, College Station, TX, USA) and Meta-Disc 1.4 (XI Cochrane Colloquium, Barcelona, Spain) softwares were used for the combination of diagnostic value of the literature included. All accuracy data from each study (true positives, false positives, true negatives, and false negatives) were extracted to obtain pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), positive predicted value, negative predicted value, diagnostic odds ratio (DOR), and their 95% CI, simultaneously, to generate the summary receiver operator characteristic (SROC) curve and calculate the area under the curve (AUC) [11]. The sensitivity, specificity, positive and negative predicted value, and diagnostic odds ratio of lncRNAs were presented as forest plots. Moreover, the heterogeneity between the studies caused by threshold effect was quantified using Spearman correlation analysis [12]. The non-threshold effect was assessed by the Cochran-Q method and the test of inconsistency index (I 2), and a low p value (≤0.1) and high I 2 value (≥50%) suggest the presence of heterogeneity caused by non-threshold effect. If p < 0.1 and I 2 > 50%, the random-effects model was used; if p > 0.1 and I 2 < 50%, the fixed-effect model was used [13]. If the non-threshold effect existed, meta-regression would be used to find out the sources. For publication bias, all eligible studies were assessed by Deek’s test using STATA 14.0 statistical software [14]. The p value with less than 0.05 shows a result of statistical significance.
Results
Literature search and study characteristics
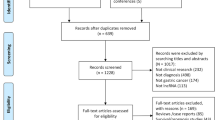
Preliminary literature search identified 246 relevant publications related to the current topic. The titles and abstracts from each article were carefully reviewed, and 138 of them were further excluded because they were review articles, letters, basic research, and so on. The retrieved 17 studies received full test review, and 7 studies were finally discarded because of the lack of sufficient data. Hence, many lncRNAs in the only 10 articles were included in this meta-analysis [15,16,17,18,19,20,21,22,23,24], such as urothelial cancer-associated 1 (UCA1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), hepatocellular carcinoma up-regulated long non-coding RNA (HULC), Linc00152, uc003wbd, AF085935, RP11-160H22.5, XLOC_014172, LOC149086, uc001ncr, AX800134, WD repeat containing antisense to TP53 (WRAP53), SPRY4 intronic transcript 1 (SPRY4-IT1), and JPX. The process of study selection is shown in Fig. 1a. Basic information of the inclused literature is shown in Tables 1 and 2. All these eligible studies were published from 2013 to 2016, accumulating 820 HCC patients and 785 healthy controls. Pathologic examination was considered as gold standard to diagnose HCC. The study characteristics included the first author, published year, country, the numbers of patients and controls, mean or median age, assay type, internal control, cut-off value, lncRNA name, sensitivity, specificity, and AUC.
The flow diagram of study selection process and quality assessment using the QUADAS checklist. a Flow diagram of study selection process. b Quality assessment of the included studies by QUADAS-2. It summarized “risk of bias” and “applicability concerns” through judging each domain for each included study
Quality assessment
QUADAS-2 quality assessment was used to assess the quality of the included studies. After the evaluation, we found that all included studies were on the upper middle quality as shown in Fig. 1b. However, there is an obvious major bias in those included studies. In other words, for these eligible studies, there are obvious shortcomings in the “index text,” which was 60% (6/10).
Heterogeneity analysis and threshold effect
To assess whether there is the heterogeneity of lncRNAs among the eligible studies, we first calculated the correlation coefficient and p value between the logit of true positive rate (TPR) and logit of false positive rate (FPR) using Spearman test to rule out the threshold effect. Spearman correlation coefficient is 0.022 (p = 0.943), suggesting that there is non-threshold effect. The Cochran-Q value of DOR is 64.21, and the inconsistency index (I 2) is 81.3%, p < 0.1, indicate that there is non-threshold effect, as shown in Fig. 2. The above results indicate that there is heterogeneity in the literature, which is considered to be related with the population, age, and so on; so, DerSimonian–Laird method should be used in the meta-analysis.
The forest plots show the pooled diagnosis index of lncRNAs for the diagnosis of hepatocellular carcinoma. The individual study symbol is shown as circle and the pooling symbol is shown as circle. Inconsistency is used to quantify the heterogeneity caused by non-threshold effect. For these studies, DerSimonian–Laird (REM) was used to pool these data. a, b Pooled sensitivity and specificity, c, d pooled PLR and NLR, e pooled DOR, and their 95% CI are illustrated separately, which shows that lncRNAs can be a potential diagnosis biomarker of HCC
Evaluation of diagnostic value
The random-effects model was used to estimate overall performance of lncRNAs in diagnosing HCC (Fig. 2). For overall lncRNAs, the diagnostic pooled sensitivity was 0.80 (95% CI 0.77–0.82), the specificity was 0.79 (95% CI 0.76–0.81), the positive likelihood ratio was 4.92 (95% CI 3.12–7.77), the negative likelihood ratio was 0.20 (95% CI 0.12–0.31), and the diagnostic odds ratio (DOR) was 27.66 (95% CI 14.26–53.63). The area under SROC was 0.91 (95% CI 0.88–0.93), as shown in Fig. 3. The above results show that lncRNAs have an advanced diagnostic value for liver cancer. However, because the heterogeneity caused by a non-threshold can be clearly observed in the forest plot of diagnosis index, we tried to explore the characteristics of the study using meta-regression, such as country, age, quality, reference gene, and cut-off value. Because the lncRNAs in the included literature were all derived from the blood of liver cancer and healthy people, specimen type and control sources were not sources of heterogeneity. If covariates have a missing value, use 0 instead of it during meta-regression. Meta-regression revealed p values more than 0.05 in all specified covariates, indicating that we could not find the sources of heterogeneity, which are shown in Table 3. Also, the overall distribution of studies is summarized in the likelihood matrix in Fig. 4a [25], and the Fagan’s nomogram described how to use diagnostic finding from lncRNA assay to calculate post-test probability of HCC (Fig. 4b) [26].
Fagan’s nomogram, likelihood matrix and publication bias from Deeks’ test. a Fagan’s nomogram describes the possibility of lncRNAs assay to confirm or exclude cancer patients. In detail, for any people with a pre-test probability of 20% to have HCC, if the lncRNAs test in cancer detection is positive, the post-test probability to have HCC will rise to 56%; while a negative result of lncRNAs assay means the post-test probability will drop to 5% for the same people. Hence, lncRNAs assay can play an important role as initial screening method for HCC. b The overall distribution of studies is summarized in the likelihood matrix. Each point corresponds to a study. Three studies, A.H. El-Tawdi et al., K.Wang et al. and M.M. Kamel et al. which reported the lncRNA-UCA1 are on the bottom left side of the matrix, indicating three sensitive “rule out” test. However, they report reasonable sensitivity with incorporation bias from knowledge of a desaturation study outcome. c Every point represents one study and the line is the regression line. The p value is of 0.54, which shows that no publication bias exists
Publication bias
The publication bias was recognized as another influencing factor to the diagnosis accuracy. Deeks’ test was used to evaluate whether the inclusive literature has publication bias. The p value of Deeks’ test was 0.54, indicating that lncRNAs had no publication bias among studies, as shown in Fig. 4c.
Discussion
With the completion of gene sequencing, it was found that only 1–2% of genes have the function of encoding protein, most of the noncoding sequences are transcribed into noncoding RNA (ncRNA) and about 80–90% have the transcriptional activity of the nonprotein coding gene, which is lncRNA [27]. Despite the large number of lncRNA, the research is far less than that for microRNA. Compared to lncRNA, microRNA was detected earlier; however, lncRNA research is still in its infancy. Recent studies suggest that lncRNA is capable of regulating important cellular signaling pathways in the regulation of transcription, posttranscriptional, and epigenetic levels [28]. Many lncRNAs have a regulatory role in cell homeostasis and proliferation, and some lncRNAs may play a regulatory role in apoptosis [29,30,31,32,33]. LncRNA regulates gene expression mainly including chromatin modification, transcription, and posttranscriptional processing. The related evidence between lncRNA and tumor is mainly derived from the differences in expression; the mechanisms of only a small number of lncRNAs are clear, maybe because of a lack of large samples of clinical research.
Although the development of diagnostic methods and surgical techniques in recent years has remarkably improved the prognosis of HCC patients, the 5-year survival rate for advanced HCC remains poor. The lack of diagnostic biomarkers accounts for the delay in early HCC detection [34]. Many noninvasive blood markers for HCC detection are available thus far. For example, AFP, gamma-glutamyl transferase (GGT), des-γ-carboxyprothrombin (DCP), HGF, HSP70, and IL-6 are currently used in detecting HCC [35,36,37]. Notwithstanding, these biomarkers are not ideal in confirming HCC because of the relatively low diagnostic accuracies. On the other hand, the pathologic diagnosis remains the most reliable diagnostic method for HCC detection, but it still yields the disadvantages of different degree of damage with relatively high surgery costs [38]. It is, therefore, necessary to identify novel diagnostic biomarkers for HCC screening. It has become increasingly apparent that the versatile lncRNA reveals a diagnostic role in various kinds of cancers including HCC [39]. Besides, many scholars are interested in making a holistic comprehensive study for the relationship between one biomarker of tumor diagnosis and prognosis by meta-analysis [40]; for instance, we have studied the relationship between alkaline phosphatase and osteosarcoma prognosis by the method of meta-analysis [13].
In this meta-analysis, we found that inconsistent expression levels of overall lncRNAs in blood have a positive statistical significance between HCC patients and the healthy control individuals. Ten papers were included in this meta-analysis by screening. Because there is no detailed description of the time interval between pathology and lncRNAs in the blood of qRT-PCR, it is possible to ignore the deviation of the detection results because of the course of disease. Heterogeneity is an important reference factor for meta-analysis, and the threshold effect is the most important factor for the heterogeneity of diagnostic meta-analysis. This paper used the correlation coefficient of Spearman model to make clear whether the ten studies’ different thresholds were not caused by heterogeneity. Forest plots and I 2 showed that heterogeneity did exist; hence, meta-regression analysis was used to determine the possible causes of the heterogeneity. Meta-regression revealed p values more than 0.1 in all specified covariates, indicating that we could not find the sources of heterogeneity. As a result, the heterogeneity of the sources of uncertainty was identified, and the random-effects model was used to merge the ten papers into the diagnostic value of the literature. The combined indices included lncRNAs’ diagnosis value in HCC, such as sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio. The summary receiver operator characteristic curve area showed that lncRNAs in liver cancer had good diagnostic value. As a conclusion, after analyzing HCC and healthy controls, the lncRNAs yielded an AUC of 0.91 with 80% pooled sensitivity and 79% pooled specificity, suggesting its potential value for noninvasive diagnosis of HCC. The diagnostic odds ratio (DOR), represented as a compact index between diagnostic efficiency and the cases, has excellent test performance with an extremely higher value. In our study, the DOR value of 27.66 prompted an advanced diagnostic accuracy for diagnosing HCC.
In conclusion, our study demonstrates that lncRNA expression profiles showed an advanced accuracy in differentiating HCC patients and cancer-free individuals. LncRNA profiling reveals promising value in the management of HCC. If validated in a large-scale study, lncRNA might be useful as a noninvasive screening tool for clinical practice of HCC. Of course, more studies are, therefore, needed to highlight the value of lncRNAs as supplemental test in the occurrence and development of HCC in the future.
References
Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–4.
Anestopoulos I, Voulgaridou GP, Georgakilas AG, Franco R, Pappa A, Panayiotidis MI. Epigenetic therapy as a novel approach in hepatocellular carcinoma. Pharmacol Therapeut. 2015;145:103–19.
Collins JF. Long noncoding RNAs and hepatocellular carcinoma. Gastroenterology. 2015;148:291–4.
Xue M, Chen W, Li X. Urothelial cancer associated 1: A long noncoding RNA with a crucial role in cancer. J Cancer Res Clin Oncol. 2016;142:1407–19.
Chang Y, Zhang K, Hu Z, Qi H, Shi Z, Han X, et al. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575:1–8.
Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: LncRNAs in cancer. J Clin Invest. 2016;126:2775–82.
Yu S, Li G, Wang Z, Wang Z, Chen C, Cai S, et al. The prognostic value of pSTAT3 in gastric cancer: a meta-analysis. J Cancer Res Clin Oncol. 2016;142:649–57.
Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15:50.
Chen J, Chen Y, Gu L, Li X, Gao Y, Lyu X, et al. LncRNAs act as prognostic and diagnostic biomarkers in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget. 2016;7(45):74325–36. doi:10.18632/oncotarget.11101.
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36.
Chu H, Guo H. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2009;10:201–3.
Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58.
Hao H, Chen L, Huang D, Ge J, Qiu Y, Hao L. Meta-analysis of alkaline phosphatase and prognosis for osteosarcoma. Eur J Cancer Care (Engl). 2016. doi:10.1111/ecc.12536.
Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–93.
Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, Hiramoto H, et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 2016;107:149–54.
Jing W, Gao S, Zhu M, Luo P, Jing X, Chai H, et al. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncol Rep. 2016;36:1085–92.
El-Tawdi AH, Matboli M, El-Nakeep S, Azazy AE, Abdel-Rahman O. Association of long noncoding RNA and c-JUN expression in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2016;10:869–77.
Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AH. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res. 2016;168:134–45.
Ma W, Wang H, Jing W, Zhou F, Chang L, Hong Z, et al. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016. doi:10.1016/j.clinre.2016.09.002 (In Press).
Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumour Biol. 2015;36:3231–6.
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, et al. HULC and linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem. 2015;37:687–96.
Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505–15.
Wang K, Guo WX, Li N, Gao CF, Shi J, Tang YF, et al. Serum LncRNAs profiles serve as novel potential biomarkers for the diagnosis of HBV-Positive hepatocellular carcinoma. PLoS ONE. 2015;10:e144934.
Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106.
Aznar-Oroval E, Mancheno-Alvaro A, Garcia-Lozano T, Sanchez-Yepes M. Likelihood ratio and Fagan’s nomogram: 2 basic tools for the rational use of clinical laboratory tests. Rev Calid Asist. 2013;28:390–1.
Fusar-Poli P, Cappucciati M, Rutigliano G, Heslin M, Stahl D, Brittenden Z, et al. Diagnostic stability of ICD/DSM first episode psychosis diagnoses: meta-analysis. Schizophr Bull. 2016;42:1395–406.
Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–910.
Esposti DD, Hernandez-Vargas H, Voegele C, Fernandez-Jimenez N, Forey N, Bancel B, et al. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget. 2016;7:31862–77.
Ouyang SS, Zhang P, Wang J, Huang ZK, Liao L. Expression of long non-coding RNA colon cancer associated transcript 2 and its clinicopathologic significance in oral squamous cell carcinoma. Zhonghua Kou Qiang Yi Xue Za Zhi. 2016;51:286–91.
Nie F, Yu X, Huang M, Wang Y, Xie M, Ma H, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2016. doi:10.18632/oncotarget.9611.
Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–52.
Qu S, Yang X, Song W, Sun W, Li X, Wang J, et al. Downregulation of lncRNA-ATB correlates with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2016;37:3933–8.
Zhao JH, Sun JX, Song YX, Chen XW, Yang YC, Ma B, et al. A novel long noncoding RNA-LOWEG is low expressed in gastric cancer and acts as a tumor suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol. 2016;142:601–9.
Chauhan R, Lahiri N. Tissue- and serum-associated biomarkers of hepatocellular carcinoma. Biomark Cancer. 2016;8:37–55.
Wang Z, Gou W, Liu M, Sang W, Chu H, Zhang W. Expression of p53 and HSP70 in chronic hepatitis, liver cirrhosis, and early and advanced hepatocellular carcinoma tissues and their diagnostic value in hepatocellular carcinoma: an immunohistochemical study. Med Sci Monit. 2015;21:3209–15.
Kudo M, Izumi N, Sakamoto M, Matsuyama Y, Ichida T, Nakashima O, et al. Survival analysis over 28 years of 173,378 patients with hepatocellular carcinoma in Japan. Liver Cancer. 2016;5:190–7.
Li Z, You K, Li J, Wang Y, Xu H, Gao B, et al. Madecassoside suppresses proliferation and invasiveness of HGF-induced human hepatocellular carcinoma cells via PKC-cMET-ERK1/2-COX-2-PGE2 pathway. Int Immunopharmacol. 2016;33:24–32.
Smekalova EM, Kotelevtsev YV, Leboeuf D, Shcherbinina EY, Fefilova AS, Zatsepin TS, et al. LncRNA in the liver: prospects for fundamental research and therapy by RNA interference. Biochimie. 2016;131:159–72.
Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78.
Cui Z, Chen Y, Xiao Z, Hu M, Lin Y, Chen Y, et al. Long noncoding RNAs as auxiliary biomarkers for gastric cancer screening: a pooled analysis of individual studies. Oncotarget. 2016;7:25791–800.
Acknowledgements
We thank Elsevier’s English Language Editing service for editing the manuscript. This work was supported by grants from the National Natural Science Foundation of China (81560475).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts to disclose.
Additional information
C. Zheng and H. Hao equally contributed to this work.
Rights and permissions
About this article
Cite this article
Zheng, C., Hao, H., Chen, L. et al. Long noncoding RNAs as novel serum biomarkers for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Clin Transl Oncol 19, 961–968 (2017). https://doi.org/10.1007/s12094-017-1626-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-017-1626-1