Abstract
Interleukin-10 (IL-10) is an anti-inflammatory cytokine with important immunoregulatory functions. It is primarily secreted by antigen-presenting cells such as activated T-cells, monocytes, B-cells and macrophages. In biologically functional form, it exists as a homodimer that binds to tetrameric heterodimer IL-10 receptor and induces downstream signaling. IL-10 is associated with survival, proliferation and anti-apoptotic activities of various cancers such as Burkitt lymphoma, non-Hodgkins lymphoma and non-small scell lung cancer. In addition, it plays a central role in survival and persistence of intracellular pathogens such as Leishmania donovani, Mycobacterium tuberculosis and Trypanosoma cruzi inside the host. The signaling mechanisms of IL-10 cytokine are not well explored and a well annotated pathway map has been lacking. To this end, we developed a pathway resource by manually annotating the IL-10 induced signaling molecules derived from literature. The reactions were categorized under molecular associations, activation/inhibition, catalysis, transport and gene regulation. In all, 37 molecules and 76 reactions were annotated. The IL-10 signaling pathway can be freely accessed through NetPath, a resource of signal transduction pathways previously developed by our group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interleukin-10 (IL-10), also known as ‘cytokine synthesis inhibitory factor’ (CSIF), is a pleiotropic cytokine with important immunoregulatory functions. It has anti-inflammatory properties and influences the activity of several cell types of the immune system. IL-10 is primarily secreted by activated T-cells, monocytes, macrophages, dendritic cells, natural killer (NK) cells and B-cells (Blanco et al. 2008; Seki et al. 1998; Chomarat et al. 1993). It is released upon activation of these cells by endogenous and exogenous mediators such as lipopolysaccharides (Barsig et al. 1995; Roach et al. 1995), catecholamines and cAMP-elevating drugs (Meisel et al. 1996; Jilg et al. 1996). In response to antigens, cells of the immune system produce pro-inflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interferon -gamma (IFN-γ), interleukin-2 (IL-2) and interleukin-1 (IL-1) (de Waal et al. 1991). They are involved in rapid pathogen clearance and cell necrosis at the site of infection. Prolonged action of pro-inflammatory cytokines can lead to excessive tissue damage, fever, inflammation, and death in extreme cases (Dinarello 2000). IL-10 dampens the inflammatory effects of pro-inflammatory cytokines in overwhelming infections which otherwise can lead to potential tissue damage (Kapur et al. 1997; Wang et al. 1994; Cassatella et al. 1993).
The human IL10 gene is 4.7 kb in size and is located on the long arm of chromosome 1. It contains five exons. Biologically functional IL-10 exists in the form of a 36 kD homodimer composed of two non-covalently bonded monomers each with 160 amino acids length (Zdanov 2010). Two disulfide bridges exist between the monomers, which are required for their biological activity and maintaining structural integrity (Windsor et al. 1993). IL-10 homodimer binds to tetrameric IL-10 receptor complex, which consists of two IL-10R-alpha and two IL-10R-beta subunits. IL-10R-alpha is known to bind to the ligand and IL-10R-beta is the accessory signaling subunit (Liu et al. 1994).
IL-10 homodimer upon binding to its receptor IL-10R-alpha, activates JAK/STAT (Janus kinase/signal transducer and activator of transcription) and Akt (also known as protein kinase B, PKB) cascades (Riley et al. 1999). IL-10 has also been involved in proliferation, survival and anti-apoptotic activities of several cancers such as Burkitt lymphoma (Kube et al. 1995), non-Hodgkins lymphoma (Cortes and Kurzrock 1997) and non-small cell lung cancer (De Vita et al. 2000b). Studies have reported that overexpression of IL-10 promotes tumor development in certain lymphomas and melanomas by suppressing the antitumor immune response (Huang et al. 1999; Boulland et al. 1998; Kruger-Krasagakes et al. 1994). Several investigations have also suggested that serum level of IL-10 may indicate disease progression. A study in advanced solid tumors has reported that IL-10 serum level returns to normal in radically resected patients. However, in case of tumor recurrence, the IL-10 level was observed to be persistently elevated (De Vita et al. 2000a). Additionally, elevated serum level of IL-10 has also been associated with autoimmune and inflammatory diseases such as systemic lupus erythematosus (SLE) (Park et al. 1998), systemic sclerosis (Hasegawa et al. 1997) and Bullous pemphigoid (Schmidt et al. 1996). Studies have reported a protective role of IL-10 in cartilage of osteoarthritis patients (Jung et al. 2013). Recombinant human IL-10 has also been tested as a promising therapeutic agent in patients with rheumatoid arthritis (Keystone et al. 1998) and Crohn’s disease (Tilg et al. 2002).
IL-10 plays a central role in survival and persistence of intracellular pathogens in vivo. Pathogens such as Leishmania donovani (Chandra and Naik 2008; Ghalib et al. 1993) Mycobacterium tuberculosis (Higgins et al. 2009), Trypanosoma cruzi (Holscher et al. 2000) and Coxiella burnetii (Ghigo et al. 2001) have evolved various mechanisms for stimulating IL-10 production in immune cells for their survival. Recent studies have indicated the correlation between IL-10 and tuberculosis (TB) susceptibility in humans and mice (Jamil et al. 2007; Bonecini-Almeida et al. 2004; Olobo et al. 2001; Verbon et al. 1999). IL-10 mediated deactivation of macrophages leads to reduced production of pro-inflammatory cytokines and reactive oxygen species which would otherwise lead to cell damage (Fiorentino et al. 1991; de Waal et al. 1991 During Mycobacterium tuberculosis infection, IL-10 contributes to survival and persistence of bacteria inside macrophage by inhibiting MHC-restricted cytotoxicity against infected macrophages (de la Barrera et al. 2004). The abundance of IL-10 in TB infected individuals, and its role in downregulation of IFN-gamma and a T-cell costimulatory molecule called cytotoxic T-lymphocyte-associated protein 4 (CTLA4), suggest an altered antigen presentation in infected individuals. In addition, the CD8+ CTL-mediated cytotoxic activity could be enhanced upon neutralization of IL-10, suggesting a direct role of IL-10 in suppressing the anti-mycobacterial response (Gong et al. 1996).
IL-10 related signaling events have been well documented in the literature. However, a well annotated pathway map for IL-10 is currently lacking in the available pathway resources. Considering their significance in immunological events with respect to disease and infection, documentation and pathway map availability in a public domain is much desired. To this end, we developed a comprehensive signaling pathway of IL-10 by manually curating the molecules associated with IL-10 signaling from published literature. The pathway map is made available through NetPath (http://www.netpath.org/reactions?path_id=NetPath_132) (Kandasamy et al. 2010), a resource for human signaling pathways. This manuscript describes the methodology of generating downstream signaling pathway by manually curating the events upon interaction of IL-10 with its cognate IL-10 receptor.
Materials and methods
Literature survey pertaining to IL-10 signaling was carried out in PubMed and Google Scholar using search terms that included - (IL-10 OR “Interleukin-10”) AND (Signaling OR Pathway OR Signal OR “Signal Transduction”) NOT ‘Review’. Studies describing the downstream signaling events induced upon binding of IL-10 to its receptor were screened for annotation of this pathway. The information was then manually curated and cataloged using PathBuilder (Kandasamy et al. 2009), an annotation tool developed in-house for the curation of signaling pathways. Annotation criteria, previously employed for generation of RANK/RANKL (Raju et al. 2011a), Interleukin-17 (Sharma et al. 2015), CRH (Subbannayya et al. 2013), Gastrin (Subbannayya et al. 2014), BDNF (Brain derived neurotrophic factor) (Subbannayya et al. 2013) and TSLP (Thymic stromal lymphopoietin) (Zhong et al. 2014) signaling pathways, were followed for mapping IL-10 signaling molecules. Besides listing of IL-10 induced molecular events, we also annotated biological information including catalytic reactions, protein-protein interactions, gene regulation events and activation/inhibition reactions corresponding to IL-10 signaling. Information such as cell lines used in experiment and experimental conditions were also documented. In case of post-translational modification (PTM), information on site and residue for PTMs were also curated. Comments on each reaction were written in brief, describing the experiment and results from which the reaction was inferred. An internal review system was followed to confirm the annotation and to avoid any error of commission. In addition, each pathway reaction was reviewed by a Pathway Authority, who has a proven expertise in the field of study (SG, co-author in this article).
IL-10 pathway map generation and visualization
The signaling events derived from manual curation are available as pathway map. We have also developed a pathway map based on the reactions by following NetSlim criteria (Raju et al. 2011b). This enabled us to shortlist high confidence reactions and develop a more stringent IL-10 pathway map. The reactions associated with IL-10 pathway can be visualized using PathVisio (van Iersel et al. 2008). The IL-10 pathway in NetSlim version can be downloaded in png, gpml and pdf formats. The pathway molecules are linked to corresponding NetPath page containing links for ‘Pathway Authority,” Curator’s details’ and ‘Comments’ section.
Results and discussion
Over 3,000 research articles were screened from PubMed and Google Scholar for curation of molecules associated with IL-10 signalling. Articles identified with the information pertaining to signalling events induced upon binding of IL-10 to its receptor were considered for curation. By manual curation, we curated 04 protein-protein interactions, 30 catalytic reactions, 87 gene regulation events, 04 activation/inhibition events and 01 protein translocation event. These pathway reactions were depicted as signalling network (Fig. 1). The reactions in the map are linked to their respective articles as listed in PubMed. The molecules are linked to their respective pages in NetPath and HPRD (Prasad et al. 2009) and the transcriptionally regulated genes are linked to their corresponding gene pages in NCBI. The data is presented in widely accepted standard formats, namely, Systems Biology Markup Language (SBML 2.1) (Hucka et al. 2003), Proteomics Standard Initiative-Molecular Interaction (PSIMI version 2.5) (Orchard and Kerrien 2010) and Biological Pathways Exchange (BioPAX level 3.0) (Demir et al. 2010). IL-10 pathway page in NetPath gives a brief description on pathway and its importance. A table is provided with pathway statistics on the NetPath page.
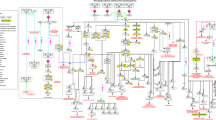
A schematic representation of reactions induced by IL-10. The signaling pathway map depicts molecules involved in protein-protein interactions, catalysis, activation/inhibition and translocation events induced upon treatment with IL-10 cytokine. Information regarding site and residue of post-translational modification is also shown in the pathway. Pathway legend is provided to identify the interactions, molecular associations and gene regulations on the map
Overall, the pathway map depicts that immune cells upon pro-inflammatory cytokine stimulation secrete IL-10, which binds to the extracellular domain of IL-10R-alpha that leads to phosphorylation of JAK1 (Janus Kinase1) associated with IL-10R-alpha and TYK2 (tyrosine kinase2) associated with IL-10R beta (Kotenko et al. 1997). These kinases further phosphorylate tyrosine residues (Y446 and Y496) located on the intracellular domain of IL-10R-Alpha. Phosphorylated residues act as temporary docking sites for STAT3 (Signal Transducer and Activator of Transcription-3) (Donnelly et al. 1999). JAK1 further phosphorylates STAT3, which then homo/hetero dimerize and translocate into the nucleus. In the nucleus, it regulates various cell cycle progression genes such as BCL2 ( Santner-Nanan et al. 2013; Levy and Brouet 1994) along with other anti-apoptotic genes (Donnelly et al. 1999). In addition, it also upregulates v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB) which is involved in regulation of macrophage deactivation (Gemelli et al. 2014). IL-10 controls the inflammatory processes by suppressing pro-inflammatory cytokines such as tumor necrosis factor alpha (TNF) (van der Poll et al. 1994), interleukin-6 (IL6) (Wang et al. 1994) and interleukin-8 (IL8) (de Waal et al. 1991). Recently, it has been shown that IL-10 prevents the differentiation of monocytes into dendritic cells, which are one of the important antigen presenting cells of immune system (Buelens et al. 1997). Studies have also reported that IL-10 down regulates co-stimulatory molecules such as CD86 (Chang et al. 1995) and intracellular adhesion molecule-1 (ICAM1 (Spittler et al. 1995) rendering the cells incapable of presenting antigens on their surface. Recent reports have demonstrated that IL-10 induces the expression of heme oxygenase-1 (HMOX1) in macrophages, which is a potent anti-inflammatory agent (Koch et al. 2009; Lee and Chau 2002). Secretion of matrix metalloproteinases (MMP1 and MMP3) is also enhanced under the influence of IL-10 (Reitamo et al. 1994). In addition, IL-10 stimulates chondrocytes proliferations and chondrogenic differentiation by activating (Bone Morphogenetic Protein) BMP signalling pathway through phosphorylation of SMAD1, SMAD5 and SMAD9 proteins (Jung et al. 2013).
IL-10 signaling pathway data can be used as a useful resource for understanding the complex mechanisms associated with the anti-inflammatory cytokines. The pathway map developed will also aid in understanding the mechanisms adopted by intracellular pathogens and cancerous cells to combat the immune system and establish their survival in the body. The users can further provide their comments about the pathway in NetPath through (http://www.netpath.org/comments).
Abbreviations
- IL-10:
-
Interleukin-10
- CSIF:
-
Cytokine synthesis inhibitory factor
- PPIs:
-
Protein-protein interactions
- PTMs:
-
Post-translational modifications
- BioPAX:
-
Biological PAthway eXchange
- SBML:
-
Systems biology markup language
- PSI-MI:
-
Proteomics standards initiative for molecular interaction
- HPRD:
-
Human protein reference database
References
Barsig J, Kusters S, Vogt K, Volk HD, Tiegs G, Wendel A (1995) Lipopolysaccharide-induced interleukin-10 in mice: role of endogenous tumor necrosis factor-alpha. Eur J Immunol 25:2888–2893
Blanco P, Palucka AK, Pascual V, Banchereau J (2008) Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor Rev 19:41–52
Bonecini-Almeida MG, Ho JL, Boechat N, Huard RC, Chitale S, Doo H, Geng J, Rego L, Lazzarini LC, Kritski AL, Johnson WD Jr, McCaffrey TA, Silva JR (2004) Down-modulation of lung immune responses by interleukin-10 and transforming growth factor beta (TGF-beta) and analysis of TGF-beta receptors I and II in active tuberculosis. Infect Immun 72:2628–2634
Boulland ML, Meignin V, Leroy-Viard K, Copie-Bergman C, Briere J, Touitou R, Kanavaros P, Gaulard P (1998) Human interleukin-10 expression in T/natural killer-cell lymphomas: association with anaplastic large cell lymphomas and nasal natural killer-cell lymphomas. Am J Pathol 153:1229–1237
Buelens C, Verhasselt V, De Groote D, Thielemans K, Goldman M, Willems F (1997) Interleukin-10 prevents the generation of dendritic cells from human peripheral blood mononuclear cells cultured with interleukin-4 and granulocyte/macrophage-colony-stimulating factor. Eur J Immunol 27:756–762
Cassatella MA, Meda L, Bonora S, Ceska M, Constantin G (1993) Interleukin 10 (IL-10) inhibits the release of proinflammatory cytokines from human polymorphonuclear leukocytes. Evidence for an autocrine role of tumor necrosis factor and IL-1 beta in mediating the production of IL-8 triggered by lipopolysaccharide. J Exp Med 178:2207–2211
Chandra D, Naik S (2008) Leishmania donovani infection down-regulates TLR2-stimulated IL-12p40 and activates IL-10 in cells of macrophage/monocytic lineage by modulating MAPK pathways through a contact-dependent mechanism. Clin Exp Immunol 154:224–234
Chang Q, Hanks S, Akard L, Thompson J, Harvey K, English D, Jansen J (1995) Maturation of mobilized peripheral blood progenitor cells: preclinical and phase I clinical studies. J Hematother 4:289–297
Chomarat P, Rissoan MC, Banchereau J, Miossec P (1993) Interferon gamma inhibits interleukin 10 production by monocytes. J Exp Med 177:523–527
Cortes J, Kurzrock R (1997) Interleukin-10 in non-Hodgkin’s lymphoma. Leuk Lymphoma 26:251–259
de la Barrera S, Aleman M, Musella R, Schierloh P, Pasquinelli V, Garcia V, Abbate E, Sasiain Mdel C (2004) IL-10 down-regulates costimulatory molecules on Mycobacterium tuberculosis-pulsed macrophages and impairs the lytic activity of CD4 and CD8 CTL in tuberculosis patients. Clin Exp Immunol 138:128–138
De Vita F, Orditura M, Galizia G, Romano C, Lieto E, Iodice P, Tuccillo C, Catalano G (2000a) Serum interleukin-10 is an independent prognostic factor in advanced solid tumors. Oncol Rep 7:357–361
De Vita F, Orditura M, Galizia G, Romano C, Roscigno A, Lieto E, Catalano G (2000b) Serum interleukin-10 levels as a prognostic factor in advanced non-small cell lung cancer patients. Chest 117:365–373
de Waal MR, Abrams J, Bennett B, Figdor CG, de Vries JE (1991) Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med 174:1209–1220
Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, Schacherer F, Martinez-Flores I, Hu Z, Jimenez-Jacinto V, Joshi-Tope G, Kandasamy K, Lopez-Fuentes AC, Mi H, Pichler E, Rodchenkov I, Splendiani A, Tkachev S, Zucker J, Gopinath G, Rajasimha H, Ramakrishnan R, Shah I, Syed M, Anwar N, Babur O, Blinov M, Brauner E, Corwin D, Donaldson S, Gibbons F, Goldberg R, Hornbeck P, Luna A, Murray-Rust P, Neumann E, Ruebenacker O, Samwald M, van Iersel M, Wimalaratne S, Allen K, Braun B, Whirl-Carrillo M, Cheung KH, Dahlquist K, Finney A, Gillespie M, Glass E, Gong L, Haw R, Honig M, Hubaut O, Kane D, Krupa S, Kutmon M, Leonard J, Marks D, Merberg D, Petri V, Pico A, Ravenscroft D, Ren L, Shah N, Sunshine M, Tang R, Whaley R, Letovksy S, Buetow KH, Rzhetsky A, Schachter V, Sobral BS, Dogrusoz U, McWeeney S, Aladjem M, Birney E, Collado-Vides J, Goto S, Hucka M, Le Novere N, Maltsev N, Pandey A, Thomas P, Wingender E, Karp PD, Sander C, Bader GD (2010) The BioPAX community standard for pathway data sharing. Nat Biotechnol 28:935–942
Dinarello CA (2000) Proinflammatory cytokines. Chest 118:503–508
Donnelly RP, Dickensheets H, Finbloom DS (1999) The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res 19:563–573
Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A (1991) IL-10 inhibits cytokine production by activated macrophages. J Immunol 147:3815–3822
Gemelli C, Zanocco Marani T, Bicciato S, Mazza EM, Boraschi D, Salsi V, Zappavigna V, Parenti S, Selmi T, Tagliafico E, Ferrari S, Grande A (2014) MafB is a downstream target of the IL-10/STAT3 signaling pathway, involved in the regulation of macrophage de-activation. Biochim Biophys Acta 1843:955–964
Ghalib HW, Piuvezam MR, Skeiky YA, Siddig M, Hashim FA, el-Hassan AM, Russo DM, Reed SG (1993) Interleukin 10 production correlates with pathology in human Leishmania donovani infections. J Clin Invest 92:324–329
Ghigo E, Capo C, Raoult D, Mege JL (2001) Interleukin-10 stimulates Coxiella burnetii replication in human monocytes through tumor necrosis factor down-modulation: role in microbicidal defect of Q fever. Infect Immun 69:2345–2352
Gong JH, Zhang M, Modlin RL, Linsley PS, Iyer D, Lin Y, Barnes PF (1996) Interleukin-10 downregulates Mycobacterium tuberculosis-induced Th1 responses and CTLA-4 expression. Infect Immun 64:913–918
Hasegawa M, Fujimoto M, Kikuchi K, Takehara K (1997) Elevated serum levels of interleukin 4 (IL-4), IL-10, and IL-13 in patients with systemic sclerosis. J Rheumatol 24:328–332
Higgins DM, Sanchez-Campillo J, Rosas-Taraco AG, Lee EJ, Orme IM, Gonzalez-Juarrero M (2009) Lack of IL-10 alters inflammatory and immune responses during pulmonary Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 89:149–157
Holscher C, Mohrs M, Dai WJ, Kohler G, Ryffel B, Schaub GA, Mossmann H, Brombacher F (2000) Tumor necrosis factor alpha-mediated toxic shock in Trypanosoma cruzi-infected interleukin 10-deficient mice. Infect Immun 68:4075–4083
Huang S, Ullrich SE, Bar-Eli M (1999) Regulation of tumor growth and metastasis by interleukin-10: the melanoma experience. J Interferon Cytokine Res 19:697–703
Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19:524–531
Jamil B, Shahid F, Hasan Z, Nasir N, Razzaki T, Dawood G, Hussain R (2007) Interferon gamma/IL10 ratio defines the disease severity in pulmonary and extra pulmonary tuberculosis. Tuberculosis (Edinb) 87:279–287
Jilg S, Barsig J, Leist M, Kusters S, Volk HD, Wendel A (1996) Enhanced release of interleukin-10 and soluble tumor necrosis factor receptors as novel principles of methylxanthine action in murine models of endotoxic shock. J Pharmacol Exp Ther 278:421–431
Jung YK, Kim GW, Park HR, Lee EJ, Choi JY, Beier F, Han SW (2013) Role of interleukin-10 in endochondral bone formation in mice: anabolic effect via the bone morphogenetic protein/Smad pathway. Arthritis Rheum 65:3153–3164
Kandasamy K, Keerthikumar S, Raju R, Keshava Prasad TS, Ramachandra YL, Mohan S, Pandey A (2009) PathBuilder--open source software for annotating and developing pathway resources. Bioinformatics 25:2860–2862
Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A (2010) NetPath: a public resource of curated signal transduction pathways. Genome Biol 11:R3
Kapur S, Jones C, DaSilva J, Wilson A, Houle S (1997) Reliability of a simple non-invasive method for the evaluation of 5-HT2 receptors using [18F]-setoperone PET imaging. Nucl Med Commun 18:395–399
Keystone E, Wherry J, Grint P (1998) IL-10 as a therapeutic strategy in the treatment of rheumatoid arthritis. Rheum Dis Clin N Am 24:629–639
Koch N, Jung M, Sabat R, Kratzschmar J, Docke WD, Asadullah K, Volk HD, Grutz G (2009) IL-10 protects monocytes and macrophages from complement-mediated lysis. J Leukoc Biol 86:155–166
Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S (1997) Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J 16:5894–5903
Kruger-Krasagakes S, Krasagakis K, Garbe C, Schmitt E, Huls C, Blankenstein T, Diamantstein T (1994) Expression of interleukin 10 in human melanoma. Br J Cancer 70:1182–1185
Kube D, Platzer C, von Knethen A, Straub H, Bohlen H, Hafner M, Tesch H (1995) Isolation of the human interleukin 10 promoter. Characterization of the promoter activity in Burkitt’s lymphoma cell lines. Cytokine 7:1–7
Lee TS, Chau LY (2002) Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat Med 8:240–246
Levy Y, Brouet JC (1994) Interleukin-10 prevents spontaneous death of germinal center B cells by induction of the bcl-2 protein. J Clin Invest 93:424–428
Liu Y, Wei SH, Ho AS, de Waal MR, Moore KW (1994) Expression cloning and characterization of a human IL-10 receptor. J Immunol 152:1821–1829
Meisel C, Vogt K, Platzer C, Randow F, Liebenthal C, Volk HD (1996) Differential regulation of monocytic tumor necrosis factor-alpha and interleukin-10 expression. Eur J Immunol 26:1580–1586
Olobo JO, Geletu M, Demissie A, Eguale T, Hiwot K, Aderaye G, Britton S (2001) Circulating TNF-alpha, TGF-beta, and IL-10 in tuberculosis patients and healthy contacts. Scand J Immunol 53:85–91
Orchard S, Kerrien S (2010) Molecular interactions and data standardisation. Methods Mol Biol 604:309–318
Park YB, Lee SK, Kim DS, Lee J, Lee CH, Song CH (1998) Elevated interleukin-10 levels correlated with disease activity in systemic lupus erythematosus. Clin Exp Rheumatol 16:283–288
Prasad TS, Kandasamy K, Pandey A (2009) Human protein reference database and human proteinpedia as discovery tools for systems biology. Methods Mol Biol 577:67–79
Raju R, Balakrishnan L, Nanjappa V, Bhattacharjee M, Getnet D, Muthusamy B, Kurian Thomas J, Sharma J, Rahiman BA, Harsha HC, Shankar S, Prasad TS, Mohan SS, Bader GD, Wani MR, Pandey A (2011a) A comprehensive manually curated reaction map of RANKL/RANK-signaling pathway. Database (Oxford) 2011:bar021
Raju R, Nanjappa V, Balakrishnan L, Radhakrishnan A, Thomas JK, Sharma J, Tian M, Palapetta SM, Subbannayya T, Sekhar NR, Muthusamy B, Goel R, Subbannayya Y, Telikicherla D, Bhattacharjee M, Pinto SM, Syed N, Srikanth MS, Sathe GJ, Ahmad S, Chavan SN, Kumar GS, Marimuthu A, Prasad TS, Harsha HC, Rahiman BA, Ohara O, Bader GD, Sujatha Mohan S, Schiemann WP, Pandey A (2011b) NetSlim: high-confidence curated signaling maps. Database (Oxford) 2011:bar032
Reitamo S, Remitz A, Tamai K, Uitto J (1994) Interleukin-10 modulates type I collagen and matrix metalloprotease gene expression in cultured human skin fibroblasts. J Clin Invest 94:2489–2492
Riley JK, Takeda K, Akira S, Schreiber RD (1999) Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem 274:16513–16521
Roach TI, Barton CH, Chatterjee D, Liew FY, Blackwell JM (1995) Opposing effects of interferon-gamma on iNOS and interleukin-10 expression in lipopolysaccharide- and mycobacterial lipoarabinomannan-stimulated macrophages. Immunology 85:106–113
Santner-Nanan B, Straubinger K, Hsu P, Parnell G, Tang B, Xu B, Makris A, Hennessy A, Peek MJ, Busch DH, da Costa CP, Nanan R (2013) Fetal-maternal alignment of regulatory T cells correlates with IL-10 and Bcl-2 upregulation in pregnancy. J Immunol 191:145–153
Schmidt E, Bastian B, Dummer R, Tony HP, Brocker EB, Zillikens D (1996) Detection of elevated levels of IL-4, IL-6, and IL-10 in blister fluid of bullous pemphigoid. Arch Dermatol Res 288:353–357
Seki S, Osada S, Ono S, Aosasa S, Habu Y, Nishikage T, Mochizuki H, Hiraide H (1998) Role of liver NK cells and peritoneal macrophages in gamma interferon and interleukin-10 production in experimental bacterial peritonitis in mice. Infect Immun 66:5286–5294
Sharma J, Balakrishnan L, Datta KK, Sahasrabuddhe NA, Khan AA, Sahu A, Singhal A, Getnet D, Raju R, Chatterjee A, Gowda H, Keshava Prasad TS, Shankar S, Pandey A (2015) A knowledgebase resource for interleukin-17 family mediated signaling. J Cell Commun Signal
Spittler A, Schiller C, Willheim M, Tempfer C, Winkler S, Boltz-Nitulescu G (1995) IL-10 augments CD23 expression on U937 cells and down-regulates IL-4-driven CD23 expression on cultured human blood monocytes: effects of IL-10 and other cytokines on cell phenotype and phagocytosis. Immunology 85:311–317
Subbannayya T, Balakrishnan L, Sudarshan G, Advani J, Kumar S, Mahmood R, Nair B, Sirdeshmukh R, Mukherjee KK, Umathe SN, Raju R, Prasad TSK (2013) An integrated map of corticotropin-releasing hormone signaling pathway. J Cell Commun Signal 4:295–300
Subbannayya Y, Anuja K, Advani J, Ojha UK, Nanjappa V, George B, Sonawane A, Kumar RV, Ramaswamy G, Pandey A, Somani BL, Raju R (2014) A network map of the gastrin signaling pathway. J Cell Commun Signal 8:165–170
Tilg H, van Montfrans C, van den Ende A, Kaser A, van Deventer SJ, Schreiber S, Gregor M, Ludwiczek O, Rutgeerts P, Gasche C, Koningsberger JC, Abreu L, Kuhn I, Cohard M, LeBeaut A, Grint P, Weiss G (2002) Treatment of Crohn’s disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut 50:191–195
van der Poll T, Jansen J, Levi M, ten Cate H, ten Cate JW, van Deventer SJ (1994) Regulation of interleukin 10 release by tumor necrosis factor in humans and chimpanzees. J Exp Med 180:1985–1988
van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C (2008) Presenting and exploring biological pathways with PathVisio. BMC Bioinforma 9:399
Verbon A, Juffermans N, Van Deventer SJ, Speelman P, Van Deutekom H, Van Der Poll T (1999) Serum concentrations of cytokines in patients with active tuberculosis (TB) and after treatment. Clin Exp Immunol 115:110–113
Wang P, Wu P, Siegel MI, Egan RW, Billah MM (1994) IL-10 inhibits transcription of cytokine genes in human peripheral blood mononuclear cells. J Immunol 153:811–816
Windsor WT, Syto R, Tsarbopoulos A, Zhang R, Durkin J, Baldwin S, Paliwal S, Mui PW, Pramanik B, Trotta PP et al (1993) Disulfide bond assignments and secondary structure analysis of human and murine interleukin 10. Biochemistry 32:8807–8815
Zdanov A (2010) Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev 21:325–330
Zhong J, Sharma J, Raju R, Palapetta SM, Prasad TS, Huang TC, Yoda A, Tyner JW, van Bodegom D, Weinstock DM, Ziegler SF, Pandey A (2014) TSLP signaling pathway map: a platform for analysis of TSLP-mediated signaling. Database (Oxford) 2014:bau007
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics. We thank the Infosys Foundation for research support to the Institute of Bioinformatics. RV is a recipient of a Junior Research Fellowship from the University Grants Commission (UGC), Government of India. AAK is a recipient of Senior Research Fellowship from Indian Council of Medical Research (ICMR), Government of India. JA is a recipient of Senior Research Fellowship from Council of Scientific & Industrial Research (CSIR), Government of India. TSKP is the recipient of the DST-IDP research grant (IDP/MED/2011/23-general) on “Development of epitope based diagnostic gadget for detection of Mycobacterium tuberculosis in the Indian population” from the Department of Science Technology, Government of India. HG is a Wellcome Trust/DBT India Alliance Early Career Fellow. SG is a Wellcome Trust/DBT India Alliance Intermediate Fellow.
Conflict of interests
The author(s) declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Verma, R., Balakrishnan, L., Sharma, K. et al. A network map of Interleukin-10 signaling pathway. J. Cell Commun. Signal. 10, 61–67 (2016). https://doi.org/10.1007/s12079-015-0302-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-015-0302-x