Abstract
Recently, microRNA-1247 (miR-1247) has been reported to function as tumour suppressor in several cancer types, including pancreatic cancer, hepatocellular cancer and lung cancer. However, the biological function of miR-1247 in bladder cancer and the underlying mechanisms have remained largely uncovered. In this study, the expression of miR-1247 was significantly downregulated, while RAB36 protein was remarkably upregulated in bladder cancer tissues and cell lines compared with that in paired adjacent normal tissues or normal cell line (SU-HUC-1). The function of miR-1247 and RAB36 in the cell viability, proliferation and invasion of bladder cancer cells (T24 and J82) was assessed by CCK-8, colony formation and Transwell assay, respectively. Gain of function studies showed that upregulation of miR-1247 significantly inhibited cell proliferation and invasion capacity of bladder cancer cells. Consistently, downregulation of RAB36 mimicked the suppressive effects of miR-1247 overexpression in bladder cancer cells. Importantly, miR-1247 was confirmed to target the 3′untranslated region (UTR) of RAB36 and downregulated its expression using luciferase reporter assay and Western blot assays. In conclusion, these results provide the first clues regarding the role of miR-1247 might be a potential therapeutic agent and diagnostic marker of bladder cancer by inhibiting RAB36 expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
As one of the most common cancer worldwide, bladder cancer is correlated with high incidence and mortality (Powles et al. 2014). There are two main long-recognized disease entity known as non-muscle-invasive and muscle-invasive bladder cancer (Rentsch et al. 2017). Non-muscle-invasive carcinoma tends to recurrence frequently, but in rare instances may also invades the muscle, while muscle-invasive carcinoma has a great potential for metastasis and migration beyond the bladder (Herr and Donat 2016). Despite the majority of newly diagnosed bladder cancer are non-muscle-invasive type (75–80% of patients), bladder cancer remains a global public health burden, and caused an estimated 150,000 deaths per year (Rentsch et al. 2017). Smoking is a major cause of bladder cancer and accounts for almost half of all cases, exposure to other chemical substances such as aromatic amines and polycyclic aromatic hydrocarbons are other potential risk factors for bladder cancer (Burger et al. 2013). Developing deep understanding of bladder cancer initiation and progression will open new avenues for exploring new treatment strategies.
MicroRNAs constitute a large family of small noncoding RNAs (~21 nucleotides) that function as guide molecules in post-transcriptional regulation of gene expression via base-pairing with their target sites (Krol et al. 2010). A surprisingly more than 1000 annotated miRNAs were identified in the human genome, as many of them are associated with human disease, particularly in cancer (Alvarez-Garcia and Miska 2005, Chiang et al. 2010). MiR-1247 were found to lie in intergenic regions and shows deviant expression in various tumour types including pancreatic cancer (Shi et al. 2014), prostate cancer (Scaravilli et al. 2015) and osteosarcoma (Zhao et al. 2015). An earlier report pointed out that miR-1247 leads to profound phenotypic impact on the alteration of SOX9, which is a master regulator of chondrogenesis (Martinez-Sanchez and Murphy 2013). Recently, miR-1247 has been reported to be down-regulated in non-small-cell lung cancer and hepatocellular carcinoma, and regulates tumour growth and invasion (Zhang et al. 2016; Chu et al. 2017). Accumulating evidences indicate that miRNAs play a crucial role in the development of bladder cancer, such as miR-150 (Lei et al. 2014), miR-143 (Lin et al. 2009), and miR-27a (Drayton et al. 2014). However, to the best of our knowledge, the relationship between miR-1247 and bladder cancer has not been reported yet.
RAB family are critical modulators of vesicle trafficking and play an important role in eukaryotic cells growth and development (Wandinger-Ness and Zerial 2014). Accumulating evidences have suggested that a number of RAB family members are crucially implicated in the development of human cancer (Ho et al. 2012). The overexpressed RAB proteins include few like RAB11A in skin carcinogenesis, RAB1A in tongue squamous cell carcinoma, and RAB5A in hepatocellular carcinomas that activates or contributes tumour progression (Gebhardt et al. 2005; Shimada et al. 2005; Fukui et al. 2007). The RABs with growth inhibitive properties include RAB37 in lung cancer and RAB25 in colorectal cancer (Wu et al. 2009; Goldenring and Nam 2011). Increasing studies have reported that the members of RAB family could be targeted by miRNAs in various tumour types. For instance, RAB27A was confirmed as a target of miR-134-3p and miR-134-3p/RAB27A might be a potential target for ovarian cancer treatment (Chang et al. 2017). RAB18 was found to be overexpressed in gastric cancer and demonstrated to be a candidate target of miR-455-5p (Liu et al. 2016). RAB36 also belongs to the RAB-family members and locates on chromosome 22q11.2 (Mori et al. 1999). Mori et al. (1999) described that depletion of RAB36 could facilitate malignant rhabdoid tumours dissemination, whereas ectopic expression of RAB36 failed to sufficiently justify the anti-tumour function of this gene.
Although the dysregulation of miR-1247 and RAB36 plays important roles in the carcinogenesis of tumour progression, there are limited reports about the correlation between miR-1247 and RAB36 in bladder cancer. In the present study, we predicted that RAB36 was a target of miR-1247. Furthermore, gain-of-function in miR-1247 and loss-of-function in RAB36 caused similar effects on proliferation and invasion of bladder cancer cells. Our investigation provides the first report of miR-1247 and RAB36 in bladder cancer growth and metastasis, also reveal a direct correlation between them, which will deepen our understanding of the pathological mechanism of bladder cancer.
2 Materials and methods
2.1 Expression datasets
To investigate the expression level of miR-1247 in bladder cancer, the mRNA expression data was downloaded from the Cancer Genome Atlas project (TCGA dataset, https://portal.gdc.cancer.gov/) dataset. Total 429 specimens were available including 410 bladder cancer and 19 adjacent tissues. The t-test method was used to compare the expression of miR-1247 between bladder cancer and adjacent normal tissues.
2.2 Tissue collection
Total 24 fresh bladder cancer tissues and paired normal adjacent tissues were obtained from patients undergoing surgical resection in Department of Urology, Ningbo No. 2 Hospital (Zhejiang, China). None of the patients received any hormones, traditional Chinese medicine, radiotherapy or chemotherapy treatments before surgery. The clinical and pathological characteristics of the patients are presented in table 1. All collected tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until further use for quantitative real-time PCR (qRT-PCR) or Western blot analysis. Written informed consent was obtained from all the patients. This study was approved by the Ethics Committee from Ningbo No. 2 Hospital.
2.3 Cell lines and transfection
The human bladder cancer cell lines (T24, HT1376, J82 and SW780) and normal bladder epithelial cell line SV-HUC-1 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). T24 and J82 cell lines were cultured in RPMI 1640 medium (Welgene, Daegu, Korea). HT1376 and SW780 cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM, Welgene). Cell line SV-HUC-1 was cultured in F12K medium (Invitrogen, Carlsbad, CA, USA). All cell culture medium were supplemented with 10% fetal bovine serum (FBS, Gibco, CA, USA). All cell lines were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
MiR-1247 mimics or the negative control (NC) mimics, small interfering RNA (siRNA) oligonucleotides targeting RAB36 (siRAB36) or scramble control (siNC) were synthesized by GenePharma (Shanghai, China). For cell transfection, T24 and J82 cells were seeded into 6-well plates until reached 70–80% confluency. Then miR-1247 overexpression was achieved by transfecting cells with miR-1247 mimics or the corresponding NC mimics, and RAB36 knockdown was achieved by transfecting either siRAB36 or siNC using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. After 48 h transfection, the cells were harvested and subjected to the following analysis.
2.4 QRT-PCR
Total RNA was extracted from bladder tissues or cells using the Trizol reagent (Invitrogen). Expression of miR-1247 was carried out by using Power SYBR qPCR and miRNA qRT-PCR detection kit (Applied Biosystems) according to the company’s instructions in a CFX96 real- time PCR detection system (Bio-Rad, USA). U6 snRNA was used as the reference gene for miRNA. The relative expression of miR-1247 was measured by the 2-ΔΔCT method. Each sample was run in three times.
2.5 Western blot
Total proteins were extracted from tissues or cells by RIPA lysis buffer (Beyotime Biotechnology, China) supplemented with protease inhibitors (Roche, China). After quantified using BCA protein assay kit (Pierce, USA), approximately 30 μg proteins were separated by 10% SDS-PAGE and transferred to PVDF membranes. Then the membranes were blocked with 5% skim milk dissolved in TBS containing 0.02% Tween 20, followed by washing and incubation with primary antibodies against RAB36 (Abcam, Cambridge, UK) and GAPDH (Cell Signaling Technology, MA, USA) at 4°C overnight. Following extensive washing with PBS, the membranes were subsequently incubated with horseradish peroxidase labeled secondary antibodies for 2 h at room temperature. Protein bands were visualized by enhanced chemiluminescence detection kit (Abcam) and the intensity of the bands was quantified using Image Lab™ Software (Bio-Rad, China).
2.6 Cell viability assay
Cell viability was assessed using a Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer’s instruction. Briefly, cells were plated in 96-well plates at a density of 5,000 cells per well. At the indicated time points (1, 2, 3, 4 and 5 day following transfection), 10 μL of CCK-8 reagent was added into each well at and the plates were incubated for 3 h at 37°C. The optical density (OD) value was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad). The experiment was performed in at least triplicate.
2.7 Colony formation assay
After 48 h transfection, cells were plated in 6-well plates at a density of 500 cells per well and cultured in an incubator containing 5% CO2 for 7 days. Then the colonies were fixed with methanol for 20 min and stained with 0.1% crystal violet for 30 min. The colonies (more than 50 cells per colony) were photographed and counted after washing with PBS. The experiment was performed in at least triplicate.
2.8 Matrigel invasion assay
The cell invasive capacity was analysed using 8-µm pore-size Transwell chambers coated with Matrigel (BD Bioscience, USA) according to the manufacturer’s instructions. In brief, cells were seeded on the upper chamber with the serum free medium while the bottom chambers were filled with complete medium containing 10% FBS used as a chemoattractant. Following 24 h incubation at 37°C, those cells that migrated to the lower membrane were fixed with 20% methanol, stained with 0.1% crystal violet, and then quantified by calculating the average cell counts in five random fields under a light microscope (Olympus, Japan).
2.9 Dual-luciferase reporter assay
The 3′UTR of RAB36 mRNA was amplified and cloned into the firefly psiCHECK2 luciferase plasmid (Promega, Madison, WI, USA) and confirmed by sequencing to generate wild-type luciferase vector (RAB36-WT). To verify the direct binding specificity, the sequences of RAB36 3′UTR that bound to the miR-1247 were mutated and also cloned into the psiCHECK2 vector to form a mutated luciferase vector (RAB36-MUT). Subsequently, T24 and J82 cells were co-transfected with RAB36-WT or RAB36-MUT vector with miR-1247 mimics or NC mimics using Lipofectamine 2000 for 48 h. The relative firefly/Renilla luciferase activity was then detected using a Dual Luciferase Reporter Assay (Promega) following the manufacturer’s instruction.
2.10 Statistical analysis
Statistical analysis was performed in the SPSS18.0 software (SPSS, Inc., IL, USA). Quantitative data was expressed as mean ± SD of at least three experiments. Student’s t-test was used to evaluate the difference between two groups. Statistical significance between three or more groups was measured by one-way analysis of variance. A value of p < 0.05 was considered to be statistically significant.
3 Results
3.1 MiR-1247 expression was downregulated while RAB36 expression was upregulated in human bladder cancer tissues and cell lines
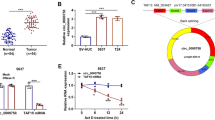
To determine whether miR-1247 was involved in the development of bladder cancer, we first downloaded the expression data of miR-1247 from TCGA to analyse the differential expression between bladder tissues and non-tumour tissues. As shown in figure 1A, the expression of miR-1247 was significantly downregulated in bladder cancer (p = 7.2E–07) in comparison with normal tissues. To further validate the expression of miR-1247 in bladder cancer, the expression level of miR-1247 was determined in twenty paired human bladder cancer tissues and several cell lines by qRT-PCR. As shown in figure 1B, the expression of miR-1247 was 2.56-fold lower in tumour tissues compared with that in paired adjacent normal tissue (p<0.001). Similarly, the expression of miR-1247 in bladder cancer cell lines including T24, HT1376, J82 and SW780, was significantly decreased compared with that in SU-HUC-1 cell line (figure 1C, p<0.01, p<0.001). In addition, we detected RAB36 expression in the remaining four paired human bladder cancer tissues and cell lines using Western blotting. Contrary to miR-1247, the expression of RAB36 protein was remarkably upregulated in representative tumour tissues and cell lines compared with that in paired adjacent normal tissues or normal cell line SU-HUC-1 (figure 1D and E). Notably, T24 and J82 cell lines were selected as the in vitro experiments due to their low expression levels of miR-1247 and high expression levels of RAB36 compared with the other bladder cancer cell lines.
Analysis of miR-1247 and RAB36 expression in bladder cancer tissues and cell lines. (A) The expression levels of miR-1247 in bladder cancer tissues from TCGA dataset (n = 429, p = 7.2E−07). Quantitative RT-PCR (qRT-PCR) analysis of expression pattern of miR-1247 in (B) 20 paired bladder cancer tissues and (C) cell lines (T24, HT1376, J82 and SW780); Relative expression of RAB36 protein was detected in (D) 4 paired bladder cancer tissues and (E) cell lines (T24, HT1376, J82 and SW780) using Western blotting analysis. Data are expressed as means ± SD of three independent experiments. **p<0.01, ***p<0.001 vs. normal tissues or the normal bladder epithelial cell (SV-HUC-1).
3.2 MiR-1247 overexpression suppressed cell proliferation and invasive activity in bladder cancer
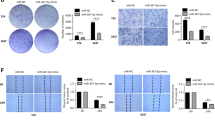
Next, we examined the effects of miR-1247 on the bladder cancer cells. The expression of miR-1247 was overexpressed in T24 and J82 cells by transfecting miR-1247 mimics, respectively. As shown in figure 2A, the transfection efficiency was confirmed by qRT-PCR (p<0.001). CCK-8 assay showed that miR-1247 mimics transfection significantly decreased cell viability compared with NC mimics transfection in both T24 and J82 cells (figure 2B, p<0.01, p<0.001). Moreover, we found miR-1247 overexpression remarkably reduced the number of colonies from 125.0 ± 2.6 to 42.3 ± 2.5 in T24 cells, from 109.3 ± 4.2 to 39.0 ± 3.0 in J82 cells (figure 2C, p<0.001). Furthermore, Matrigel invasion assay indicated that the number of invasive cells in the miR-1247 mimics transfected group was significantly decreased compared that in the NC mimics group in both T24 and J82 cells (figure 2D, p<0.001).
The effects of miR-1247 overexpression on the proliferation and invasion of bladder cancer cells. T24 and J82 cells were transfected with the miR-1247 mimics or NC mimics, respectively. (A) Quantitative real time PCR analysis of miR-1247 expression in cells after 48 h transfection. (B) CCK-8 assay was performed to analyse cell viability. (C) The proliferation capacity was determined by colony formation assay. (D) Transwell analysis of invading cells treated with miR-1247 mimics or NC mimics. Data are expressed as means ± SD of three independent experiments. **p<0.01, ***p<0.001 vs. NC mimics.
3.3 RAB36 knockdown inhibited bladder cancer cell proliferation and invasive activity
RAB36, a novel Rab-family gene, was shown to be significantly upregulated in bladder cancer, but its function remains unclear. Thus, we further performed loss-of-function studies to investigate its functional role in bladder cancer cells. As shown in figure 3A, siRAB36 transfection obviously downregulated the expression of RAB36 protein in both T24 and J82 cells. Consistently with miR-1247 overexpression, RAB36 knockdown significantly reduced cell viability (figure 3B, p<0.001), impaired colony formation ability (figure 3C, p<0.001) and inhibited invasive capacity (figure 3D, p<0.001) in both T24 and J82 cells.
The effects of RAB36 knockdown on the proliferation and invasion of bladder cancer cells. T24 and J82 cells were transfected with the siRAB36 or siNC mimics, respectively. (A) Western blotting analysis of RAB36 expression in cells after 48 h transfection. (B) CCK-8 assay was performed to analyse cell viability. (C) The proliferation capacity was determined by colony formation assay. (D) Transwell analysis of invading cells treated with siRAB36 or siNC. Data are expressed as means ± SD of three independent experiments. **p<0.01, ***p<0.001 vs. siNC.
3.4 MiR-1247 directly targeted the 3’UTR of RAB36 and inhibited its expression in bladder cancer cells
To test whether miR-1247 could directly interact with RAB36 in bladder cancer cells, we found that the 3′UTR of RAB36 contained a potential miR-1247 binding site by the online miRDB (http://www.mirdb.org/), miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/) (figure 4A). Therefore, the RAB36-WT and RAB36-MUT luciferase vector were constructed and co-transfected into T24 and J82 cells together with miR-1247 mimics or NC mimics. As shown in figure 4B, the relative luciferase activity in the RAB36-WT group was obviously suppressed compared with the RAB36-MUT group after transfected with miR-1247 mimics (p < 0.001). Meanwhile, the Western blotting demonstrated that the protein levels of RAB36 were obviously downregulated after transfection with miR-1247 mimics in T24 and J82 cells (figure 4C).
RAB36 was identified as a direct target gene of miR-1247 in the bladder cancer cell. (A) TargetScan software was used to predict the binding sequences of miR-1247 in the 3′UTR of RAB36. (B) Luciferase reporter assays in T24 and J82 cells. (C) The protein expression of RAB36 was determined by Western blotting in T24 and J82 cells. Data are expressed as means ± SD of three independent experiments. ***p<0.001 vs. siNC.
4 Discussion
Abnormal expression of miRNAs can destroy the network of protein-coding RNA, it is not surprising that they have been found to affect diverse biological processes (Murchison and Hannon 2004). A large number of studies have characterized the regulation of miRNAs profiles in malignant tumours and identified multiple miRNAs that are increased or decreased in tumour cells and tissues (Calin and Croce 2006). Several genes including oncogenes and anti-oncogenes were discovered as targets of dysregulated miRNAs in bladder cancer (Hanke et al. 2010). Such as, miR-16 suppresses cell viability in bladder cancer cells by its action on Cyclin D1, which function as a cell cycle modulatory switch for cell proliferation (Jiang et al. 2013). MiR-124-3p and miR-409-3p has been shown to be implicated in growth and tumorigenesis of bladder cancer cells though targeting ROCK1 and c-Met (Xu et al. 2013a, b). Therefore, identification of miRNAs differentially expressed between normal and bladder cancer tissues (cell lines), and their target genes are of great help in revealing the occurrence and development mechanism of bladder cancer.
MiR-1247 is reported to be downregulated in pancreatic cancer (Shi et al. 2014), osteosarcoma(Zhao et al. 2015), and non-small-cell lung cancer (Zhang et al. 2016), and its expression associates with tumor proliferation and invasion. In the current study, we have also observed reduced expression of miR-1247 in bladder cancer tissues and cell lines compared to normal controls, indicate that miR-1247 exert its anti-tumour activity in bladder cancer. Subsequent studies succeed to confirm this speculation that hyperexpression of miR-1247 attenuated proliferation and invasion of bladder cancer cells. Contrary to miR-1247, a novel member of the RAB family, RAB36 was overexpressed in bladder cancer tissues and cell lines and silencing of RAB36 was linked to the suppression of malignant biological behaviors. Interestingly, RAB36 was negatively regulated at the translational level by miR-1247. Luciferase reporter assay validated that RAB36 was a direct target of miR-1247. These data suggest that miR-1247 regulate tumour proliferative activity and invasiveness though targeting RAB36.
Small GTPase RAB36, encoding a protein homologous to RAB34, they share more than 50% amino acid sequence identity (Chen et al. 2010). By a similar mechanism, RAB36 and RAB34 were shown to mediate redistribution of late endosomes and lysosomes (Chen et al. 2010). Unexpectedly, Wu et al. (2017) found that RAB34 are correlated with clinicopathological features in hepatocellular carcinoma, including large tumour size, vessel invasion and advanced tumour stage. Moreover, depletion of RAB34 resulted in G1 cell cycle arrest and disruption of mesenchymal-epithelial transition (Wu et al. 2017). Hence, we hypothesized that RAB36 regulated proliferation and invasion possibly via a similar mechanism to RAB34 in bladder cancer with miR-1247 overexpression. Besides, other RAB family members like RAB25 and RAB5A are oncogenes in ovarian cancer (Cheng et al. 2005; Zhao et al. 2010). Briefly, APPL1-related epidermal factor signaling might be responsible for the pro-tumour properties of RAB5A in ovarian cancer (Zhao et al. 2010). Increases in RAB25 could promotes AKT phosphorylation in ovarian cancer, it provides an indication of PI3K pathway activation, which roles in oncogenesis and implicated cell proliferation, growth and motility (Lu et al. 2003). In other situations, overexpression of miR-1247 contributed to proliferation and invasion of bladder cancer cells, might though alteration of APPL1-related epidermal factor and/or PI3K/AKT signallings, via targeting RAB36. Our next study will focus on the concrete-mechanism of miR-1247/RAB36 pathway responsors in bladder cancer.
In conclusion, miR-1247 was significantly downregulated in bladder cancer tissues and cell lines in comparison with adjacent normal tissues or normal cell line. It could attenuate the tumour growth and metastasis by interaction with RAB36. Our results may provide useful information for uncovering the occurrence and development of bladder cancer. However, further studies on the regulatory mechanisms remain to be elucidated.
References
Alvarez-Garcia I and Miska EA 2005 MicroRNA functions in animal development and human disease. Development 132 4653–4662
Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La C Vecchia, Shariat S and Lotan Y 2013 Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 63 234–241
Calin GA and Croce CM 2006 MicroRNA signatures in human cancers. Nat. Rev. Cancer 6 857–866
Chang C, Liu T, Huang Y, Qin W, Yang H and Chen J 2017 MicroRNA-134-3p is a novel potential inhibitor of human ovarian cancer stem cells by targeting RAB27A. Gene 605 99–107
Chen L, Hu J, Yun Y and Wang T 2010 Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol. Membr. Biol. 27 23–30
Cheng KW, Lahad JP, Gray JW and Mills GB 2005 Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 65 2516–2519
Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C and Bartel DP 2010 Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 24 992–1009
Chu Y, Fan W, Guo W, Zhang Y, Wang L, Guo L, Duan X, Wei J and Xu G 2017 miR-1247-5p functions as a tumor suppressor in human hepatocellular carcinoma by targeting Wnt3. Oncol. Rep. 38 343–351
Drayton RM, Dudziec E, Peter S, Bertz S, Hartmann A, Bryant HE and Catto JW 2014 Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin. Cancer Res. 20 1990–2000
Fukui K, Tamura S, Wada A, Kamada Y, Igura T, Kiso S and Hayashi N 2007 Expression of Rab5a in hepatocellular carcinoma: Possible involvement in epidermal growth factor signaling. Hepatol. Res. 37 957–965
Gebhardt C, Breitenbach U, Richter KH, Furstenberger G, Mauch C, Angel P and Hess J 2005 c-Fos-dependent induction of the small ras-related GTPase Rab11a in skin carcinogenesis. Am. Pathol. J. 167 243–253
Goldenring JR and Nam KT 2011 Rab25 as a tumour suppressor in colon carcinogenesis. Br. J. Cancer 104 33–36
Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM and Sczakiel G 2010 A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 28 655–661
Herr H and Donat SM 2016 Commentary: Does asymptomatic bacteriuria reduce the risk of recurrence of non-muscle-invasive bladder tumors? Urology 98 1–3
Ho JR, Chapeaublanc E, Kirkwood L, Nicolle R, Benhamou S, Lebret T, Allory Y, Southgate J, Radvanyi F and Goud B 2012 Deregulation of Rab and Rab effector genes in bladder cancer. PLoS One 7 e39469
Jiang QQ, Liu B and Yuan T 2013 MicroRNA-16 inhibits bladder cancer proliferation by targeting Cyclin D1. Asian Pac. J. Cancer Prev. 14 4127–4130
Krol J, Loedige I and Filipowicz W 2010 The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 597–610
Lei Y, Hu X, Li B, Peng M, Tong S, Zu X, Wang Z, Qi L and Chen M 2014 miR-150 modulates cisplatin chemosensitivity and invasiveness of muscle-invasive bladder cancer cells via targeting PDCD4 in vitro. Med. Sci. Monit. 20 1850–1857
Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C and Huang L 2009 MicroRNA-143 as a Tumor Suppressor for Bladder Cancer. J. Urol. 181 1372–1380
Liu J, Zhang J, Li Y, Wang L, Sui B and Dai D 2016 MiR-455-5p acts as a novel tumor suppressor in gastric cancer by down-regulating RAB18. Gene 592 308–315
Luo J, Manning BD and Cantley LC 2003 Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4 257–262
Martinez-Sanchez A and Murphy CL 2013 miR-1247 functions by targeting cartilage transcription factor SOX9. J. Biol. Chem. 288 30802–30814
Mori T, Fukuda Y, Kuroda H, Matsumura T, Ota S, Sugimoto T, Nakamura Y and Inazawa J 1999 Cloning and characterization of a novel Rab-family gene, Rab36, within the region at 22q11.2 that is homozygously deleted in malignant rhabdoid tumors. Biochem. Biophys. Res. Commun. 254 594–600
Murchison EP and Hannon GJ 2004 miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16 223–229
Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, Shen X, Boyd Z, Hegde PS, Chen DS and Vogelzang NJ 2014 MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515 558–562
Rentsch CA, Muller DC, Ruiz C and Bubendorf L 2017 Comprehensive molecular characterization of urothelial bladder carcinoma: A step closer to clinical translation? Eur. Urol. 72 960–961
Scaravilli M, Porkka KP, Brofeldt A, Annala M, Tammela TL, Jenster GW, Nykter M and Visakorpi T 2015 MiR-1247-5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate 75 798–805
Shi S, Lu Y, Qin Y, Li W, Cheng H, Xu Y, Xu J, Long J, Liu L, Liu C and Yu X 2014 miR-1247 is correlated with prognosis of pancreatic cancer and inhibits cell proliferation by targeting neuropilins. Curr. Mol. Med. 14 316–327
Shimada K, Uzawa K, Kato M, Endo Y, Shiiba M, Bukawa H, Yokoe H, Seki N and Tanzawa H 2005 Aberrant expression of RAB1A in human tongue cancer. Br. J. Cancer 92 1915–1921
Wandinger-Ness A and Zerial M 2014 Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6 a022616
Wu CY, Tseng RC, Hsu HS, Wang YC and Hsu MT 2009 Frequent down-regulation of hRAB37 in metastatic tumor by genetic and epigenetic mechanisms in lung cancer. Lung Cancer 63 360–367
Wu J, Lu Y, Qin A, Qiao Z and Jiang X 2017 Overexpression of RAB34 correlates with poor prognosis and tumor progression in hepatocellular carcinoma. Oncol. Rep. 38 2967–2974
Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu X, Zhu Y, Li S, Zheng X and Xie L 2013 MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol. Cells 36 62–68
Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, Xu X, Wu J, Zhu Y, Zheng X, Luo J and Xie L 2013 MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J. Transl. Med. 11 276
Zhang J, Fu J, Pan Y, Zhang X and Shen L 2016 Silencing of miR-1247 by DNA methylation promoted non-small-cell lung cancer cell invasion and migration by effects of STMN1. Oncol. Targets Ther. 9 7297–7307
Zhao F, Lv J, Gan H, Li Y, Wang R, Zhang H, Wu Q and Chen Y 2015 MiRNA profile of osteosarcoma with CD117 and stro-1 expression: miR-1247 functions as an onco-miRNA by targeting MAP3K9. Int. Clin. J. Exp. Pathol. 8 1451–1458
Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni PH, Chen XH and Fan QS 2010 Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci. 101 1454–1462
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Rita Mulherkar.
Corresponding editor: R ITA M ULHERKAR
Rights and permissions
About this article
Cite this article
Zhu, Y., Liang, S., Pan, H. et al. Inhibition of miR-1247 on cell proliferation and invasion in bladder cancer through its downstream target of RAB36. J Biosci 43, 365–373 (2018). https://doi.org/10.1007/s12038-018-9755-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-018-9755-4