Abstract
Brain abnormalities and congenital malformations have been linked to the circulating strain of Zika virus (ZIKV) in Brazil since 2016 during the microcephaly outbreak; however, the molecular mechanisms behind several of these alterations and differential viral molecular targets have not been fully elucidated. Here we explore the proteomic alterations induced by ZIKV by comparing the Brazilian (Br ZIKV) and the African (MR766) viral strains, in addition to comparing them to the molecular responses to the Dengue virus type 2 (DENV). Neural stem cells (NSCs) derived from induced pluripotent stem (iPSCs) were cultured both as monolayers and in suspension (resulting in neurospheres), which were then infected with ZIKV (Br ZIKV or ZIKV MR766) or DENV to assess alterations within neural cells. Large-scale proteomic analyses allowed the comparison not only between viral strains but also regarding the two- and three-dimensional cellular models of neural cells derived from iPSCs, and the effects on their interaction. Altered pathways and biological processes were observed related to cell death, cell cycle dysregulation, and neurogenesis. These results reinforce already published data and provide further information regarding the biological alterations induced by ZIKV and DENV in neural cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Zika virus (ZIKV) outbreak in 2015, which in Brazil and other American countries peaked in 2016, was an international concern due to the increased number of cases of congenital brain abnormalities such as microcephaly, known as congenital Zika syndrome [1]. The World Health Organization (WHO) has declared an international public health emergency in 2016 due to those effects on newborns, first observed in Brazil [2,3,4]. After 2016, the number of ZIKV infections has declined to under 30,000 cases reported and the low-level circulation of the ZIKV was observed in 2018 [5, 6]. Although transmission of ZIKV has decreased in the Americas, more than 3700 cases of congenital birth defects associated with ZIKV infection have been described [7].
Microcephaly is a developmental malformation and the virus is considered to be a causative agent [1, 8, 9]; it severely impairs neurodevelopment and its virus-independent pathophysiology is mainly related to changes in the proliferative processes of neural progenitor cells (NPCs) and migratory processes during neurodevelopment [10,11,12,13]. ZIKV has a distinctive ability, between flaviviruses, to trespass the placental barrier, being able to infect a fetus’ brain in development during pregnancy [9, 14, 15]. As a result, congenital Zika infection causes neurological complications, such as delayed development, seizures, and hearing and visual impairment [16]. Therefore, several studies sought to understand the relationship between ZIKV infection and the development of neurological changes and brain malformations using animal models, human neurospheres, and brain organoids [17,18,19,20,21,22,23].
Previous studies have used pluripotent stem cell models to understand mechanistic effects of ZIKV on neurodevelopment, which includes deregulation of cell cycle and proliferation, oxidative damage, and neural cell death [17, 22,23,24]. In these models, pluripotent stem cells differentiated into 2D or 3D neural systems to represent different brain regions [18]. While 2D models of neural stem cells provide insight into the direct infection of a monolayer cell culture, 3D structural models, neurospheres, and brain organoids constitute more complex structures, allowing the study of tissue infection and essential cell communication and other mechanisms during neurodevelopment, closer to that which is observed in vivo [18, 24, 25]. ZIKV-infected neurospheres revealed impaired growth and disrupted pathways, affecting neurogenesis during brain development [19, 24].
Two major strains of the Zika virus, the African and the Asian, are distinguished by sequencing analysis. Those isolates found circulating in Brazil were matched to the Asian genotype [9]. Comparisons between African and Asian ZIKV strains have shown specific changes in infected neural progenitors. While the African ZIKV strain induced gene alterations leading to impairments related to the cell cycle and cell death [19, 26], the Asian ZIKV strains, including those circulating in Brazil, are connected to changes in oxidative stress, DNA replication and repair mechanisms, chromosome instability, and interruption of the neurogenic program, additionally affecting genes related to IFN signaling [23, 24, 26]. Moreover, it has been reported that the Brazilian isolate can induce a delayed innate immune response, unlike the African and others from the Asian strain [27].
Although the African ZIKV strain infection presented more rapid viral replication in the mosquito C6/36 cell line [28], and also resulted in higher mortality of chicken embryos [28], and in addition to immunocompetent mouse embryos [29], it has been shown to be less neuroinvasive [30] than infection with Asian strain isolates; thus, the congenital microcephaly is more associated with the Asian ZIKV strains [31]. Therefore, it is important to understand the mechanism underlying the ZIKV strains, not only for the development of treatment strategies but also for the comprehension of drastic increase in congenital syndromes associated with the Br ZIKV since the 2015 outbreak.
The more the temporal and geographical differences of ZIKV strains are studied, the more is understood about how specific aspects of the infection process can be altered by modifying different metabolic pathways. Nonetheless, the molecular mechanisms behind such differences remain largely unexplored. Here we investigated the molecular responses to two ZIKV strains (Br and MR766) in iPS-derived neural stem cells and neurospheres using label-free, mass spectrometry-based proteomics, and compared them to another mosquito-borne flavivirus, a strain of Dengue II serotype (DENV2 16,681). ZIKV and DENV virus strains share several protein structures [32, 33], yet DENV has not been found able to cross the placental barrier, while is able to cause neurological complications in adults [34]. Here, we observe altered patterns of expression when comparing both ZIKV strains and DENV, with the modulation of pathways involved with cellular transport, glucose metabolism, neurogenesis, and mRNA metabolism. Thus, molecular differences between the infection processes of these strains were focused on gathering data for the discovery of potential biomarkers, as well as to help understand and treat brain malformations triggered by ZIKV viral infection.
Methods
Ethics Statement
The experimental protocols and procedures were carried out in partnership with the Rehen Lab at the D’Or Institute for Research and Education (IDOR) and the Institute of Biomedical Sciences ICB-UFRJ. The experimental procedures were approved by the Institutional Research Ethics Committee of Hospital Copa D’Or (CEPCOPADOR) under protocols #727.269 and #1.269.816. All experiments were performed following relevant guidelines and regulations.
Culture of Neural Stem Cells and Neurospheres
Three different control iPS cell lines (CF1CL10, CF2CL2, and GM23679A) were differentiated into neural stem cells (NSCs) using PSC neural induction medium (Thermo Fisher Scientific, USA), containing Neurobasal medium and PSC supplement, according to the manufacturer’s protocol, and as previously described in [24]. Briefly, NSCs were cultured in monolayers on neural induction medium (Advanced DMEM/F12 and Neurobasal medium (1:1), supplemented with PSC supplement, Thermo Fisher Scientific, USA), and media was changed every other day, and maintained at 37 °C in humidified air with 5% CO2.
Neurospheres were differentiated from NSCs cultured until 80% confluence and split with Accutase (Merck-Millipore, Germany) [24]. Resuspended cells were then grown under a 90-rpm rotation on Advanced DMEM/F12 and Neurobasal medium (1:1), supplemented with 1 × N2 and 1 × B27 supplements (Thermo Fisher Scientific, USA) for 3 days after virus or MOCK infections, and maintained at 37 °C in humidified air with 5% CO2.
NSC Infection with ZIKV and DENV
DENV (DENV2 16,681) and ZIKV (BrZIKV (KU497555) and MR766) strains were propagated in C6/36 or Vero (ATCC #CCL-81) cells, respectively. Briefly, cells were either MOCK or virus-infected and cultured in the appropriate medium supplemented with 2% FBS. Conditioned medium was collected, frozen in aliquots, and titred as previously described in [19, 24]. NSCs were then inoculated with ZIKV (Br or MR766) or DENV2 at a multiplicity of infection (MOI) of 0.025. After incubation for 2 h, the medium was replaced with advanced DMEM/F12 and neurobasal media (1:1), supplemented with PSC supplement in NSCs, or suspended and cultured according to the neurosphere protocol. Cells were then maintained in culture for 3 days after infection, the same for both monoculture and suspension cell culture, enabling the formation of neurospheres.
Protein Extraction and Digestion
After viral infection and incubation, cells were washed with ice-cold PBS, scraped, and collected into a tube. Then, cells were centrifuged (200 × g for 5 min), and PBS was removed and fresh RIPA lysis buffer (300 mM NaCl; 50 mM Tris, pH 7.4; 0.5% Triton X-100) containing cOMPLETE protease inhibitor cocktail (Roche) was added. After lysis for approximately 30 min on ice, samples were kept frozen at − 80 °C.
Just prior to the quantification and digestion steps, cells were thawed and centrifuged at 10,000 × g for 10 min to separate debris. The proteins obtained during the extraction were quantified by fluorescence in a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific), and 50 μg of protein from each sample was submitted to SDS-PAGE polyacrylamide gel digestion [35]. Briefly, SDS-gel slices containing protein samples were reduced with 100 mM DTT (30 min at 60 °C) and alkylated with 200 mM iodoacetamide for 30 min at room temperature. Finally, samples were digested at a ratio of 1:100 (trypsin:protein w/w) in Ambic buffer and optimal temperatures (50 mM ammonium bicarbonate, at 37 °C) for 16 h. Extracted peptides were kept frozen (− 20 to − 80 °C) until mass spectrometry analyses were carried out.
Proteomics Analysis
Peptide loads (500 pg/μL) were injected into a nano-LC system (ACQUITY UPLC; 2D-RP/RP) coupled to the mass spectrometer (Synapt G2-Si, Waters Corporation), ionized with a nano-electrospray ionization source in positive mode (ESI +). Chromatographic separation was performed on an HSS T3 Column (1.8 µm, 75 µm × 150 mm, Waters Corporation, USA), eluted with an acetonitrile gradient from 7 to 40% (v/v) for 95 min at a flow rate of 0.4 μL/min directly into a Synapt G2-Si. Fragmentation spectra (MS/MS mode) were obtained through data-independent acquisition (DIA), with the optimization of collision energies of ionic precursors across their drift-time (UDMSE, Waters Co.). This method results in a more complex spectrum but favoring the identification of proteins, with high efficiency in the fragmentation of peptide precursor ions, regardless of peak intensity [36]. The mass spectrometer operated in resolution mode with an m/z resolving power of about 20,000 FWHM, using ion mobility with a cross-section resolving power of at least 40 Ω/ΔΩ. Injection was performed by nano-electrospray ionization in positive ion mode nano-ESI ( +) and a NanoLock Spray (Waters, UK) ionization source. The lock mass channel was sampled every 30 s. The MS/MS spectrum of [Glu1]-Fibrinopeptide B human (Glu-Fib) from the NanoLock Spray source was used for calibration of the mass spectrometer. Biological samples were run in triplicates.
The spectra were deconvolved using the Ion Accounting algorithm by Progenesis QI for Proteomics version 3.0 (Waters Co.). For the assignment of spectra, we used the Homo sapiens database from Uniprot (revised, October 2019, 20,365 entries), and identified and quantified proteins using the following parameters: 1% false discovery rate (FDR), 2 fragments/peptide, 5 fragments/protein, and 1 peptide/protein. The 3 most intense peptides of each protein were used for relative quantitation (Hi-3). Finally, peptides with a mass error greater than 20 ppm were discarded, along with any contaminating proteins common in shotgun proteomics. Proteins were considered to be differentially expressed when one-way analysis of variance (ANOVA) returned a p-value < 0.05.
In Silico Analysis
Protein data were then submitted to bioinformatic analysis using tools, databases, and predictions in systems biology for in silico evaluation of protein interactions and altered metabolic pathways, including Metascape [37]. Protein–protein interaction enrichment analysis was performed using the Molecular Complex Detection (MCODE) algorithm to identify network components and connections [38]. All Metascape analyses were carried out with the update of 2020–09-16.
Other bioinformatic tools and databases include String [39], Reactome Knowledgebase [40], CORUM [41], and the KEGG and KO (KEGG ORTHOLOGY) Databases [42].
Immunofluorescence Staining
Neural stem cells (NSCs) and neurospheres were fixed in 4% paraformaldehyde solution (Sigma-Aldrich) for 20 min and 1 h, respectively. Additionally, neurospheres were cryopreserved in 30% sucrose solution overnight, embedded in O.C.T compound (Sakura Finetek, USA), and frozen at − 80 °C, until the sectioning at 20-μm slices with a Leica CM1860 cryostat. After washing with buffer solution, NSC and neurosphere sections were permeabilized with 0.3% Triton X-100 solution for 20 min, followed by incubation with PBS-BSA 3% blocking solution for 1 h. All the primary antibodies were incubated overnight at 4 °C. For characterization, anti-Vimentin (1:500, Abcam, UK), anti-Pax-6 (1:200, Santa Cruz, USA), and anti-MAP-2 (1:100, Invitrogen, USA) for neurospheres and anti-Nestin (1:500, Neuromics, USA), anti-Sox-2 (1:100, Merck-Millipore, Germany), and anti-Pax-6 for NSC were used. For virus staining, anti-double-stranded RNA (dsRNA) rJ2 (1:60, Merck-Millipore) was used. Then, NSC and neurosphere sections were re-blocked with PBS-BSA 3% solution for 20 min and incubated with the secondary antibodies (goat anti-Mouse IgG [H + L] Secondary Antibody, Alexa Fluor® 488 conjugate [1:400, Invitrogen], and goat anti-Rabbit IgG [H + L] Secondary Antibody, Alexa Fluor® 594 conjugate [1:400, Invitrogen]) for 1 h. After washing with buffer saline, nuclei were counterstained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. The neurosphere slides and the NSC glass coverslips were mounted with Aqua-Poly/Mount (Polysciences, USA). Images were acquired on a Leica TCS-SP8 confocal microscope with the 63 × objective.
Results and Discussion
NSCs and neurospheres are two different steps in cell organization and differentiation. Using human iPSC-derived brain cells at different stages, we could explore and compare different molecular aspects of ZIKVs and DENV infection (Fig. 1). Previous report has shown the ability of ZIKV and DENV to infect NSCs [19, 24]; thus, the infection with Br ZIKV, ZIKV 766, and DENV were first performed in NSCs in monolayers. After infection, NSCs, which are one of the main targets of ZIKV [22], are cultured for 3 days in a medium designed for the expansion of NSCs in monolayers, keeping their progenitor state of differentiation, with most cells expressing PAX6 and SOX2, typical markers of progenitor cells, in addition to nestin (Supplementary Fig. 1A). Whereas neurospheres, on the other hand, are the 3D differentiation of those infected NSCs which immediately after infection were allowed to differentiate for a short period of time (3 days in vitro) in a medium that promotes differentiation of NSCs into neurons and glial cells. Due to differentiation process and more complex cell–cell contact communication, neurospheres present some cells expressing MAP2, a typical microtubule-associated protein found in neuronal cells, and vimentin, an intermediate filament typically found in glial cells, in addition to cells expressing PAX6 progenitors (Supplementary Fig. 1B).
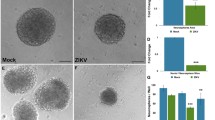
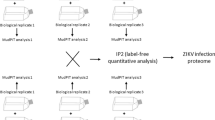
Flavivirus infection of neural cells. A Proteomic analysis workflow. The three different viral strains infected NSCs for 2 h. Then, two different protocols were performed: the culture of NSCs for 3 days and the protocol for NSCs cultivated in suspension, as neurospheres, for 3 days. The cells were harvested and the samples were prepared for proteomic analysis, followed by in silico analysis. B Representative images of neurospheres infected with Br ZIKV, ZIKV 766, and DENV and immunostained for double-stranded RNA (dsRNA) to detect viral particles in cells. Scale bar = 50 μm
All three viruses are able to infect cells (Fig. 1B) and we observe that these infections are present in neurosphere cells during the differentiation process. Therefore, after 3 days, the effects of infection of NSCs and neurospheres with ZIKV and DENV were evaluated using proteomics and resulted in a total of 792 differentially regulated proteins (p value < 0.05). The whole-cell proteomic analyses of NSCs identified and quantified a total of 1265 proteins (Supplementary Table 1), 149 of which were differentially regulated (p value < 0.05) in ZIKV MR766, 53 in Br ZIKV, and 121 in DENV, a combined modulation of 323 different proteins, as shown in Fig. 2A. A comparative analysis of those proteins revealed 10 proteins in common between Br ZIKV and MR766, 7 between Br ZIKV and DENV, and 28 between MR766 and DENV (Fig. 2A), and several biological pathways are shared among the viruses. A Pearson correlation of total protein changes in NSCs shows differences of ZIKV strains in comparison to DENV. While Br ZIKV has a higher correlation with ZIKV MR766 (range from 0.19 to 0.51) (Fig. 2B) than DENV (range from − 0.01 to 0.2), ZIKV MR766 has a similar correlation between Br ZIKV (range from 0.19 to 0.51, average of 0.38) and DENV (range from 0.20 to 0.48, average of 0.34).
Comparison of regulated proteins found in neural stem cells (NSC) and neurospheres infected by ZIKV (Br and MR766 strains) and DENV. A Overlap of proteins differentially regulated in NSCs, represented by chord diagram lines in purple (ZIKV MR766 and DENV), green (Br ZIKV and DENV), pink (Br ZIKV and MR766), and black for commonly found in all three viral strains. Light gray lines on the background are relative to the overlap of pathways and GO terms enriched by regulated proteins in NSCs. B Pearson correlation of the effects of ZIKV strains and DENV on NSCs. C Overlap of proteins differentially regulated in neurospheres, represented by chord diagram lines in purple (ZIKV MR766 and DENV), green (Br ZIKV and DENV), pink (Br ZIKV and MR766), and black for commonly found in all three virus strains. Light gray lines on the background are relative to the overlap of pathways and GO terms enriched by regulated proteins in neurospheres. D Pearson correlation of the effects of ZIKV strains and DENV on neurospheres
As neural stem cells differentiated into neurospheres, we observed a shift in the effects of the viruses on neural cell proteomes. In neurospheres, we identified and quantified a total of 1068 proteins (Supplementary Table 2), 127 of which were differentially regulated (p value < 0.05) in ZIKV MR766, 182 in Br ZIKV, and 160 in DENV. Analysis of proteins common among different strains revealed that Br ZIKV had 25 proteins in common with ZIKV MR766 and 34 with DENV, while DENV and ZIKV MR766 had 25 other proteins in common, with a total of 469 differentially regulated proteins in comparison to MOCK-infected cells (Fig. 2C). Despite having few proteins in common, several pathways are shared. In contrast to NSCs, the correlation in neurospheres between both ZIKV strains was higher (range from 0.43 to 0.71) than those with DENV, which ranged from 0.12 to 0.39 for Br ZIKV and from − 0.02 to 0.21 for ZIKV MR766 (Fig. 2D). Given that neurospheres represent more complex cellular organization than NSCs, the proteomic differences observed may reflect the complexity of the models that can be affected by these viruses.
The proteomic data thus revealed that ZIKV strains, though similar in some regards, have a distinct signature that can change in response to neurodevelopmental cell conditions. Recently, others have explored the proteomic signature differences among ZIKVs and DENV in serum samples, considering recent and past infections, aimed at distinguishing those signatures [43], finding several connections to brain proteins. Here, the similarities and differences observed among the strains — comparing the two ZIKV strains as well as ZIKV vs. DENV — will be further explored in the following sections.
ZIKV Hijacks Multiple Pathways of Cellular Maintenance in NSCs and Neurospheres
When exploring how different ZIKV strains can target neural cells and what some of their phenotypic consequences might be [21, 27, 44], several of the molecular dysregulations were varied among different models, strains, and cell types. Previous indications that Br ZIKV effects could deplete neural stem cells [24] left unanswered questions regarding when the molecular effects of infections leading to major phenotypic changes first appear: in neural progenitor cells, or when those cells start to differentiate. Therefore, we looked deeper into the modulation occurring in neural cells by exploring affected proteins and pathways.
We identified a series of pathways enriched in both NSCs and neurospheres which revealed that a major target effect of ZIKV MR766 and Br ZIKV is a common viral effect to hijack the cellular machinery to perform viral transcription and replication. Beginning at the NSC stage, the cellular metabolism of RNA, nuclear transport, RNA degradation, protein folding, translation, protein processing in the endoplasmic reticulum (ER), and destabilizing microtubule-based movement are pathways constituting the main targets of ZIKVs (Fig. 3A). These effects are essential for cell proliferation and growth and dysregulation culminates in cell cycle arrest of progenitor cells [21, 22, 24]. During the neurosphere differentiation process, defects in cellular maintenance are enhanced, leading to a major cellular response to stress, including mitochondrial permeabilization, apoptosis, and DNA change and repair, among other pathways (Fig. 3B).
Comparison of pathways and proteins associated with NSCs and neurospheres infected by ZIKV (Br and MR766 strains) and DENV. Heatmap hierarchical clustering of enriched ontology terms and pathways altered due to differentially regulated proteins in A NSCs and B neurospheres. C Differential expression data of commonly deregulated proteins among the ZIKV MR766, Br ZIKV, and DENV infections in NSCs and neurospheres; data is represented by Log fold change average ± SD of three biological replicates
Although ZIKV and DENV infections shared several pathways, there were few differentially regulated proteins common to all three strains tested. In NSCs, the only protein in common was cytoplasmic dynein 2, heavy chain 1 (DYNC2H1) (Fig. 3C), a cellular motor protein with ATP binding activity that plays an important role in cell transport and division. DYNC2H1 showed consistent, downregulated expression in all viral strains compared to MOCK-infected cells. DYNC2H1 variants have been associated with a lethal perinatal skeletal disorder [45] and a congenital malformation known as hypothalamic hamartoma [46]. Hence, those studies are in line with the association between DYNC2H1 and fetal malformations as a possible role in brain abnormalities induced by the Flaviviridae viruses, such as congenital malformations, including microcephaly, caused by ZIKV [9, 17], and neurological alterations linked to DENV (reviewed in [47]).
Regarding the neurospheres, seven proteins were differentially regulated in both ZIKVs and DENV infections (Fig. 3C), all of which showed a similar regulation in response to the viruses. Downregulated proteins were involved in ATP synthesis, cell signaling, and proliferation, including eukaryotic initiation factor 4A-II (EIF4A2), ribosomal protein L30 (RPL30), mostly when related to RNA metabolism, and 14–3-3 protein beta/alpha (YWHAB), ATP synthase peripheral stalk-membrane subunit B (ATP5PB), Reticulocalbin-1 (RCN1), and serine/threonine-protein phosphatase 2A (PPP2R2D). These proteins are associated with the Hippo signaling pathway, which promotes the inactivation of YAP/TAZ transcription factors modulating the expression of genes related to cell growth, proliferation, and apoptosis [48]. Previous studies have already linked ZIKV infection to the Hippo signaling [49], specifically regarding the control of cell proliferation, responses to stress, differentiation, and renewal of stem cells in development and the apoptosis signaling pathway [50]. In contrast, the only protein found to be upregulated in all strains was glycylpeptide N-tetradecanoyltransferase 1 (NMT1; also known as N-myristoyltransferase 1), as shown in Fig. 3C. NMT1 is a protein that performs N-terminal protein modifications [51, 52] and is thought to be associated with mechanisms affecting viral replication of flaviviruses, as its inhibition impaired DENV replication [53].
Although modulation of viral replication cellular machinery was present in ZIKVs and DENV, the direction of modulation diverged among the viruses. Our results revealed increased levels of FANCA (FA Complementation Group A) in ZIKV MR766-infected NSCs (Fig. 3C), indicating a possible association between this strain and DNA damage, as FANCA activation is associated with the blockade of replication due to DNA damage [54]. In Br ZIKV, however, this protein was downregulated. Downregulation of FA Complementation Group proteins have been associated with ZIKV infection, as a mechanism involved with viral replication and selective autophagy [55].
In NSCs, the neuronal migration protein doublecortin, encoded by the gene DCX, was identified as downregulated in both ZIKV strains. This microtubule-associated protein is a key player in neuronal differentiation and migration [56] and its downregulation in ZIKV MR766 and Br ZIKV supports the evidence that both strains impact neurogenesis [22]. DCX has been previously linked to ZIKV infection and was also reported downregulated in transcriptomic and proteomic analyses of NPCs [57], while upregulated in the in a stem cell-derived neuronal differentiation model [58]. In neurosphere, similarly modeling early neuronal differentiation, DCX was not significantly modified in our dataset (Supplementary Table 2). This shows that ZIKV infection is able to disrupt and reorganize signaling regulation throughout the infected cell since the early progenitors, influencing their outcome. The interactome of ZIKV to host proteins have revealed several cellular proteins involved in neurodevelopment [58, 59]. And ZIKV-NS4B alone, for instance, have shown to be responsible to downregulate proteins involved in neurodifferentiation, such as microtubule-associated proteins 2 and 4 [58].
The main divergent protein between the two ZIKV strains in neurospheres is already known to be essential for viral replication and intracellular transport of viruses. WASHC2C (WASH Complex Subunit 2C) (Fig. 3C) is associated with the endolysosomal system, lysosomal degradation, and receptor recycling [60, 61] and was strongly downregulated in both ZIKV strains. WASH-depleted cells have been shown to display a collapsed endolysosomal system [62]. Modifications in the aforementioned pathways suggest impaired cellular trafficking, affecting viral replication [61]. Potentially aiming to escape from these host-cell mechanisms, ZIKV-induced downregulation of WAHS2C might be associated with assured viral replication. These changes in turn deplete progenitors and disrupt the neuronal differentiation program due to interference in important pathways in NSCs and neurospheres by ZIKV infection.
Divergent Modulation of Pathways by ZIKV Strains
ZIKV and DENV have shown distinct alterations on NSCs and neurospheres. ZIKV MR766 and DENV revealed a common modulated pattern of downregulated pathways in NSCs, whereas Br ZIKV revealed upregulated pathways (Fig. 4A). This pattern, however, was not observed in neurospheres, where Br ZIKV was found to downregulate several pathways similarly to ZIKV MR766 and DENV in this 3D model (Fig. 4B).
Despite being common to the three viruses, proteins involved in mRNA processes were, on average, upregulated in Br ZIKV infection of NSCs (Fig. 4A), though DENV and ZIKV MR766 had major downregulation of proteins in this pathway. Another interesting aspect is the regulation of mRNA splicing, which was found upregulated in both ZIKV strains in NSCs and downregulated in DENV. However, in neurospheres, mRNA splicing was found upregulated in DENV (Fig. 4A), indicating potential differences due to the distinct complexity of the models evaluated. The viral NS5 protein was found to be responsible to modulate the mRNA splicing machinery, as DENV-NS5 directly interacts with U5 snRNP particle, CD2BP2 and DDX23 [63], while ZIKV-NS5 has been shown to interact with components of the Cajal body, required for spliceosomal snRNP maturation, in addition to other transcriptional and RNA processing proteins [59].
In addition to the previously mentioned dynein subunit DYNC2H1, in regards to cellular transport-related pathways, we also highlight biological processes of retrograde vesicle-mediated transport, Golgi to ER transport and Golgi organization, which might signal altered protein conformation from the ER and have been observed in the ZIKV strains in NSCs and neurospheres. Moreover, NSCs and neurospheres showed downregulation of Ras-related protein Rab-6A (RAB6A) and RAB6B (Supplementary Tables 1 and 2), which are involved with cellular transport and secretory and endocytic pathways [64], and RAB6B also plays a role in retrograde transport in neuronal cells [65]. RAB GTPases have been linked with viral replication mechanisms, especially within the late stages of viral replication [66].
NSCs showed an upregulation of Ras-related protein Rap-1B (RAP1B), which exhibits GTPase activity [67] and is involved in biological processes such as the interleukin-12-mediated signaling pathway, neutrophil degranulation, and negative regulation of synaptic vesicle exocytosis. Furthermore, RAP1B variants have been associated with congenital malformations [68]. This might indicate a potential association involving ZIKV-induced altered expression of Ras GTPase and the development of congenital malformations.
Also regarding cellular transport alterations in NSCs, AKAP9 was found upregulated in ZIKV MR766 and downregulated in Br ZIKV (Supplementary Table 1), suggesting the existence of different mechanisms involving cellular transport between both viral strains. AKAP9 is a protein from the family of A-kinase anchor proteins (AKAPs), which are known for binding to the regulatory subunit of protein kinase A (PKA), arresting the holoenzyme to determined locations within the cell, including the centrosome and the Golgi apparatus [69, 70]. These results are in line with the fact that ZIKV and other flaviviruses are known to replicate in the ER [71]; however, the specifics regarding viral assembly within the ER remain elusive [72]. On the other hand, the centrosome assembly disturbances, in addition to other proteins, have been known to contribute to significant developmental defects in genetic microcephaly [25, 73], or caused by ZIKV [44]. Altering division plane in progenitor cells, core centrosomal proteins, such as CEP128, are upregulated in Br ZIKV-infected neurospheres (Supplementary Table 2). In addition, several centrosomal proteins were found to specifically interact with the NS3 ZIKV helicase, disturbing proliferation [59].
In ZIKV MR766-infected NSCs, differentially regulated proteins highlighted the following biological processes: translation initiation, viral transcription, nuclear-transcribed mRNA catabolic process, SRP-dependent cotranslational protein targeting membrane, and nonsense-mediated mRNA decay (NMD). We also identified pathways involving ribosome and RNA transport as shown in Fig. 5A. Moreover, downregulated proteins were mostly involved in L13a-mediated translational silencing of Ceruloplasmin expression, GTP hydrolysis, and joining of the 60S ribosomal subunit (Supplementary Fig. 2), evidencing potential alterations regarding translation and associated processes.
Protein–protein interaction network from the differentially regulated proteins in NSCs infected by ZIKV. A Clustered interactions of proteins modulated by ZIKV MR766 in comparison to MOCK. In orange, SRP-dependent cotranslational protein targeting the membrane (Log10(P) − 16.3) and ribosome activity (Log10(P) − 12.9). In blue, cytoplasmic translational initiation (Log10(P) − 6.3) and RNA transport (Log10(P) − 6.0). In gray, RNA splicing, via transesterification reactions with bulged adenosine as a nucleophile (Log10(P) − 11.2), mRNA splicing, via spliceosome (Log10(P) − 11.2). In purple, proton transmembrane transport (Log10(P) − 6.1). Green nodes represent downregulated proteins and red nodes upregulated proteins. Node circumference size is determined by p-value. B Clustered interactions of proteins modulated by Br ZIKV in comparison to MOCK. The cluster in orange represents the enrichment of biosynthesis of amino acids (Log10(P) − 9.1) and glucose metabolism (Log10(P) − 6.2); in gray, mRNA splicing (Log10(P) − 6.6). Green nodes represent downregulated proteins and red nodes upregulated proteins. Node circumference size is determined by p-value
Concerning the pathways related to Br ZIKV in infected NSCs, we observed deregulation of mRNA metabolic processes, RNA stability processes, and mRNA splicing (Fig. 5B), all with the majority of proteins in each pathway found to be upregulated (Supplementary Fig. 2A). In both NSCs and neurospheres infected with Br ZIKV, mRNA stability and binding were enriched pathways, and the protein AUF1 (hnRNP D0). Moreover, we also observed pathways related to metabolic alterations including biosynthesis of amino acids and glucose metabolism, as observed in Fig. 5B. Altered glucose metabolism has been previously associated with ZIKV infection in relation to important aspects involving viral replication [74, 75]. L-lactate dehydrogenase-like protein LDHAL6A, for instance, was found upregulated in ZIKV MR766-infected NSCs, indicating increased pyruvate metabolism, though it was downregulated in Br ZIKV-infected NSCs, evidencing potential metabolic differences caused by ZIKV strains.
Between NSCs and neurospheres, there are shared pathways involved with neurodevelopment, such as regulation of expression of SLITs and ROBO receptors (Fig. 4). In NSCs, SLITs and ROBO signaling, along with connection with the transcription regulation mediated by RUNX3, presented the majority of proteins identified to be upregulated in NSCs, showing an activated state in Br ZIKV, while downregulated in neurospheres (Fig. 4). SLITs and ROBO signaling play a role in axonal guidance, cell migration, and proliferation [76, 77], and RUNX3 acts in nervous system development, including the development of sensory neurons [78, 79]. The activation state indicates an initial function in neurogenesis, which is not sustained when cells enter the differentiation path. Progenitors are a main target for ZIKV, impairing their proliferation [21, 80]. It was previously observed that ZIKV tropism is modulated by the state of differentiation of progenitor cells and that the infection affects important pathways involved in neurogenesis [81]. Our results highlight this divergence when the infection of NSCs and neurosphere proteomes were compared, due to the intrinsic differences of the models studied, which indicates that the ZIKV infection alters processes related to neurogenesis and neural cell differentiation, such as the ROBO signaling.
Neurosphere infection by ZIKV MR766 affected several cytoskeleton and ribosomal proteins in addition to voltage-dependent channels which participate in a plethora of pathways related to cell cycle regulation, as well as signaling involving activation of NMDA receptors (NMDAr), metabolism of proteins, selective autophagy, cellular response to stress, and nervous system development (Fig. 6A and Supplementary Table 2). Most proteins are downregulated, and disturbances or even small modulation in assemble and activation of NMDAr may be associated with neuronal damage induced by ZIKV, as its blockade has been shown to prevent neuronal death induced by ZIKV infection [82]. Br ZIKV infection, however, showed cell cycle regulation to be enriched by the role of GTSE1 in G2/M progression after G2 checkpoint (Fig. 6B) in a balance with the regulation of apoptosis. In neurospheres, ZIKV infection had effects on the metabolism of RNA, and differentially regulated proteins related to proteasomes — which presented opposite patterns of expression when comparing ZIKV MR766 with Br ZIKV (Supplementary Fig. 1B) — are a major cellular response to stress. Proteins of the ubiquitin–proteasome systems have been reported to interact with the NS3 helicase from ZIKV [59]. These biological pathways can lead to effects on proteins controlling growth and proliferation, such as selenophosphate synthetase 1 (SEPHS1), found downregulated in both strains of ZIKV-infected neurospheres (Fig. 3C and Supplementary Table 2). SEPHS1 is essential for cellular proliferation and differentiation, and its knockout in Drosophila results in reduced brain size [83]. This evidence strengthens the association between ZIKV and congenital malformations.
Protein–protein interaction network identified in the differentially regulated proteins in neurospheres infected by ZIKV. A Clustered interaction of proteins modulated by ZIKV MR766 in comparison to MOCK. The purple cluster was enriched for L13a-mediated translational silencing of Ceruloplasmin expression (Log10(P) − 25.3), eukaryotic translation initiation (Log10(P) − 24.9), and Cap-dependent translation initiation (Log10(P) − 24.9). The blue cluster was enriched for the role of GTSE1 in G2/M progression after G2 checkpoint (Log10(P) − 22.0), separation of sister chromatids (Log10(P) − 17.9), G2/M transition (Log10(P) − 17.9), nervous system development (Log10(P) − 11.2), and activation of AMPK downstream of NMDARs (Log10(P) − 10.3). The gray one was enriched for RAB GEFs to exchange GTP for GDP on RABs (Log10(P) − 5.3). And the orange one for COPI-mediated anterograde transport (Log10(P) − 7.3), Golgi-to-ER retrograde transport (Log10(P) − 7.0), and ER to Golgi Anterograde Transport (Log10(P) − 6.8). Green nodes represent downregulated proteins and red nodes upregulated proteins. Node circumference size is determined by p-value. B Clustered interaction of proteins modulated by Br ZIKV in comparison to MOCK. The cluster in red was enriched for the cellular amino acid metabolic process (Log10(P) − 19), the role of GTSE1 in G2/M progression after G2 checkpoint (Log10(P) − 17.6), and proteasome (Log10(P) − 17.1). The cluster in blue was enriched for the HSP90 chaperone cycle for steroid hormone receptors (SHR) (Log10(P) − 5.6), mitochondrial protein import (Log10(P) − 5.4), and cellular responses to stress (Log10(P) − 5.2). The cluster in green was enriched for SRP-dependent cotranslational protein targeting to the membrane (Log10(P) − 16.6), influenza Viral RNA Transcription and Replication (Log10(P) − 15.9), and influenza Infection (Log10(P) − 15.4). Green nodes represent downregulated proteins and red nodes upregulated proteins. Node circumference size is determined by p-value
Conclusion
Understanding ZIKV strains and their divergent and similar mechanisms is important not only for the development of treatment strategies but also for the comprehension of congenital syndromes associated with the Br ZIKV since the outbreak of 2015. We compared ZIKV infection in two different models, a two-dimensional (2D) NSC culture and a 3D neurosphere culture. Although longer and more complex 3D models are available, our study was able to compare how a similar infection would develop considering those models. Long-term infection of viral strains leads to major cell stress and cell death effects in neural cells [19, 24]. Thus, we aimed at understanding what are the first pathways disturbed after a 3-day infection to associate to possible effects on development. This allowed us to gather information regarding differentially regulated proteins and biochemical pathways shared between ZIKV strains and DENV. Br ZIKV show increase in DNA damage and consequent cell cycle arrest in G2 checkpoint, disruption in mRNA translation, and proteasome organization, leading to cell signaling stress and microtubule disarray, pointing to a decrease in neuronal program differentiation. The proteomic comparison after ZIKV (Br ZIKV and MR766) and DENV infection in a two-dimensional (2D) NSC culture and a 3D neurosphere culture revealed a proteomic fingerprint of ZIKV infection depending on the developmental state of neural cells, pointing to dysregulation of pathways leading to an impairment of the nervous system development.
Data Availability
The proteomic datasets generated for this study can be found in the PRIDE proteomics data repository (https://www.ebi.ac.uk/pride/archive/) with the accession numbers PXD026825 and PXD026909.
References
World Health Organization (2019) Zika epidemiology update. 1–14.
Brasil P, Pereira JP, Moreira ME et al (2016) Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. https://doi.org/10.1056/NEJMoa1602412
de Araujo TVB, Rodrigues LC, de Alencar Ximenes RA et al (2016) Association between Zika virus infection and microcephaly in Brazil, January to May, 2016: preliminary report of a case-control study. Lancet Infect Dis 16:1356–1363. https://doi.org/10.1016/S1473-3099(16)30318-8
Golubeva VA, Nepomuceno TC, de Gregoriis G et al (2020) Network of interactions between ZIKA virus non-structural proteins and human host proteins. Cells 9:153. https://doi.org/10.3390/cells9010153
Liu Z-Y, Shi W-F, Qin C-F (2019) The evolution of Zika virus from Asia to the Americas. Nat Rev Microbiol 17:131–139. https://doi.org/10.1038/s41579-018-0134-9
Musso D, Ko AI, Baud D (2019) Zika virus infection — after the pandemic. N Engl J Med 381:1444–1457. https://doi.org/10.1056/NEJMra1808246
Pan American Health Organization (2018) Zika cases and congenital syndrome associated with Zika virus reported by countries and territories in the Americas, 2015–2018: cumulative cases. 1–1.
Bayer A, Lennemann NJ, Ouyang Y et al 2016 Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection Cell Host Microbe 1 9 https://doi.org/10.1016/j.chom.2016.03.008
Calvet G, Aguiar RS, Melo ASO et al 2016 Articles Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study Lancet Infect Dis 1 8 https://doi.org/10.1016/S1473-3099(16)00095-5
Woods CG, Bond J, Enard W (2005) Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. The American Journal of Human Genetics 76:717–728. https://doi.org/10.1086/429930
Woods CG, Parker A (2013) Investigating microcephaly. Arch Dis Child 98:707–713. https://doi.org/10.1136/archdischild-2012-302882
Ming G-L, Tang H, Song H (2016) Advances in Zika virus research: stem cell models, challenges, and opportunities. Stem Cell 19:690–702. https://doi.org/10.1016/j.stem.2016.11.014
McGrath EL, Rossi SL, Gao J et al (2017) Differential responses of human fetal brain neural stem cells to Zika virus infection. Stem Cell Reports 8:715–727. https://doi.org/10.1016/j.stemcr.2017.01.008
Martines RB, Bhatnagar J, Keating MK et al (2016) Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses — Brazil, 2015. MMWR Morb Mortal Wkly Rep 65:159–160. https://doi.org/10.15585/mmwr.mm6506e1
Pierson TC, Diamond MS (2018) The emergence of Zika virus and its new clinical syndromes. Nature 560:573–581. https://doi.org/10.1038/s41586-018-0446-y
de Miranda-Filho D, B, Martelli CMT, Ximenes RA de A, et al (2016) Initial description of the presumed congenital Zika syndrome. Am J Public Health 106:598–600. https://doi.org/10.2105/AJPH.2016.303115
Cugola FR, Fernandes IR, Russo FB et al (2016) The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. https://doi.org/10.1038/nature18296
Dang J, Tiwari SK, Lichinchi G et al (2016) Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Stem Cell 19:258–265. https://doi.org/10.1016/j.stem.2016.04.014
Garcez PP, Loiola EC, Madeiro da Costa R et al (2016) Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. https://doi.org/10.1126/science.aaf6116
Li C, Xu D, Ye Q et al (2016) Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. https://doi.org/10.1016/j.stem.2016.04.017
Qian X, Nguyen HN, Song MM et al (2016) Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165:1238–1254. https://doi.org/10.1016/j.cell.2016.04.032
Tang H, Hammack C, Ogden SC et al (2016) Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18:1–22. https://doi.org/10.1016/j.stem.2016.02.016
Ledur PF, Karmirian K, Pedrosa CDSG et al (2020) Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci Rep 10:1218–1314. https://doi.org/10.1038/s41598-020-57914-x
Garcez PP, Nascimento JM, de Vasconcelos JM, et al (2017) Zika virus disrupts molecular fingerprinting of human neurospheres. Sci Rep 7:40780 EP –. https://doi.org/10.1038/srep40780
Lancaster MA, Renner M, Martin C-A et al (2013) Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. https://doi.org/10.1038/nature12517
Zhang F, Hammack C, Ogden SC et al (2016) Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res 44:8610–8620. https://doi.org/10.1093/nar/gkw765
Esser-Nobis K, Aarreberg LD, Roby JA et al (2019) Comparative analysis of African and Asian lineage-derived Zika virus strains reveals differences in activation of and sensitivity to antiviral innate immunity. J Virol 93:e00640-e719. https://doi.org/10.1128/JVI.00640-19
Willard KA, Demakovsky L, Tesla B et al (2017) Zika virus exhibits lineage-specific phenotypes in cell culture, in Aedes aegypti mosquitoes, and in an embryo model. Viruses 9:383. https://doi.org/10.3390/v9120383
Aubry F, Jacobs S, Darmuzey M et al (2021) Recent African strains of Zika virus display higher transmissibility and fetal pathogenicity than Asian strains. Nat Comms 12:916–1014. https://doi.org/10.1038/s41467-021-21199-z
Annamalai AS, Pattnaik A, Sahoo BR et al (2017) Zika virus encoding nonglycosylated envelope protein is attenuated and defective in neuroinvasion. J Virol. https://doi.org/10.1128/JVI.01348-17
Anfasa F, Siegers JY, van der Kroeg M, et al (2017) Phenotypic differences between Asian and African lineage Zika viruses in human neural progenitor cells. mSphere
Sirohi D, Chen Z, Sun L, et al. (2016) The 3.8 Å resolution cryo-EM structure of Zika virus. Science. https://doi.org/10.1126/science.aaf5316
Zhang X, Ge P, Yu X et al (2013) Cryo-EM structure of the mature Dengue virus at 3.5-Å resolution. Nat Struct Mol Biol 20:105–110
Carod-Artal FJ, Wichmann O, Farrar J, Gascón J (2013) Neurological complications of Dengue virus infection. Lancet Neurol 12:906–919. https://doi.org/10.1016/S1474-4422(13)70150-9
Shevchenko A, Tomas H, Havlis J et al (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. https://doi.org/10.1038/nprot.2006.468
Cassoli JS, Brandao-Teles C, Santana AG et al (2017) Ion mobility-enhanced data-independent acquisitions enable a deep proteomic landscape of oligodendrocytes. Proteomics 17:1700209. https://doi.org/10.1002/pmic.201700209
Zhou Y, Zhou B, Pache L et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Comms 10:1523. https://doi.org/10.1002/sim.4780090710
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2–27. https://doi.org/10.1186/1471-2105-4-2
Szklarczyk D, Gable AL, Lyon D et al (2018) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Jassal B, Matthews L, Viteri G et al (2020) The reactome pathway knowledgebase. Nucleic Acids Res 48:D498–D503. https://doi.org/10.1093/nar/gkz1031
Giurgiu M, Reinhard J, Brauner B et al (2019) CORUM: the comprehensive resource of mammalian protein complexes—2019. Nucleic Acids Res 47:D559–D563. https://doi.org/10.1093/nar/gky973
Kanehisa M, Furumichi M, Tanabe M et al (2016) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. https://doi.org/10.1016/j.febslet.2013.06.026
Allgoewer K, Maity S, Zhao A et al (2021) New proteomic signatures to distinguish between Zika and Dengue infections. Mol Cell Proteomics 20:100052. https://doi.org/10.1016/j.mcpro.2021.100052
Gabriel E, Ramani A, Karow U et al (2017) Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20:397-406.e5. https://doi.org/10.1016/j.stem.2016.12.005
Chen LS, Shi SJ, Zou PS, et al (2016) Identification of novel DYNC2H1 mutations associated with short rib-polydactyly syndrome type III using next-generation panel sequencing. Genetics and molecular research : GMR. https://doi.org/10.4238/gmr.15028134
Fujita A, Higashijima T, Shirozu H et al (2019) Pathogenic variants of DYNC2H1, KIAA0556, and PTPN11 associated with hypothalamic hamartoma. Neurology 93:e237–e251. https://doi.org/10.1212/WNL.0000000000007774
Li G-H, Ning Z-J, Liu Y-M, Li X-H (2017) Neurological manifestations of Dengue infection. Front Cell Infect Microbiol 7:449. https://doi.org/10.3389/fcimb.2017.00449
Buttitta LA, Edgar BA (2007) How size is controlled: from Hippos to Yorkies. Nat Cell Biol 9:1225–1227
Garcia GJ, Paul S, Beshara S et al (2020) Hippo signaling pathway has a critical role in Zika virus replication and in the pathogenesis of neuroinflammation. Am J Pathol 190:844–861. https://doi.org/10.1016/j.ajpath.2019.12.005
Mo J-S, Park HW, Guan K-L (2014) The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep 15:642–656. https://doi.org/10.15252/embr.201438638
Giglione C, Fieulaine S, Meinnel T (2015) N-terminal protein modifications: bringing back into play the ribosome. Biochimie 114:134–146. https://doi.org/10.1016/j.biochi.2014.11.008
Thinon E, Serwa RA, Broncel M et al (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Comms 5:4919–4919. https://doi.org/10.1038/ncomms5919
Suwanmanee S, Mahakhunkijcharoen Y, Ampawong S et al (2019) Inhibition of N-myristoyltransferase1 affects Dengue virus replication. MicrobiologyOpen 8:e00831. https://doi.org/10.1002/mbo3.831
Deans AJ, West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11:467–480
Tiwari SK, Dang JW, Lin N et al (2020) Zika virus depletes neural stem cells and evades selective autophagy by suppressing the Fanconi anemia protein FANCC. EMBO Rep 21:e49183. https://doi.org/10.15252/embr.201949183
Gleeson JG, Minnerath SR, Fox JW et al (1999) Characterization of mutations in the gene doublecortin in patients with double cortex syndrome. Ann Neurol 45:146–153. https://doi.org/10.1002/1531-8249(199902)45:2%3c146::aid-ana3%3e3.0.co;2-n
Jiang X, Dong X, Li S-H et al (2018) Proteomic analysis of Zika virus infected primary human fetal neural progenitors suggests a role for doublecortin in the pathological consequences of infection in the cortex. Front Microbiol 9:1067–1067. https://doi.org/10.3389/fmicb.2018.01067
Scaturro P, Stukalov A, Haas DA et al (2018) An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561:253–257. https://doi.org/10.1038/s41586-018-0484-5
Coyaud E, Ranadheera C, Cheng D et al (2018) Global interactomics uncovers extensive organellar targeting by Zika virus. Mol Cell Proteomics 17:2242–2255. https://doi.org/10.1074/mcp.TIR118.000800
Bartuzi P, Billadeau DD, Favier R et al (2016) CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat Comms 7:10961–11011. https://doi.org/10.1038/ncomms10961
Hashimoto Y, Sheng X, Murray-Nerger LA, Cristea IM (2020) Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat Comms 11:806
Gomez TS, Gorman JA, Artal-Martinez de Narvajas A et al (2012) Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol Biol Cell 23:3215–3228. https://doi.org/10.1091/mbc.e12-02-0101
De Maio FA, Risso G, Iglesias NG et al (2016) The Dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog 12:e1005841. https://doi.org/10.1371/journal.ppat.1005841
Martinez O, Goud B (1998) Rab proteins. Biochem Biophys Acta 1404:101–112. https://doi.org/10.1016/s0167-4889(98)00050-0
Wanschers BFJ, van de Vorstenbosch R, Schlager MA et al (2007) A role for the Rab6B Bicaudal-D1 interaction in retrograde transport in neuronal cells. Exp Cell Res 313:3408–3420. https://doi.org/10.1016/j.yexcr.2007.05.032
Spearman P (2018) Viral interactions with host cell Rab GTPases. Small GTPases 9:192–201. https://doi.org/10.1080/21541248.2017.1346552
Scrima A, Thomas C, Deaconescu D, Wittinghofer A (2008) The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J 27:1145–1153. https://doi.org/10.1038/emboj.2008.30
Niemann JH, Du C, Morlot S et al (2020) De novo missense variants in the RAP1B gene identified in two patients with syndromic thrombocytopenia. Clin Genet 98:374–378. https://doi.org/10.1111/cge.13807
Colledge M, Scott JD (1999) AKAPs: from structure to function. Trends Cell Biol 9:216–221. https://doi.org/10.1016/s0962-8924(99)01558-5
Larocca MC, Shanks RA, Tian L et al (2004) AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol Biol Cell 15:2771–2781. https://doi.org/10.1091/mbc.e03-10-0757
Reid CR, Airo AM, Hobman TC (2015) The virus-host interplay: biogenesis of +RNA replication complexes. Viruses 7:4385–4413. https://doi.org/10.3390/v7082825
Sirohi D, Kuhn RJ (2017) Zika virus structure, maturation, and receptors. J INFECT DIS 216:S935–S944. https://doi.org/10.1093/infdis/jix515
Garcez PP, Diaz-Alonso J, Crespo-Enriquez I et al (2015) Cenpj/CPAP regulates progenitor divisions and neuronal migration in the cerebral cortex downstream of Ascl1. Nat Comms 6:1–14. https://doi.org/10.1038/ncomms7474
Sanchez EL, Lagunoff M (2015) Viral activation of cellular metabolism. Virology 479–480:609–618. https://doi.org/10.1016/j.virol.2015.02.038
Gilbert Jaramillo J, Garcez P, James W et al (2019) The potential contribution of impaired brain glucose metabolism to congenital Zika syndrome. J Anat 235:468–480. https://doi.org/10.1111/joa.12959
Borrell V, Cárdenas A, Ciceri G et al (2012) Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76:338–352. https://doi.org/10.1016/j.neuron.2012.08.003
Blockus H, Chédotal A (2016) Slit-Robo signaling Development 143:3037–3044. https://doi.org/10.1242/dev.132829
Dykes IM, Lanier J, Raisa Eng S, Turner EE (2010) Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev 5:3–18. https://doi.org/10.1186/1749-8104-5-3
Wang JW, Stifani S (2017) Roles of Runx genes in nervous system development. Adv Exp Med Biol 962:103–116. https://doi.org/10.1007/978-981-10-3233-2_8
Brault J-B, Khou C, Basset J et al (2016) EBioMedicine EBIOM 10:71–76. https://doi.org/10.1016/j.ebiom.2016.07.018
Ferraris P, Cochet M, Hamel R et al (2019) Zika virus differentially infects human neural progenitor cells according to their state of differentiation and dysregulates neurogenesis through the Notch pathway. Emerg Microbes Infect 8:1003–1016. https://doi.org/10.1080/22221751.2019.1637283
Costa VV, Del Sarto Juliana L, Rocha RF, et al (2017) N-methyl-d-aspartate (NMDA) receptor blockade prevents neuronal death induced by Zika virus infection. mBio 8:e00350–17. https://doi.org/10.1128/mBio.00350-17
Serras F, Morey M, Alsina B et al (2001) The Drosophila selenophosphate synthetase (selD) gene is required for development and cell proliferation. BioFactors (Oxford, England) 14:143–149. https://doi.org/10.1002/biof.5520140119
Acknowledgements
The authors thank Gabriela Vitoria, Ismael Gomes, Paulo Baldasso, and Erick Loiola for their excellent technical support and Bradley Smith, MSc for the critical comments and English review support during the process.
Funding
Financial support was provided by the São Paulo Research Foundation (D. G. J., J. M. N., G. S. Z., C. B. T., J. S. C., A. S. L. M. A. and D. M. S. are supported by FAPESP grant numbers 2014/14881-1, 2014/21035-0, 2017/25055-3, 2018/25439–9, 2018/14666–4, 2020/10282-7, 2017/25588–1, 2019/00098-7), and the National Council of Scientific and Technological Development (CNPq), in addition to intramural grants from D’Or Institute for Research and Education.
Author information
Authors and Affiliations
Contributions
J. M. N., P. P. G., S. K. R., and D. M. S. conceived and designed the study. D. J. G. and J. M. N. performed in silico proteomic analyses. J. M. N. and J. S. C. performed the mass spectrometry experiments. G. S. Z. performed the pathway analysis and interpretation. P. P. G., C. S. G. P., K. K., J. A. S., A. S. L. M. A., C. B. T., L. M. H., G. F. S., S. P. M., and F. C. cultured iPS cells and/or virus strains, performed cell-based assays and/or infection, and contributed to discussion. A. T. and J. L. P. M. performed the data interpretation. D. J. G. and J. M. N. interpreted the data, wrote, edited, and revised the manuscript. D. M. S. and S. K. R. coordinated the study. All authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nascimento, J.M., Gouvêa-Junqueira, D., Zuccoli, G.S. et al. Zika Virus Strains and Dengue Virus Induce Distinct Proteomic Changes in Neural Stem Cells and Neurospheres. Mol Neurobiol 59, 5549–5563 (2022). https://doi.org/10.1007/s12035-022-02922-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02922-3