Abstract
A series of neutral hexacoordinated nickel(II) complexes of formula [Ni II(L)X2] (where L =3,4-bis(2-pyridylmethylthio)toluene with tetradentate N2S2 donor set and X = chloride (1), azide (2), cyanate (3) and isothiocyanate anion (4)) have been synthesized and isolated in pure form. The complexes were characterized by physicochemical and spectroscopic methods along with detailed structural characterization of 1,2 and 3 by single crystal X-ray diffraction analyses. The structural study showed that the nickel(II) ion has a distorted octahedral geometry being chelated by the tetradentate N2S2 ligand and bound to cis- located choride or pseudohalide anions. In dimethylformamide solution the complexes showed quasi-reversible Ni II/NiIII redox couples in cyclic voltammograms with E1/2 values of + 0.723, + 0.749, + 0.768 and + 0.868 V for 1, 2, 3 and 4, respectively. The study of interaction of the complexes with calf thymus DNA, bovine serum albumin (BSA) and human serum albumin (HSA) using spectroscopic and physicochemical tools clearly indicates that the complexes interact with DNA via groove binding mode.

Four new neutral distorted octahedral nickel(II) complexes of general formula, [NiII(L)X2] have been synthesized and structurally characterized. The spectroscopic and physicochemical studies of interaction of the complexes with calf thymus DNA, BSA and HSA have also been included.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
A considerable interest has been shown to biologically active transition metal complexes, largely due to their ability to interact with DNA.[1] The complexes, which can bind or cleave DNA at specific sites, may play an important role in genomic research and in photodynamic therapy against cancer.[2] It is well known that some coordination compounds can inhibit the multiplication of cancer cells by binding and damaging DNA.[3] In particular, complexes of nickel(II) ion with nitrogen based heterocyclic ligands have been widely investigated for their potential applications in bioinorganic chemistry,[4] medicine,[5] catalysis[6,7] and great variety of biological activities as antimalarial, antibacterial, antitumoral, antiviral agents, etc.[8,9]
Pursuing our continuing interest in nitrogen-sulphur polydentate chelators[10–14] and considering the importance of the coordination chemistry of nitrogen-sulfur donor ligands towards nickel ion, herein, we report an account of coordination chemistry of nickel(II) ion with a N2S2 donor site (L = 3,4-bis (2-pyridylmethylthio)toluene) and chloride or different pseudohalide anions. The binding interaction of these complexes with calf thymus-DNA (CT-DNA), bovine serum albumin (BSA) and human serum albumin (HSA) have been studied systematically to explore the mode of biological activity which is also a part of our interest.[14–16]
2 Experimental
2.1 Materials and physical measurements
All chemicals and reagents were obtained from commercial sources and used as received, unless otherwise stated. Solvents were distilled from an appropriate drying agent. Tris-HCl buffer solution and CT-DNA were purchased from Bangalore-Genie and ethidium bromide (EB) from Sigma-Aldrich and were used as received. The organic molecule 3,4-bis(2-pyridyl-methylthio)toluene (L), used as ligand, was synthesized following the reported procedure.[11]
The elemental (C, H, N) analyses were performed on a Perkin Elmer model 2400 elemental analyzer. Nickel analysis was carried out with the Varian atomic absorption spectrophotometer (AAS) model-AA55B, GTA using graphite furnace. Electronic absorption spectra were recorded on a JASCO UV-Vis/NIR spectrophotometer model V-570. IR spectra (KBr discs, 4000-400 cm −1) were recorded using a Perkin-Elmer FTIR model RX1 spectrometer. The room temperature magnetic susceptibility measurements were performed by using a vibrating sample magnetometer PAR 155 model. Molar conductances (ΛM) were measured in a Systronics conductivity meter 304 model using ∼10 −3 mol.L −1 solutions in appropriate organic solvent. Electrochemical measurements were performed using computer-controlled CH-Instruments (Model No- CHI620D). The fluorescence spectra of EB bound to DNA were obtained with Fluorimeter Hitachi-4500. The stock solutions of proteins (1.00 × 10 −4 mol L −1) were prepared by dissolving the solid HSA and BSA in 0.05 M phosphate buffer at pH 7.4 and stored at 0-4 ∘C in the dark. The concentrations of BSA and HSA were determined from optical density measurements, using the values of molar absorptivity of ε 280= 44,720 and 35,700 M −1 cm −1 for BSA and HSA, respectively. Stock solutions of complexes were prepared in DMF-water.
2.2 Preparation of complexes 1-4
The complex [Ni(L)(Cl)2] (1) was prepared by performing the reaction using a methanolic solution (10 mL) of L (338.0 mg, 1.0 mmol) with nickel(II) chloride hexahydrate (238.0 mg, 1.0 mmol) in methanol under stirring condition. The mixture was then refluxed for 4 h. The product was precipitated out and was collected by filtration followed by washing with cold methanol and water. Then the product was dried in vacuo. The pure crystalline product of 1 was obtained from methanol solution. The complex 1 was also synthesized by the following method.
The complexes 1-4 were synthesized following a common procedure, as described below. A methanolic solution of nickel(II) acetate tetrahydrate (249.0 mg, 1.0 mmol) was added to 10 mL of methanolic solution containing the organic compound (L) (338.0 mg, 1.0 mmol). The resulting solution was stirred for 1 h at ambient temperature and then refluxed for 3 h. To this mixture an aqueous solution of sodium chloride (2.0 mmol) (for 1), or sodium azide (2.0 mmol) (for 2) or sodium cyanate (2.0 mmol) (for 3) or potassium thiocyanate (2.0 mmol) (for 4) was added and stirring was continued for another 1.0 h. The volume of the solution was reduced at room temperature by slow evaporation. The product was collected by washing with cold methanol and water and dried in vacuo and the pure crystallized product was obtained from methanol.
Complex 1: [Ni(L)(Cl)2]: C19 H 18 N 2S2NiCl2: Anal. Calc. (%): C, 48.72; H, 3.82; N, 5.90; Ni, 12.48; Found; C, 48.69; H, 3.75; N, 5.98; Ni, 12.16. IR (cm −1): ν C=N, 1476; ν C−S, 758. Magnetic moment (μ, B.M.): 3.06. Conductivity (Λo, ohm −1 cm 2 mol −1) in DMF: 46. Yield 80-85%.
Complex 2: [Ni(L)(N3)2]: C19 H 18 N 8NiS2 : Anal. Calc. (%): C, 47.48; H, 3.78; N, 23.38; Ni, 12.13; Found; C, 47.39; H, 3.72; N, 23.55; Ni; 11.99. IR (cm −1): ν C=N, 1478; ν C−S, 756, ν N3, 2027. Magnetic moment (μ, B.M.): 3.10. Conductivity (Λo, ohm −1 cm 2 mol −1) in DMF: 44. Yield 80-85%.
Complex 3: [Ni(L)(NCO)2]: C21 H 18 N 4NiO2S2: Anal. Calc. (%): C, 52.58; H, 3.78; N, 11.58; Ni, 12.24; Found; C, 52.53 H, 3.73; N, 11.69; Ni, 12.02. IR (cm −1): ν C=N, 1479; ν C−S, 754, ν NCO, 2182. Magnetic moment (μ, B.M.): 3.08. Conductivity (Λo, ohm −1 cm 2 mol −1) in DMF: 48. Yield 75-80%.
Complex 4: [Ni(L)(NCS)2]: C21 H 18 N 4NiS4: Anal. Calc. (%): C, 49.51; H, 3.58; N, 10.93; Ni, 11.51; Found; C, 49.43 H, 3.49; N, 11.02; Ni, 11.31. IR (cm −1): ν C=N, 1476; ν C−S, 754, ν NCS, 2084. Magnetic moment (μ, B.M.): 3.09. Conductivity (Λo, ohm −1 cm 2 mol −1) in DMF: 46. Yield 65-70%.
2.3 X-ray crystal structure analysis
Suitable single crystals for X-ray diffraction analysis of complexes 1-3 were grown at ambient temperature by slow evaporation of a methanolic solution. Diffraction data for all compounds were collected at room temperature on a Nonius DIP-1030H system, by using Mo-K α radiation (λ= 0.71073 Å). Cell refinement, indexing and scaling of the data sets were performed using programs Denzo and Scalepack.[17] The structures were solved by direct methods and subsequent Fourier analyses and refined by the full-matrix least-squares method based on F 2 with all observed reflections.[18] The methyl group of molecule B in both compounds 2 and 3 was found disordered over two positions, one trans located to S4 the other to S3 (occupancies refined at 0.686(8)/0.314(8) and 0.614(9)/ 0.386(9), respectively). The contribution of hydrogen atoms at calculated positions were included in final cycles of refinement. All the calculations were performed using the WinGX System, Ver 1.80.05.[19] Crystal data and details of data collection and refinement for the structures reported were summarized in table 1.
2.4 DNA binding experiments
UV-Vis absorption spectra were recorded by maintaining the CT-DNA concentration as constant with increasing amounts of the metal complex. Before measuring, equal quantities of the complex were added to both the CT-DNA and the reference solutions to eliminate the absorbance of the complex itself. The DNA complex solutions were incubated for 2 h before recording the absorption spectra. In the fluorescence-quenching experiments, the complex was added to a CT-DNA solution treated with EB, and the fluorescence spectrum was measured for each sample after 2 h excited at λex = 522 nm to record the emission spectra.
Viscosities of CT-DNA with different concentrations of the nickel(II) complex in buffer solution were measured using a viscometer at a temperature of 30.0 ± 0.1 ∘C in a thermostatic water bath. The flow time was recorded with a digital stopwatch. Each experiment measurement was repeated three times and an average flow time was calculated. Data are presented as (η/η 0)1/3 versus the binding ratio, where η is the viscosity of CT-DNA in presence of the complex and η 0 is the viscosity of DNA alone. Viscosity values were calculated from the observed flow time of CT-DNA containing solutions (t) duly corrected for that of the buffer alone (t0), η= t - t0. The control experiments were carried out on EB and methyl green using the same method as described above.
2.5 Bovine and human serum albumin (BSA and HSA) binding experiments
The quantitative analyses of the interaction between nickel(II) complex and serum albumins were performed by fluorimetric titration. The stock solutions of each of the serum albumins (∼1.0 × 10 −5 M) were prepared by dissolving BSA/HSA in MiliQ water. A 3.0 mL portion of aqueous solution of albumin was titrated by addition of the appropriate concentration of nickel(II) complex solution (to give a final concentration of 4.0 × 10 −6 mol L −1 of the complexes). For each addition, the mixture solution was shaken and allowed to stand for 20 min and then the fluorescence intensities were measured with an excitation wavelength of 280 nm.
3 Result and Discussion
3.1 Synthesis and characterization
The organic ligand L was synthesized by the reaction of 2-picolyl chloride hydrochloride with toluene-3,4-dithiol in presence of sodium ethoxide (viz. scheme 1). Complex 1 was obtained from the reaction of the nickel(II) chloride with equimolar amount of organic moiety in the methanol medium and all the complexes (1-4) were obtained in the good yield from the reaction of nickel(II) acetate and organic ligand (L) in the methanol medium followed by addition of the respective aqueous solution of the chloride/pseudohalides ions (scheme 1). In all the complexes the organic molecule L acts as tetradentate neutral ligand through N2S2 donor centres. All the complexes are monomeric in nature and soluble in common organic solvents. Conductivity measurement of the complexes in N,N-dimethyl formamide show conductance values in the range of 44-48 (Λo, ohm−1 cm2 mol−1) which suggests that all complexes are non-electrolytes. The magnetic moments (μ) at room temperature of these complexes are 3.06, 3.10, 3.08 and 3.09 which indicate all complexes are high spin distorted octahedral geometry.
3.2 Structural studies
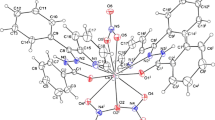
The molecular structures of complexes 1, 2 and 3 with the atom numbering scheme are shown in figures 1 and 2, respectively. Complexes 2 and 3 are isomorphous and comprise two independent molecules in the unit cell. The X-ray crystal structure analyses of all compounds confirm the formation of neutral complexes with the nickel(II) ion coordinated by the neutral tetradentate ligand having N2S2 donor set. The ligand embraces the metal ion in such a way that pyridine rings are in trans positions, while the two thioether-S donors are located in the equatorial plane trans located to the chloride (in complex 1), azide (in 2) and cyanate anions (in 3), completing a distorted octahedral coordination sphere around the metal ion. The coordination geometrical parameters (reported in tables 2 and s1) are virtually comparable in 1-3 and have typical values for this type of complexes.[14] The Ni-N(py) bond distances fall in a range from 2.109(5) to 2.144(3) Å) with the pyridine mean planes that form a dihedral angle of ca. 70 ∘. The N(py)-Ni-N(py) bond angles, in the range from 164.18(19) to 166.59(14) ∘, evidence the deviation from the ideal octahedral geometry and confirm the strain of the ligand upon coordination. The Ni-S bond lengths (falling within a narrow range from 2.3887(18) to 2.4249(17) Å) do not appear to be affected by the trans chloro or pseudohalide ligands. The two independent molecules of 2 and 3 show small differences only in the orientation of the pseudohalides, which bind the metal with a N-N-Ni bond angle of 122.2(4)-135.5(5) ∘ in 2 and somewhat larger C-N-Ni angle values of 137.7(8)-155.8(7) ∘ in 3.
The present data agree with those of previous structures reported of nickel(II) complexes of 1,3-bis(2-pyridylmethylthio)-propane (L\(^{\prime }\)).[12,13] However, this more flexible ligand induces some difference in the coordination geometry dictated by the chelating requirements. In the [Ni(L\(^{\prime }\))Cl2] complex,[12] in the [Ni(L\(^{\prime }\))(N3)2] and [Ni(L\(^{\prime }\))(NCS)2] derivatives,[13] slightly longer Ni-S bond lengths (range 2.451(1)-2.4604(14) Å) and larger S-Ni-S bond angles (92.87(6)-96.56(4) ∘) were measured. In addition, in the azide and isothiocyanate complexes shorter Ni-N bond lengths were observed (2.031(3) - 2.097(3) Å), while here all values are greater than 2.10 Å. The crystal packing of all crystals evidences π- π interactions between aromatic rings. This feature is particularly marked in compound 1 where short centroid-to-centroid distances of 3.580(4) and 3.499(4) Å are measured between symmetry related pyridines N(1) and N(2), respectively.
3.3 Spectroscopic analysis
The infrared spectral data of all the complexes exhibit characteristic strong to medium intensity bands in the region of 1468-1472 cm−1 and 758-761 cm−1, which are assigned to ν C=N stretching and ν C−S, respectively. An intense band centred at 2027 cm−1 is assignable to ν N3 (2), that at 2183 cm −1 assignable to ν NCO (3), and an intense band centred at 2081 assignable to ν NCS (4).[12] This observation supports the presence of the ligand frame containing the pyridine ring and the C-S bond, ν C−S is generally observed in free ligand in the range of 780-790 cm−1.
The electronic absorption spectra of complexes 1-4 were recorded at room temperature using DMF as solvent and the data are tabulated in table 3. All the bands at wavelength lower than 400 nm are due to intramolecular π→π* and n →π* transitions for the aromatic ring. In octahedral nickel(II) complexes, three spin allowed transitions are generally expected from the energy level diagram for d 8 ions due to 3 A 2g→3 T 1g(P), 3 A 2g→3 T 1g(F), 3 A 2g→3 T 2g transitions, which are observed from low to high wavelengths, respectively. Here, two bands at around 472 nm and around 654 nm assignable to 3 A 2g→3 T 1g(P) and 3 A 2g→3 T 2g transition, respectively, were observed, but the 3 A 2g→3 T 1g(F) transition was absent due to the probable merger of \(^{\mathrm {3}}\textit {T}_{\mathrm {1g}}\)(P) and \(^{\mathrm {3}}\textit {T}_{\mathrm {1g}}\)(F) due to distortion. This observation suggests a distorted octahedral geometry for the nickel(II) ion.
3.4 Electrochemistry
Redox properties of the complexes were studied by cyclic voltammetry using a Pt-disk working electrode and a Pt-wire auxiliary electrode in dry dimethylformamide using [n-Bu4N]ClO4 (0.1 M) as the supporting electrolyte. The potentials expressed with reference to Ag/AgCl electrode are tabulated in table 3 and all the complexes displayed a quasi-reversible voltammogram having ipc/ipa ratio close to unity. During voltammetric study the anodic current increases and a well defined anodic response resulting in a cyclic quasi-reversible voltammogram corresponding to Ni II/Ni III redox couple was observed in the potential range 0.6 – 1.0 V but no cathodic response expected for Ni II→Ni I reduction was obtained for the complexes up to −1.50 V. The E1/2 values (versus Ag/AgCl at 298 K) were found to be + 0.723, + 0.749, + 0.768 and + 0.868 V for 1, 2, 3 and 4, respectively. Here, the E1/2 value for complex 4 be positioned at the relatively higher anodic side of the range generally measured for the Ni II/Ni III oxidation potential data for thioether containing nickel(II) complexes[12,20–23] indicating that Ni II is reasonably stabilized in 4 and this feature may be due to the presence of the soft isothiocyanate ligand compared to chloride, azide and cyanate co-ligand. Here, the peak potential showed a small dependence on the scan rate but the ratio between the cathodic peak current and the square root of the scan rate (Ipc/ ν 1/2) is almost constant, from which it can be comprehended that the redox couple is related to a quasi-reversible one-electron transfer process controlled by diffusion.
3.5 DNA-binding studies
The mode of interaction of all the mononuclear octahedral nickel(II) complexes (1-4) with calf thymus DNA (CT-DNA) presents similar features and the investigation by using absorption and emission spectra indicates a groove binding interaction.[24,25] For this reason, only the interaction of complex 1 and 2 with DNA has been studied in detail and discussed as follows.
Spectrophotometric study: To examine the binding mode of metal complexes with DNA, electronic absorption spectroscopy has been used. An increase of the absorption band of complex 1 observed at ca. 264 nm is considered for the binding of the nickel(II) complex to the CT-DNA helix. This increasing trend in absorbance indicates that a strong interaction is involved between 1 and the base pairs of DNA.[26] The absorption spectra of the complexes 1 and 2 in the absence and presence of CT-DNA are given in figure 3. The extent of the hyper-chromism in the charge transfer band is generally consistent with the strength of interaction.[27–29]
In order to further elucidate the binding strength of the complexes with CT-DNA, the intrinsic binding constant Kb was determined from the spectral titration data using the following equation.[30]
where [DNA] is the concentration of DNA, ε a, ε f and ε b correspond to Aobsd/[nickel(II) complex], the extinction coefficient for the free nickel(II) complex (1, 2, 3 or 4), and the extinction coefficient for the nickel(II) complex (1, 2, 3 or 4) in the fully bound form, respectively. From the plot of [DNA]/(ε a- ε f)versus [DNA] (figure 4) the apparent binding constant Kb were estimated as the ratio of slope to the intercept. The values of Kb for the complexes were listed in the table 4 indicating in term of groove binding mode.
3.5.1 Spectrofluorimetric study
A decreasing trend was observed for the fluorescence intensity of EB bound to CT-DNA (at λ ex= 522 nm) with the increasing concentration of complexes. Results for complexes 1 and 2 are shown in figure 5. The quenching phenomenon by these nickel complexes (1, 2, 3 and 4) is in agreement with the linear Stern-Volmer equation.[31]
where I0 and I represent the fluorescence intensities in the absence and presence of quencher, respectively. Ksv is a Stern-Volmer quenching constant, [Q ] is the concentration of quencher. The Ksv value calculated from the plot (figure 6) of I0/I versus [complex] is tabulated in table 5. All the values suggest a strong affinity of the complexes to CT-DNA.
Emission spectra of the CT-DNA-EB system upon titration with complexes 1 (left) and 2 (right) in Tris-HCl buffer. λ ex= 522 nm; [EB] = 9.6 × 10 −5 M, [DNA] × 10 −5 M = 1.58; [complex] × 10 −5= (a) 0.0, (b) 1.28, (c) 2.56, (d) 3.84, (e) 5.12, (f) 6.40 mol L −1. The arrow denotes the gradual increase of complex concentration.
The number of binding sites can also be calculated from fluorescence titration data using the following equation:[32]
where K is the binding constant and n the binding sites of complexes to CT-DNA. The number of binding sites (n) was determined from the intercept of log[(I0 - I)/I] vs log[Q] and tabulated in table 5. This value indicates a low association of complexes to the DNA bases but suggests the strong affinity of this species through surface or groove binding.
3.5.2 Viscosity measurements of CT-DNA
To explore the nature of the binding of the complexes with DNA, we measured the viscosity of CT-DNA by adding the complex at different concentrations, as the hydrodynamic measurements (namely viscosity and sedimentation) are critical tests to assess the binding mode in solution. In fact the intrinsic viscosity of DNA is sensitive to the length changes of nucleic acids, and so a classical intercalation mode should be indicated by a lengthening of the DNA double chain, being base pairs separated to accommodate the binding ligand.[33] On the other hand, non-classical intercalation could bend or kink the DNA helix, thereby decreasing its length and viscosity. From the viscosity measurements no change on the relative viscosity of the DNA solution was observed by increasing the concentration of the complex, thus suggesting that complex 1 interacts with CT-DNA through groove binding mode.
3.6 BSA and HSA binding experiments
The absorption spectra of BSA and HSA in the absence and presence of the complex 1at different concentrations were obtained in phosphate buffer at pH 7.4. From this study, we observed that the absorbance of BSA or HSA increases regularly upon increasing the concentration of the complex (figure 7).
The enhancement of absorbance may be explained by assuming the adsorption of the complex on the surface of BSA or HSA. The apparent association constants (Kapp) of the complex with BSA or HSA were determined using the Benesi-Hildebrand equation:[34]
where Aobs is the observed absorbance of the solution at different concentrations of the complex at 280 nm, A0 and Ac are the absorbances of BSA or HSA and the complex at 280 nm, respectively, and Kapp represents the apparent association constant.
Based on the linear relationship between 1/(Aobs- A0) vs the reciprocal concentration of the complex (with a slope corresponding to 1/Kapp(Ac- A0) and an intercept to 1/(Ac - A0) (figure 8), the values of the apparent association constants (Kapp) with BSA and HSA were found to be 2.73 × 104 M−1 (R = 0.98362 for four points) and 3.75 × 103 M−1 (R = 0.99713 for four points), respectively.
3.6.1 Fluorescence quenching of BSA and HSA
The interaction of complex 1 with BSA and HSA was also studied by recording the fluorescence emission spectra of BSA or HSA with increasing concentration of 1. The emission spectra of BSA or HSA in presence of different concentrations of 1, recorded in the wavelength range 300-500 nm by exciting the albumin at 280 nm, are given in figure 9.
In both cases it was found that the fluorescence intensities of the proteins (BSA/HSA) regularly decreased with increasing the concentration of the added complex 1. This strongly supports the binding of the metal complex with BSA or HSA.[35] The fluorescence quenching of BSA or HSA is in accord with the Stern-Volmer equation (figure 10). From the linear plot of I0/I versus [complex], KSV values for BSA and HSA were calculated to be 3.62 × 104 M−1 (R = 0.98562 for four points) and 2.69 × 105 M−1 (R = 0.98946 for four points), respectively.
4 Conclusion
The synthesis and characterization of four new mononuclear nickel(II) complexes with a tetradentate ligand (L) having N2S2 chelator have been carried out. The detailed solid state structures of three of these complexes have been established by single crystal X-ray diffraction. The data indicate that the nickel(II) ion exhibits an octahedral geometry with pyridine rings in trans positions, while the two thioether-S donors are located in the equatorial plane together with the chloride anions (in complex 1), azide (in 2) and cyanate (in 3), which are at mutual cis positions. The interaction of complexes with calf thymus DNA and serum albumins (BSA and HSA) has been investigated using spectroscopic methods which indicate a strong groove binding mode of the metal complexes with CT-DNA.
References
Li Y, Yang Z Y and Wang M F 2009 Eur. J. Med. Chem. 44 4585
Margiotta N, Marzano C, Gandin V, Osella D, Ravera M, Gabano E, Platts J A, Petruzzella E, Hoeschele J D and Natile G 2012 J. Med. Chem. 55 7182
Tian X, Han X J, Feng J N, Liu J L and Zou Y Z 2012 Inorg. Chem. Commun. 15 5
Kretzer R M, Ghiladi R A, Lebeau E L, Liang H C and Karlin K D 2003 Inorg. Chem. 42 3016
Marzano C, Pellei M, Colavito D, Alidori S, Lobbia G G, Gandin V, Tisato F and Santini C 2006 J. Med. Chem. 49 7317
Solomon E I, Jones P M and Maj A 1993 Chem. Rev. 93 2623
Suganthy P K, Prabhu R N and Sridevi V S 2013 Tetrahedron Lett. 54 5695
Casas J S, Castellans E E, Louce M D, Ellena J, Sanchez A, Sordo J and Taboada C 2006 J. Inorg. Biochem. 11 1858
Tian Y P, Duan C Y, Lu Z L, You X Z, Fun H K and Kandasamy S 1996 Polyhedron 15 2263
Dhara P K, Das B, Lo J M and Chattopadhyay P 2005 Appl. Radiat. Isotopes 62 729
Sarkar S, Sen S, Dey S, Zangrando E and Chattopadhyay P 2010 Polyhedron 29 3157
Patra A, Sen S, Sarkar S, Zangrando E and Chattopadhyay P 2012 J. Coord. Chem. 65 4096
Chattopadhyay P, Chui Y H, Lo J M, Chung C S and Lu T H 2000 Appl. Radiat. Isot. 52 217
Patra A, Sen B, Sarkar S, Pandey A, Zangrando E and Chattopadhyay P 2013 Polyhedron 31 156 and refs. therein
Mukherjee T, Marek J and Chattopadhyay P 2012 Trans. Met. Chem. 37 155
Mukherjee T, Sen B, Patra A, Banerjee S, Hundal G and Chattopadhyay P 2014 Polyhedron 69 127 and refs. therein
Otwinowski Z and Minor W In Processing of X-ray Diffraction Data Collected in Oscillation Mode, Methods in Enzymology 1997 C W Carter Jr. and R M Sweet (Eds.) (New York: Academic Press) 276 307
Sheldrick G M 2008 Acta Cryst. A 64 112
Farrugia L J 1999 J. Appl. Crystallogr. 32 837
Karmakar S, Choudhury S B, Ray D and Chakravorty A 1993 Polyhedron 12 291
Mukhopadhyay S and Ray D 1995 J. Chem. Soc., Dalton Trans. 265
Bhunia P, Sardar D, Sarker K K, Ray U S, Wu J -S, Lu T -H and Sinha C 2009 J. Coord. Chem. 62 552
Patra A, Sarkar S, Drew M G B, Zangrando E and Chattopadhyay P 2009 Polyhedron 28 1261
Raman N, Pothiraj K and Baskaran T 2011 J. Coord. Chem. 64 4286
Manjunatha M, Naik V H, Kulkarni A D and Patil S A 2011 J. Coord. Chem. 64 4264
Xu J, Chen Y F, Zhou H and Pan Z Q 2011 J. Coord. Chem. 64 1626
Tysoe S A, Morgan R J, Baker A D and Strekas T C 1993 J. Phys. Chem. 97 1707
Barton J K, Danishefsky A T and Goldberg J 1984 J. Am. Chem. Soc. 106 2172
Vijayalakshmi R, Kanthimathi M, Subramanian V and Nair B U 2000 Biochem. Biophys. Acta 157 1475
Pyle A M, Rehmann J P, Meshoyrer R, Kumar C V, Turro N J and Barton J K 1989 J. Am. Chem. Soc. 111 3051
Stern O and Volmer M 1919 Z. Phys. 20 183
Kathiravan A and Renganathan R 2009 Polyhedron 28 1374
Satyanarayana S, Dabrowiak J C and Chaires J B 1992 Biochemistry 31 9319
Benesi H A and Hildebrand J H 1949 J. Am. Chem. Soc. 71 2703
Zhang X P, Hou Y H, Wang L, Zhang Y Z and Liu Y 2013 J. Lumin. 138 8
Acknowledgements
Financial support from Council of Scientific and Industrial Research (CSIR), New Delhi, India is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supporting Information
Crystallographic data for the structural analyses have been deposited with the Cambridge Crystallographic Data Centre, CCDC Nos. 953033, 953035 and 953034 for compounds 1, 2 and 3, respectively. Copies of this information are available on request at free of charge from CCDC, 12 Union Road, Cambridge, CB21EZ, UK (fax: + 44-1223-336-033; e-mail: deposit@ccdc.ac.uk or http://www.ccdc.cam.ac.uk). Selected bond distances (Å) and angles (∘) for complexes 2 and 3 are given in Table S1 available at www.ias.ac.in/chemsci.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
PATRA, A., MONDAL, B., SEN, B. et al. Nickel(II) complexes of N2S2 donor set ligand and halide/pseudohalides: Synthesis, crystal structure, DNA and bovine/human serum albumin interaction. J Chem Sci 127, 1883–1893 (2015). https://doi.org/10.1007/s12039-015-0962-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12039-015-0962-3