Abstract
This study was conducted to evaluate the ambient water bacterial dynamics and its influence on the gut of resident teleosts; Oreochromis niloticus. The bacterial communities in the ambient water and the gastrointestinal tract (GIT) of O. niloticus were profiled using a culture-dependent method and followed by the 16S rRNA gene sequencing. The results indicated bacterial phyla of Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes respectively between the two microbial consortia. However, the relative abundance among the bacterial phyla notably differed between the two consortia. The relative abundance of Proteobacteria (≤ 67%) was dominant in both consortia, but contrarily, Actinobacteria and Bacteroidetes were higher in the ambient water relative to the GIT which indicated Proteobacteria and Bacilli as the highest diversity. Nevertheless, the relative abundance of γ-proteobacteria and Bacilli remarkably dominated both consortia at the class level, with Bacillus and Pseudomonas being the most abundant operational taxonomic units (OTUs).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aquaculture is a fast-growing department in the animal production sector and will eventually subdue the capture fisheries. During the last three decades, the global aquaculture production contributed substantially about 17% of the total animal protein supply [1]. However, lack of sustained effective disease control measures has been the major limiting factor for the expansion and profitability of the sector which often results to the unauthorized or unregulated use of antibiotics, especially in developing countries where regulations are not properly defined or enforced [2]. Coupled with growing concerns about the presence of antibiotic compounds in the foods and animal production systems, indicating the significance of antibiotic resistance in the bacterial communities; a serious threat to consumer safety [3]. For example, the European Union in 2006 widely banned the use of antibiotics as growth promoters (EU Regulation No. 1831/2003), making it possible for the development and application of functional feeds with selected additives (examples: probiotics, prebiotics, and phytogenics) to foster an enhanced animal health [4]. This ultimately promoted research interests in alternative—antibiotics (biotherapeutics) which includes the functional use of probiotics and prebiotics during treatment of diseases and prophylaxis [5] in aquaculture [6, 9].
Probiotics and prebiotics are microbial feed supplements which beneficially influence hosts intestinal balance by the manipulation of the gut microbiota for the enhancement of host-fitness through the exclusion of opportunistic pathogens [12]. The gut microbiota improves host health by helping the development of gut epithelium, biosynthesization of essential nutrients, and stimulating the innate immune system [8, 10]. Thus, augmentation of the gut bacterial communities with probiotics may enhance disease prevention through a variety of mechanisms [7].
Generally, aquatic environments exposed fishes to higher microbial loads compared to their terrestrial animal counterparts. In aquatic environments, the fish gut-microbiota is initiated at an early stage during growth, but advanced at later stages of growth by feed-associated microflora, suggesting that the environmental food-sources are the major source of fish gut-microbiota [11]. Contrarily, aquaculture management systems (e.g. recirculatory and active suspension systems) control microbial proliferation through nutrient and solid regulation and bioflocs formulation to maintain good water quality.
In aquaculture, the characterization of gut microbiota of relevant fish species resulted in the definition of the core gut-microbiota in many species which serve as a guide for choosing potential probiotic or prebiotic candidates [6]. Nevertheless, lactic acid bacteria and Bacillus are predominantly used as probiotics in aquaculture [1]. However, there are cases where exogenous bacteria appeared significantly beneficial in fish against lactic acid bacteria or Bacillus [13, 14]. Meanwhile, the microbiomes of the fish-gut or their growth environments has been extensively studied, there are no reports on the concurrent study between the microbiota or microbiomes of fish-gut and their growth environment. Similarly, Yan et al. recently profiled the bacterial community associated with the growth environment of Pacific white shrimp [15] but irrespective of their gut-microbiota which was also independently reported by other studies [16,17,18].
Since the ambient bacterial community influences the gut of aquatic animals [6], the concurrent characterization of both gut and environmental microbiota is necessary for the identification and selection of more robust functional probiotic candidates for the general improvement of aquaculture welfare and production [15].
Considering that tilapia feeds on wide range of foods [11] including zooplankton, phytoplankton, detritus, etc. we hypothesized that all microorganisms closely associated with these food-sources might colonize the GIT of tilapia, whereby the gut acidic condition might eventually provide some selective background suitable for the survival of only a few to perpetuate, serving as a guide for the selection and functional application of potential probiotics. To test this hypothesis, we profiled the cultivable-bacterioplankton dynamics between the ambient water and GIT of tilapia using the culture-dependent approach for the pure-culture isolations and the 16S rRNA gene sequencing to identify the bacterial isolates for prospective application studies.
Materials and Methods
Sample Collection
A total of fifteen O. niloticus (Nile tilapia) were sampled at Ibusuki city (31° 16′ 27″ N, 130° 37′ 07″ E) in Kagoshima prefecture of Japan (Fig. 1); one of the major tilapia reservoirs in Japan [19, 20] using baited hooks and lines and scoop nets. Fishes [weight = 112 ± 3.2 g (mean ± standard error)] were kept alive in aerated coolers filled with lake water until processing. Similarly, 500 ml of habitat water (ecological niche) was sampled independently at the sampling location and stored at 4 °C until processed.
Recovery of Bacterial Microbiota by Culture-Dependent-Approach
Upon arrival at the laboratory, fish were immediately euthanized with an overdose of MS-222 (250 mgl−1). The lower one-third of the intestine was aseptically removed, and the contents were squeezed into a sterile Erlenmeyer flask containing 10 ml PBS (-KCl); intestinal content from all individuals was pooled together to form a single homogenized sample (stock) after 5 min of homogenization. Aliquots (1000 μl) of both the homogenate (stock) and habitat water samples were subjected to tenfold serial dilutions in sterile PBS (-KCl) and subsequently cultivated in triplicates on nutrient agar using the spread plate method.
The standard media (nutrient agar) for the culture-dependent analysis was prepared to contain 0.3% (w/v) beef extract, 0.5% (w/v) peptone, and 1.5% (w/v) agar in a liter of distilled water and final pH adjusted to 7.4 ± 0.2 and autoclaved (sterilized) at 121 °C for 15 min. The sterilized media was cooled to 55 °C and approximately 20 ml each was dispensed into disposable Petri dishes and allowed to solidify at room temperature overnight. Subsequently, aliquots (100 μl) of the serially diluted samples were spread onto the nutrient agar plates and incubated at 28 °C for ≤ 72 h. Bacterial colony examination was performed by a comprehensive culture-dependent approach (available at http://www.microbelibrary.org/component/resource/laboratory-test/3136-colony-morphology-protocol) to extract colonies of difference for pure culture preparations. Extracted bacterial pure cultures were further stored in 50% glycerol at −8 0 °C.
Bacterial Composition by Sequencing
Genomic DNA was extracted by suspending a loopful of 2–3 days old pure-cultured bacterial isolates in proteinase K (10% KOH) solution (Macherey–Nagel, Düren, Germany) using NucleoSpin® Tissue kit (Macherey–Nagel, Düren, Germany) extraction protocols. The quality and concentration of eluted DNA were estimated by spectrophotometer (NanoDrop 2000/200c, Thermo Fisher Scientific Inc., U.S.A). Extracted genomic DNAs were used as templates for PCR amplification of about 1500 base pair (bp) fragment of 16S rRNA using two universal bacteria-specific primers: 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The PCR amplification reaction was set to a volume of 50 µl, containing 36.5 µl of sterilized Milli-Q water, 5.0 µl each of 10 × PCR Buffer and 2 mM dNTPs, 1.0 µl each of 10 pmolµl−1 forward and reverse primers, 0.5 µl of 2.5U µl−1 Blend Taq® (TOYOBO Co. Ltd., Japan), and 10 ngµl−1 of template DNA. The cycling conditions consisted of an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 45 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 15 min. Samples were then cooled and kept at 4 °C until use. PCR products were examined for yield and size on 1.0% (w/v) agarose gel in 1 × TAE buffer loaded into a MUPID-2 plus Submarine Electrophoresis System. Each 5.0 μl of PCR product mixed with 1.0 μl loading buffer was dispensed into each well, and 100 V electrical current was applied for 30 min. The gel was stained in ethidium bromide fluorogenic dye for 25 min and examined under UV light at 100 nm. Successful amplicons with expected band sizes were purified using NucleoSpin Gel and PCR Clean-up kits (Macherey–Nagel, Düren, Germany) and sequenced at FASMAC Co. Ltd. (Kanagawa, Japan) using a 3730xl Genetic Analyzer (Applied Biosystems, USA).
Sequence Analysis
The quality of the 16S rRNA gene sequences was analyzed in BioEdit by confirming the presence of variable sites by manual inspection of the chromatograms and subsequent manually editing. The sequences were then analyzed using the Basic Local Alignment Search Tool (BLAST) against the non-redundant database at the National Center for Biotechnology Information (NCBI). Subsequently, identification of bacterial taxonomy relied on the 16S rRNA gene as a conventional method for routine bacterial identification at ≥ 97% sequence similarity.
Results
Bacterial Composition and Diversity
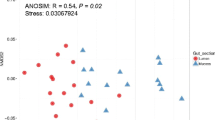
A total of 278 bacterial isolates were retrieved and subsequently screened subject to colony morphology eventually yielded 44 pure culture isolates and their corresponding sequences based on the 16S rRNA pyrosequencing which ultimately generated a total of 4 phyla (Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria), 7 classes (Actinobacteria, Flavobacteria, Sphingobacteria, Bacilli, α-proteobacteria, β-proteobacteria, and γ-proteobacteria), 17 families (Nocardiaceae, Microbacteriaceae, Flavobacteriaceae, Sphingomonadaceae, Bacillaceae, Paenibacillaceae, Caulobacteraceae, Rhizobiaceae, Phyllobacteriaceae, Comamonadaceae, Enterobacteriaceae, Pseudomonadaceae, Morganellaceae, Vibrionaceae, Xanthomonadaceae, and Moraxellaceae), and 23 genera (Rhodococcus, Microbacterium, Chryseobacterium, Pedobacter, Bacillus, Paenibacillus, Lysinibacillus, Brevundimonas, Rhizobium, Aminobacter, Aquamicrobium, Novosphingobium, Diaphorobacter, Aeromonas, Pseudomonas, Klebsiella, Plesiomonas, Buttiauxella, Enterobacter, Providencia, Vibrio, Pseudoxanthomonas, and Acinetobacter) of relevance for the definition of the OTUs (Table 1). The relative uniqueness of associated bacterioplankton OTUs indicated 56.52% and 21.74% for the environment and GIT respectively. Nevertheless, the environments and GIT shared OTUs indicated 21.74% (Fig. 2; Table 1). On the other hand, the environment indicated a higher abundance of percentage compared to the GIT. Ultimately, the OTU identification at ≥ 97% indicated the microbial richness and evenness between the environment and GIT (Table 1).
Relative Microbiota Composition
Four bacterial phyla each were identified from the environment and GIT (Fig. 3, Table 1) respectively, such that, the phylum Proteobacteria (α, β, and γ) made up the majority of all sequences both from the environment (60%) and GIT (67%). Within the Proteobacteria, the fish GIT microbiota was dominantly composed of γ-proteobacteria, followed by α-proteobacteria and β-proteobacteria. Actinobacteria (20%) was the second most predominant phylum from the environment, followed by Bacteroidetes (10%) and Firmicutes (10%) respectively. On the other hand, the second and third most common phylum from the GIT is the Firmicutes (17%) and Actinobacteria (11%) respectively, followed by the phylum Bacteroidetes (5%). The common phyla composition between the environment and GIT differed in relative abundances and evenness with some degree of uniqueness for each community. The microbiota between the environment and GIT indicated 5 shared and 5 unique genera from the environment, and 13 unique genera from the GIT (Table 1). Comparatively, the most dominant sequences were identified as Bacillus. Relatively, other abundant genera included Pseudomonas and Aeromonas. The 21.74% shared genera (i.e. Rhodococcus, Microbacterium, Bacillus, Aeromonas, and Pseudomonas) identified from all sequences from both microbial communities, suggested some degree of similarity in the bacterial composition of the environment and GIT of resident fish fauna at the genus level.
Comparison of the dominant bacterial populations at genus level between the ambient water (ecological niche) and GIT of resident O. niloticus. The operational taxonomic units (OTUs) were clustered at genus level based on the lowest taxon information. Pie charts indicate the relative abundance of each dominant OTU (> 1%) between the two microbial consortia. (green and blue indicated the relative bacterial abundance in the ambient water and GIT respectively); and the size of the pie chart represent the total proportional dominance of OTUs between the two microbial consortia (color figure online)
The main differences between the two microbial communities were due to the varying genus composition of Proteobacteria and Firmicutes, whose species composition cannot be readily established in this study. Contrarily, the high abundance in the genus of Bacillus, Pseudomonas, and Aeromonas as identified by the 16S rRNA sequences indicated high sequence similarities in the environment relevant to these genera. Additionally, the high diversity of the phylum proteobacteria (for example, γ-proteobacteria) indicated high sequence dissimilarity in the GIT compared to the environment. Most of the sequences from the phylum proteobacteria had a relatively equal abundance. However, the relatively higher abundance of Bacillus, Pseudomonas, and Aeromonas suggested bacterioplankton separation between the aquatic environment and GIT of resident fish fauna; an indication that fish–guts are selective of the microbial–ecology.
Discussion
Studying the microbiota associated with fish are often limited to either the gut of fish species [15] or the growth environment [16,17,18], but results from relative studies between several fish species from the same environment showed that gut-microbiota specificity between fish species is significantly correlated with environmental food-sources and feeding habit of fishes [11]. Considering the feeding habit of tilapia, our results indicated a comparative difference in bacterial evenness between the gut microbiota of O. niloticus and the same growth environment. Although, the tilapia is known to have varying diet preferences and feeding behaviors in their natural environments, majority of which have microbial association within a balanced ecosystem, but our results was limited to the cultivable-bacterial flora irrespective of the Viable-But-Not-Cultivables (VBNCs) and slow growing bacterial components of the two examined consortia.
The feeding habit of tilapia is omnivorous and detritivorous, feeding on phytoplankton, zooplankton, nematode, and detritus [21]. As all the tilapia samples used in this study were ≥ 112 ± 3.2 g they were likely consuming phytoplankton, zooplankton, and detritus as their main diet within the natural environment, and therefore allowing diverse bacterial flora associated with food to colonize their gut [22]. Although stomach content was not analyzed in this study, our results potentially reflect these diet differences.
Nevertheless, the concept of “forward microbiomics” implies the regulation of gut flora for the advancement of animal health, which of course is very crucial in aquaculture [6, 24]. The relevance of the microbiota on its host has a plentitude of functional significance on behavior and functional physiology. It was acknowledged that the characterization of fish-gut associated microbiota [23] showed some remarkable specificity for fish species [11, 15], meanwhile, some members of the microbiota have been shown to produce bioactive substances that subdue opportunistic pathogens [4, 9, 22]. Moreover, previous studies have indicated the microbiota diversity in fish gut relative to diet preferences and feeding behaviors with increased diversity from carnivores to omnivores to herbivores in mammals [8, 10, 14, 18]. However, our study indicated a significant number of OTUs associated with herbivory and detritivory [23, 24]; consequently, species proportionality was higher in the GIT as compared to the ambient water.
The 16S rRNA gene sequencing identified differences between the ambient water and fish-gut microbiota in terms of bacterial abundances at each taxonomic level. The phylum Proteobacteria was the main component of the consortia followed by Firmicutes. Previous studies have shown Proteobacteria as dominant member of the gut microbiota of freshwater fishes [4, 6, 18, 23, 24]. Generally, the representative OTUs identified by our results indicated an obvious difference in bacterial flora. Nevertheless, the microbial structure of a particular ecological niche is moderated by factors, such as salinity, temperature, organic matter, and pH. However, the constant modification in water quality as a result of anthropogenic or natural events might cause the microbial structure to change and ultimately influence the gut microbiota of resident fish fauna repeatedly. Thus, the gut-microbiota of inhabitant fish fauna may continue to respond by the recruitment of specific microbial taxa in correspondence to the effective distribution of both abundant and rare niche microbiota [15]. Additionally, the fish-gut can be segmented into three components, thus, fore-gut, mid-gut (stomach), and hind-gut, which is believed to influence gut selectivity during fish-gut colonization relative to the of gastric fluids in each section of the gut.
Related sequences of Bacilli and γ-proteobacteria dominated both microbial consortia, suggesting that such bacterial communities are more resilient to environmental stress. However, the availability of sequences closely related to the functional groups of Rhodococcus, Pedobacter, Rhizobium, Aminobacter, and Aquamicrobium suggested their usefulness in the nitrogen cycles and biosynthesizing of protein from indirect sources of protein within the environment and gut of fishes. Similarly, the availability of Rhodococcus, Bacillus, Acinetobacter, and Paenibacillus suggested their usefulness in biodegradation and fermentation activities. To some extent, some of these microbes could be actively performing principal ecosystem services, particularly, nutrient cycling, bioremediation, and maintaining good water quality [9, 18, 24].
Notably, closely related sequences classified as the γ-proteobacteria featured potential pathogenic isolates (i.e. Providencia, Pseudomonas, Klebsiella, Plesiomonas, Buttiauxella, Enterobacter, Aeromonas, and Vibrio). The pathogenic relatives of these isolates are ubiquitous and could associate with different disease conditions in aquatic and terrestrial animals. The pathogenicity of Aeromonas, Vibrio, and Pseudomonas is well known in aquaculture, however, during outbreaks or mass mortalities, unauthorize or unregulated use of antibiotics [2] could trigger the development of antibiotic development among these microbial communities which ultimately might induce different zoonotic and multi-drug resistant conditions [3] among different consumers.
However, some of these seems to be a normal component of the fish-gut microbiota, but the coexistence of pathogens (Vibrio, Klebsiella, Plesiomonas, Pseudomonas, Aeromonas, Enterobacter, and Providencia) amidst medically bioactive bacteria (Rhodococcus, Acinetobacter, and Paenibacillus) suggested a kind of complex interplay of microbial activities that could competitively, sequentially, or symbiotically regulate pathogenic actions and ultimately reduce disease incidences [7, 12]. Even though several studies were done on fish-gut and environmental microbiomes [6, 13,14,15], which is serving as the lead guide for choosing probiotics candidates [6, 25], this is the first study to focus on cultivable bacterial consortia concurrently between the ecological niche and GIT of resident teleost.
In conclusion, this is the first concurrent study to characterize the culturable bacterial microbiota between the environment and GIT of an economically important teleost (O. niloticus). Despite the high proportional taxonomic differences observed between ambient water and GIT of resident tilapia species, a good proportion of same bacterial genera in microbiota composition was shared, sparking a myth about microbial cohabitation impact of these bacterial isolates in fish gut. Prospective studies are scheduled to ascertain the cohabitation impact of these bacterial isolates towards the selection of potential probiotic candidates and for further application studies in aquaculture.
References
FAO (2016) World aquaculture production by species groups. In FAO Yearbook. Fish Aquac Stat 2014:52–53
Chuah L, Effarizah ME, Goni AM, Rusul G (2016) Antibiotic application and emergence of multiple antibiotic resistance (MAR) in global catfish aquaculture. Curr Environ Health Rep 3:118–127
Muziasari WI, Parnanen K, Johnson TA, Lyra C, Karkman A, Stedtfeld RD, Tamminen M, Tiedje JM, Virta M (2016) Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol Ecol 92(4):fiw052. https://doi.org/10.1093/femsec/fiw052
Dawood MAO, Koshio S, Esteban MA (2018) Beneficial roles of feed additives as immunostimulants in aquaculture: a review. Rev Aquacult 10:950–974. https://doi.org/10.1111/raq.12209
Pauly D, Zeller D (2016) Catch reconstructions reveal that global marine fisheries catches are higher than reported and declining. Nat Commun 7:10244
Tarnecki AM, Burgos FA, Ray CL, Arias CR (2017) Fish intestinal microbiome: diversity and symbiosis unraveled by metagenomics. J Appl Microbiol 123:2–17. https://doi.org/10.1111/jam.13415.4
Price LB, Hungate BA, Koch BJ, Davis GS, Liu CM (2017) Colonizing opportunistic pathogens (COPs): the beasts in all of us. PLoS Pathog 13:e1006369. https://doi.org/10.1371/journal.ppat.1006369
Kumar R, Sood U, Gupta V, Singh M, Scaria J, Lal R (2019) Recent advancements in the development of modern probiotics for restoring human gut microbiome dysbiosis. Indian J Microbiol. https://doi.org/10.1007/s12088-019-00808-y
Comte J, Langenheder S, Berga M, Lindström ES (2017) Contribution of different dispersal sources to the metabolic response of lake bacterioplankton following a salinity change. Environ Microbiol 19:251–260. https://doi.org/10.1111/1462-2920.13593
Durack J, Lynch SV (2019) The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med 216:20–40. https://doi.org/10.1084/jem.20180448
Ringø E, Zhou Z, Vecino JLG, Wadsworth S, Romero J, Kroghdal A, Olsen RE, Dimitoglou A (2016) Effect of dietary components on the gut microbiota of aquatic animals: the never-ending story. Aquac Nutr 22:219–282
Sood U, Bajaj A, Kumar R, Khurana S, Kalia VC (2018) Infection and microbiome: impact of tuberculosis on human gut microbiome of indian cohort. Indian J Microbiol 58:123–125. https://doi.org/10.1007/s12088-018-0706-4
Yue H, Huang X, Ruan R, Ye H, Li Z, Li C-J (2019) Effects of dietary protein levels on the growth, body composition, serum biochemistry and digestive enzyme activity in Chinese rice field eel (Monopterus albus) fingerlings. Aquac Res 51:400–409
Buret AG, Motta JP, Allain T, Ferraz J, Wallace JL (2019) Pathobiont release from dysbiotic gut microbiota biofilms in intestinal inflammatory diseases: a role for iron? J Biomed Sci 26:1. https://doi.org/10.1186/s12929-018-0495-4
Yan M, Zhang X, Hu L, Huang X, Zhou Q, Zeng G, Zhang J, Xiao G, Chai X, Chen J et al (2020) Bacterial community dynamics during nursery rearing of pacific white shrimp (Litopenaeus vannamei) revealed via high-throughput sequencing. Indian J Microbiol 60:214–221. https://doi.org/10.1007/s12088-019-00853-7
Fan JQ, Chen LM, Mai GQ, Zhang HR, Yang JF, Deng D, Ma YF (2019) Dynamics of the gut microbiota in developmental stages of Litopenaeus vannamei reveal its association with body weight. Sci Rep 9:734. https://doi.org/10.1038/s41598-018-37042-3
Xiong J, Xuan L, Yu W, Zhu J, Qiu Q, Chen J (2019) Spatiotemporal successions of shrimp gut microbial colonization: high consistency despite distinct species pool. Environ Microbiol 21:1383–1394. https://doi.org/10.1111/1462-2920.14578
Cornejo-Granados F, Gallardo-Becerra L, Leonardo-Reza M, Ochoa-Romo JP, Ochoa-Leyva A (2018) A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ 6:e5382. https://doi.org/10.7717/peerj.5382
Fatsi PSK, Hashem S, Kodama A, Appiah EK, Saito H, Kawai K (2020) Population genetics and taxonomic signatures of wild Tilapia in Japan based on mitochondrial DNA control region analysis. Hydrobiol 847(6):1491–1504. https://doi.org/10.1007/s10750-020-04203-3
Fatsi PSK, Hashem S, Appiah EK, Mensah ET-D, Setufe SB, Saito H, Kawai K (2021) Morphological divergence within the largest genetically consistent group of wild Tilapia. Environ Biol Fish. https://doi.org/10.1007/s10641-021-01098-4
Ganesan K, Chung SK, Vanamala J, Xu B (2018) Causal relationship between diet-induced gut microbiota changes and diabetes: a novel strategy to transplant faecalibacterium prausnitzii in preventing diabetes. Int J Mol Sci 19:3720. https://doi.org/10.3390/ijms19123720
Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T (2017) Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358:359–365. https://doi.org/10.1126/science.aan4526
Zhu HJ, Qiang J, Tao YF, Ngoepe TK, Bao JW, Chen DJ, Xu P (2020) Physiological and gut microbiome changes associated with low dietary protein level in genetically improved farmed tilapia (GIFT, Oreochromis niloticus) determined by 16S rRNA sequence analysis. Microbiol 9:e1000
Ray C, Bujan N, Tarnecki A, Allen DD, Browdy C, Cr A (2017) Analysis of the gut microbiome of Nile tilapia Oreochromis niloticus L. fed diets supplemented with previda®and saponin. J Fish Sci 11:36
Liu W, Wen H, Luo Z (2018) Effect of dietary protein levels and feeding rates on the growth and health status of juvenile genetically improved farmed tilapia (Oreochromis niloticus). Aquac Int 26:153–167
Acknowledgements
We extend our gratitude to the Japanese Government, through the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and the Graduate School of Integrated Sciences for Life (under the Sustainable Food Production program; S.F.P.P), Hiroshima University, Japan, for financially supporting this study. We are equally grateful to the Hiroshima University’s Oceanography Cruise (Toyoshio-Maru) Team for their valuable support during field sampling.
Funding
This study was funded by the Japanese Government, through the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and the Graduate School of Integrated Sciences for Life, Hiroshima University, Japan; under the Sustainable Food Production program (S.F.P.).
Author information
Authors and Affiliations
Contributions
All authors have contributed significantly, have read the manuscript, attested to the validity and legitimacy of the data and its interpretation, and agree to its submission.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declared that there is no conflict of interest regarding the publication of this paper.
Data Availability
The datasets generated during and/or analyzed in this study are available from the corresponding author on reasonable request.
Ethical Approval
All efforts made to minimize the suffering of experimental animals were under the animal care 111, and experimental procedures were conducted according to the Ethical practices approved in 112 Animal Experimentation at Hiroshima University (Permit Number: G13-3), even though such recommendations are not strictly enforced for teleost research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fatsi, P.S.K., Appiah, E.K., Ogasawara, C. et al. 16S rRNA Gene Sequence Identification of Cultivable-Bacterioplankton Between Ambient Water and Gastrointestinal Tract (GIT) of Resident Teleost. Indian J Microbiol 62, 187–194 (2022). https://doi.org/10.1007/s12088-021-00992-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-021-00992-w