Abstract
Hairy and enhancer of split homolog-1 (HES1), regulated by the Notch, has been reported to play important roles in the immune response and cancers, such as leukemia. In this study, we aim to explore the effect of HES1-mediated Notch1 signaling pathway in chronic lymphocytic leukemia (CLL). Reverse transcription quantitative polymerase chain reaction and Western blot assay were conducted to determine the expression of HES1, Notch1, and PTEN in B lymphocytes of peripheral blood samples of 60 CLL patients. We used lentivirus-mediated overexpression or silencing of HES1 and the Notch1 signaling pathway inhibitor, MW167, to detect the interaction among HES1, Notch1, and PTEN in CLL MEC1 and HG3 cells. MTT assay and flow cytometry were employed for detection of biological behaviors of CLL cells. HES1 and Notch1 showed high expression, but PTEN displayed low expression in B lymphocytes of peripheral blood samples of patients with CLL in association with poor prognosis. HES1 bound to the promoter region of PTEN and reduced PTEN expression. Overexpression of HES1 activated the Notch1 signaling pathway, thus promoting the proliferation of CLL cells, increasing the proportion of cells arrested at the S phase and limiting the apoptosis of CLL cells. Collectively, HES1 can promote activation of the Notch1 signaling pathway to cause PTEN transcription inhibition and the subsequent expression reduction, thereby promoting the proliferation and inhibiting the apoptosis of CLL cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic lymphocytic leukemia (CLL) is regarded as the most frequently occurring peripheral blood as well as bone marrow cancer in developed countries [1]. It is a mature B-cell malignancy relying on microenvironmental factors for survival as well as proliferation [2]. Elevated lymphocyte counts were present in a large proportion of cases prior to the diagnosis of CLL. CLL is genetically complicated and afflicts a heterogeneous patient population [3]. It is known that the elderly are more vulnerable to CLL, with a mean age of more than 70 years at diagnosis [4]. Only patients with CLL showing active or symptomatic disorder or those of advanced Binet or Rai stages are in need of therapy [5]. The treatment strategies for CLL have gained advancements in the past decades, with the use of targeted agents in lieu of chemoimmunotherapy [6]. Against such backdrop, it is of significance to explore promising targets for the treatment of CLL.
Our bioinformatics analysis predicted hairy/enhancer of split 1 (HES1) to be a differential gene in CLL. HES1 is identified as an important helix-loop-helix transcriptional suppressor [7]. Intriguingly, silencing of HES1 was reported to inhibit chronic myelogenous leukemia cell growth, thereby functioning as a crucial molecule in the transition of blast crisis into chronic myelogenous leukemia [8]. It is well known that HES1, a suppressor of cell cycle inhibitors, is a transcription factor triggered by the Notch signaling pathway [9]. The bioinformatics analysis conducted in this study also identified the notch receptor 1 (Notch1) signaling pathway as a pathway closely related to HES1 in CLL and that HES1 is correlated with phosphatase and tensin homologue (PTEN)-related proteins. The crucial role of abnormal Notch signaling in CLL has been highlighted [10]. The activating mutations of Notch1 have been detected in 10% of patients with CLL, which is accountable for an unsatisfactory prognosis [11]. Moreover, Gianfelici unfolded the activated Notch1 pathway in CLL [12]. Increased PTEN expression due to microRNA-26a (miR-26a) and miR-214 inhibition induces the apoptosis of CLL cells in vitro [13]. Additionally, PTEN displays lower mRNA expression in CLL patients and is negatively correlated with Binet staging [14]. Of note, activated Notch1 pathway stimulated by B-cell receptor can contribute to transcription of HES1 and c-Myc in CLL [15]. Rosati et al. have proved that Notch signaling plays a critical role in B-CLL cell survival and apoptosis resistance [16]. Another previous study has implicated that c-myc as a developmentally regulated direct downstream target of Notch1 that contributes to the growth of T-ALL cells [17]. Tottone et al. indicated that GSKJ4 may be a promising therapeutic agent in Notch1- and TAL1-dependent T-ALL [18]. Moreover, Notch1 has been found to upregulate the PI3K-AKT pathway via HES1, which causes decreased PTEN expression, thereby favoring the cell growth of T-cell acute lymphoblastic leukemia [19]. Based on the above findings, we conducted this study aiming to explore whether the Notch1/HES1/PTEN signaling participates in the development of CLL.
Materials and Methods
Ethics Statement
This study was approved by the Ethics Committee of our Hospital and performed in strict accordance with the Declaration of Helsinki. All specimens used in this study were obtained with the informed consent of patients and their families.
Bioinformatics Analysis
CLL-related dataset GSE26725 was retrieved from the Gene Expression Omnibus database, followed by differential analysis using “limma” package of R language. GSE26725 dataset includes 12 CLL samples and 5 normal samples. HES1 expression profile analysis was performed using GraphPad Prism. A protein–protein interaction network of differentially expressed genes was plotted on STRING. HES1-related signaling pathways were predicted on Wikipathways.
Study Subjects
We collected 60 peripheral blood samples from patients with CLL admitted at our hospital from March 2014 to March 2017. In addition, 30 peripheral blood samples collected from age-matched healthy volunteers were used as controls whose indicators were detected to be normal in our physical examination center. All CLL patients were diagnosed according to the 2008 World Health Organization diagnostic criteria. All patients were newly diagnosed or had not received any treatment within 6 months before specimen collection. Of the 60 CLL patients, 39 cases were male and 21 cases were female; 25 cases were aged 60 years or older and 35 cases were younger than 60 years. Based on Rai stage, 14 cases were in Rai stage 0, 12 in stage I, 14 in stage II, 10 in stage III, and 10 in stage IV; and based on Benit stage, 26 cases in Benit stage A, 16 in stage B, and 18 in stage C. Besides, 23 cases were with elevated lactate dehydrogenase (LDH) and 37 with normal LDH; 16 cases with elevated β2 microglobulin (β2-MG) and 44 with normal β2-MG.
Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
PBMCs were isolated by density gradient centrifugation (Ficoll-Paque; Pharmacia LKB Biotechnology) [20]. The collected PBMCs were resuspended in 1 mL of normal saline and counted using a counting plate under a microscope.
Immunomagnetic Bead Separation (MACS)
The magnetic activating-sorting column was placed in a magnetic field and washed with 500 μL of phosphate buffer saline (PBS). Next, PBMCs were resuspended in 10 mL normal saline in a 10 mL centrifuge tube and then centrifuged for 10 min, followed by removal of the supernatant. Afterward, every 5 × 106 PBMCs were added with 10 μL of CD19 magnetic beads and 90 μL of PBS. After 15 min of incubation, every 1 × 107 cells were supplemented with 1–2 mL of PBS and centrifuged for 5 min, with the supernatant subsequently removed. The washed cells were resuspended in 50 μL of PBS and the cell suspension was slowly added to the MS sorting column with a syringe. The cells flowing out of the sorting column, namely CD19+ cells, were sorted out. After further cell sorting, a cell flow cytometer was applied to ensure the proportion of CD19+ cells above 95%. The sorted cells were resuspended in 1 mL of normal saline, after which 20 μL of suspension was counted under a microscope. Cells were separately placed at a density of 1 × 107 cells per tube at − 80 °C in a refrigerator.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted utilizing the one-step method according to the instructions of the TRIzol kit (15596-026, Invitrogen, Carlsbad, CA). RNA was reversely transcribed into complementary DNA (cDNA) following the instructions of the reverse transcription kit (K1621, Fermentas, Glen Burnei, Maryland). Fluorescence PCR was performed using a fluorescence qPCR instrument (ABI 7500, ABI, Oyster Bay, NY) following the instructions of the SYBR® Premix Ex Taq™ II kit (RR820A, Xingzhi Biotechnology Co., Ltd., Huai’an, Jiangsu, China). Primer sequences are shown in Table S1 and synthesized by Shanghai Genechem Co., Ltd. (Shanghai, China). With glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serving as the internal reference, the relative expression of each target gene is calculated using the 2−ΔΔCt method.
Western Blot Assay
Total protein was extracted from tissues or cells in strict accordance with the manual of highly efficient radioimmunoprecipitation assay lysis buffer (R0010, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). The concentration of total protein in the collected supernatant was determined using a bicinchoninic acid kit (20201ES76, Yeasen Biotech Co., Ltd., Shanghai, China). After that, the proteins were separated by 10% SDS-PAGE and transferred onto a nitrocellulose membrane via the wet transfer method, followed by blockade with 5% skim milk and 1 × Tris -Buffered Saline with Tween 20 (pH 7.6) at room temperature for 2 h. Then, the membrane was incubated with the diluted primary rabbit anti-human antibodies against HES1 (1: 500; ab71559), Notch1 (1: 1000; ab52627), PTEN (1: 10,000; ab32199), c-Myc (1:1000; ab32072), c-Fos (1:2000; ab190289), caspase-3 (1:500; ab13847), Bcl-2-associated X protein (Bax) (1: 1000; ab32503), and B-cell lymphoma 2 (Bcl-2) (1: 500; ab59348) overnight at 4 °C. Afterward, the membrane was incubated with the secondary goat anti-rabbit polyclonal antibody (ab205718) diluted with 5% skim milk at room temperature for 1 h. The used antibodies were all purchased from Abcam (Cambridge, MA). The Bio-Rad gel imaging system (MG8600, Beijing Thmorgan, Beijing, China) was applied for development. Quantity One v4.6.2 software was applied for analysis. The ratio of the gray value of the target band to that of internal reference GAPDH band was recorded as the relative protein expression. The experiment was repeated three times and the average values were obtained.
Cell Culture and Screening
Five CLL cell lines (CLL-AAT, MEC2, 183-E95, MEC1, and HG3) were selected (Cell Bank of Chinese Academy of Sciences, Shanghai, China) and cultured with Iscove’s Modified Dulbecco’s Medium (HyClone Company, Logan, UT) supplemented with 10% fetal calf serum in an incubator at 37 °C with 5% CO2 under saturated humidity conditions. The medium was renewed every 1–2 days, and subculture was performed when the cell confluence reached 80–90%. The culture medium was renewed every 24 h, and the passage was conducted every 2–3 days. RT-qPCR was used to screen the two cell lines with the highest and lowest HES1 expression for subsequent experiments.
Cell Treatment
According to the HES1 (NC_000003.12) information in the National Center for Biotechnology Information database, a lentiviral infection vector and an overexpression vector of HES1 were designed and packaged. In this experiment, the construction, identification, and sequencing of the lentiviral interference vector (GV118 [U6-MCSUbi-EGFP]) and overexpression vector of the HES1 gene (GV320 [Ubi-MCS-3FLAG-SV40-Cherry]) [21], as well as the extraction and packaging of the lentivirus-mediated RNA interference were performed by Shanghai SunBio Biomedical Technology (Shanghai, China). Cells were seeded into a 6-well plate 24 h before transfection. When reaching about 50% confluence, CLL cell lines were transfected using Lipofectamine 2000 reagent (11668027, Invitrogen) with blank vector, negative control (NC; the sequence is 5′-CACCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAA-3′), small hairpin RNA (sh)-HES1 (HES1 interference vector; the sequence is 5′-CCGGCCGGACTCTAAACAGGAACTTCTCGAGAAGTTCCTGTTTAGAGTCCGGTTTTT-3′), oe-HES1 (HES1 overexpression plasmid completed by Shanghai SunBio Biomedical Technology, Shanghai, China), MW167 (Notch1 signaling pathway inhibitor, Sigma-Aldrich Chemical Company, St Louis, MO, treatment concentration of 50 µM), or sh-HES1 + MW167 (HES1 interference vector and Notch1 signaling pathway inhibitor MW167).
Chromatin Immunoprecipitation (ChIP)
Cells were treated with 4% formaldehyde (a final concentration of 1%), sonicated, and incubated with anti-HES1 antibody (1: 50; ab71559; Abcam) to bind to the PTEN promoter (FW: TTTGAGAGTTGAGCCGCTGT; RV: ATGCTTTGAATCCAAAAACCTTACT; product length = 992). Cells were treated with protein A-agarose/salmon sperm DNA to bind to HES1 antibody-HES1-PTEN promoter complex, followed by precipitation of the complex. The precipitated complex was washed to remove non-specific binding. After elution, enriched HES1-PTEN promoter complex was obtained and the cross-linking was relieved. After purification and enrichment of PTEN promoter fragments, qRT-PCR was performed to analyze the level of PTEN promoter [22].
3-[4,5-Dimethylthiazol-2-yl]-2,5-Diphenyltetrazolium Bromide (MTT) Assay
The cells were seeded in a 96-well culture plate with 0.2 mL of cell suspension (1 × 104 cells/mL) per well and three duplicated wells were designed for each. After seeding, the cells were cultured in an incubator with 5% CO2 at 37 °C for 24, 48, 72, and 96 h; at each time point, 10 μL 5 mg/mL of MTT solution (M1020, Solarbio) was added and incubated in an incubator at 37 °C for 4 h in the dark. After removal of the supernatant, 200 μL of dimethyl sulfoxide (D5879-100ML, Sigma-Aldrich) was supplemented to each well to dissolve the purple crystals formed by the cells. The optical density value (490 nm) of each well was recorded for data analysis.
Flow Cytometry
Propidium iodide (PI) staining was performed to detect cell cycle distribution. Cells were collected based on the manual of a cell cycle detection kit (CA1510, Solarbio). The cells were fixed in pre-chilled 70% ethanol solution overnight at 4 °C and washed twice with PBS. Then, 100 μL of the cell suspension (1 × 105 cells) was stained with 1 mL of 50 mg/L PI staining solution containing RNAase. After 30 min of staining in the dark at 4 °C, the cells were filtered through a 300-mesh nylon mesh. Finally, a flow cytometer (BD FACSCalibur) was employed to record the red fluorescence at 488 nm.
Annexin V-fluorescein isothiocyanate (FITC)/PI dual-label staining was used to detect cell apoptosis. Cells were resuspended in 500 μL binding buffer solution with reference to the manual of cell apoptosis detection kit (556547, BD Bioscience). Afterward, 5 μL FITC and 5 μL PI solution were gently mixed and incubated with the cells at room temperature for 15 min in the dark. Cell apoptosis was detected with a flow cytometer (BD FACSCalibur).
Statistical Analysis
Data analysis was performed using SPSS 21.0 (IBM Corp., Armonk, NY). Measurement data were presented as mean ± standard deviation. Data between two groups were compared by unpaired t test, while those among multiple groups were compared by one-way analysis of variance (ANOVA) with Tukey’s post hoc tests. Data among multiple groups at different time points were compared by two-way ANOVA with Tukey’s post hoc test. p < 0.05 was considered to be statistically significant difference.
Results and Discussion
The Significance of HES1, Notch1 Signaling Pathway, and PTEN in the CLL Development
We screened out 173 upregulated genes and 211 downregulated genes in the GSE26725 dataset downloaded from the GEO database. A heat map of the top 30 differential genes is shown in Fig. 1A and Table S2, among which the top 10 differentially expressed genes were mainly enriched in the Notch signaling pathway (Fig. 1A). HES1 has been reported as the main component of the Notch1 signaling pathway in association with multiple malignancies [23,24,25] yet its implication in CLL remains understudied. Next, analysis in the GSE26725 dataset showed that HES1 gene was significantly upregulated in patients with CLL (Fig. 1B). Prediction results on Wikipathways website revealed a close correlation between HES1 and the Notch1 signaling pathway (Supplementary Fig. 1A). Afterward, the STRING website was used to plot the protein–protein interaction network of the first 30 differentially expressed genes in the GSE26725 dataset, results of which identified that HES1 was related to Notch1 and PTEN (Supplementary Fig. 1B). Therefore, we speculated that HES1 may affect CLL progression by mediating the Notch1 signaling pathway.
Bioinformatics prediction of key genes involved in CLL. A A heat map of the CLL-related GSE26725 dataset. The X-axis represents sample number, the Y-axis represents gene name, the dendrogram represents sample type clustering, the right color histogram represents gene expression, and the left dendrogram represents gene expression clustering. Each box in the figure represents gene expression in one sample. B HES1 gene expression profile retrieved from the GSE26725 dataset. The green represents the gene expression in normal control samples, and red represents the gene expression in CLL samples. *p < 0.05
Notch1/HES1 Signaling was Activated in CLL
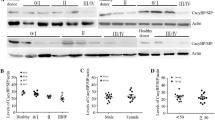
Subsequently, we detected the mRNA and protein expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in human normal B lymphocytes and B lymphocytes of CLL patients by RT-qPCR and Western blot assay. Results revealed that the mRNA expression and protein levels of HES1, Notch1, Bcl-2, c-Myc, and c-Fos were higher in B lymphocytes of CLL patients than those in human normal B lymphocytes, while the mRNA expression and protein levels of PTEN, caspase-3, and Bax displayed significant declines (Fig. 2). These data demonstrated that Notch1/HES1 signaling pathway was activated in CLL.
NOTCH1/HES1 signaling pathway is activated in CLL. A mRNA expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in B lymphocytes of normal controls (N = 30) and patients with CLL (N = 60) as detected by RT-qPCR. B Protein expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in B lymphocytes of normal controls (N = 30) and patients with CLL (N = 60) as detected by Western blot assay. *p < 0.05 vs. human normal B lymphocytes. The experiment was repeated 3 times independently
HES1 Expression was Positively Correlated with Clinicopathological Staging of Patients with CLL
In addition, we analyzed the relationship between the mRNA expression of HES1 and the clinicopathological characteristics of patients with CLL. From the results (Table S3), the mRNA expression of HES1 in patients with CLL of Rai stage III and IV was notably higher than that in patients with CLL of Rai stage 0, I, or II. Moreover, the more advanced the Benit staging was, the higher the expression of HES1 mRNA was. LDH concentration was positively correlated with HES1 mRNA expression. HES1 mRNA expression showed no significant correlation with the age, gender, or β2-MG content.
HES1 Showed the Highest Expression in the MEC1 Cell Line and the Lowest Expression in the HG3 Cell Line
Our experimental results of RT-qPCR demonstrated that in five CLL cell lines (CLL-AAT, MEC2, 183-E95, MEC1, and HG3), the MEC1 cell line showed the highest mRNA expression of HES1 and the HG3 cell line showed the lowest mRNA expression of HES1 (Supplementary Fig. 2A). In addition, Notch1 expression was lower while PTEN expression was higher in HG3 cell line than in MEC1 cell line (Supplementary Fig. 2B). The above results showed the highest expression of HES1 in the MEC1 cell line and the lowest in the HG3 cell line.
HES1 Bound to the Promoter Region of PTEN
The results of ChIP assay showed that the enrichment of HES1 in the PTEN promoter region was reduced in MEC1 and HG3 cell lines treated with sh-HES1, while it was enhanced in the presence of oe-HES1 in addition to no significant difference in cells incubated with anti-IgG antibody (Fig. 3). The aforementioned results suggested that HES1 could bind to the promoter region of PTEN.
HES1 binds to the promoter region of PTEN. A Binding of HES1 to the PTEN promoter region in MEC1 cell line analyzed by ChIP assay. B Binding of HES1 to the PTEN promoter region in HG3 cell line analyzed by ChIP assay. *p < 0.05 vs. MEC1 or HG3 cell line treated with sh-NC. #p < 0.05 vs. MEC1 or HG3 cell line treated with Vector. The experiment was repeated 3 times independently
HES1 Activated the Notch1 Signaling Pathway in CLL Cells
We then performed RT-qPCR and Western blot assay (Fig. 4) to detect the mRNA and protein expression of HES1, Bcl-2, c-Myc, c-Fos, caspase-3, PTEN, and Bax in MEC1 and HG3 cell lines treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167, and the trend between MEC1 and HG3 cell lines was the same. The mRNA and protein expression of HES1, Bcl-2, c-Myc and c-Fos were decreased upon treatment with sh-HES1, but that of caspase-3, PTEN, and Bax was increased. In contrast, oe-HES1 contributed to an increase in the mRNA and protein expression of HES1, Bcl-2, c-Myc, and c-Fos yet a decrease in that of caspase-3, PTEN, and Bax. MW167 treatment alone led to diminished levels of Notch1, HES1, Bcl-2, c-MYC, and c-fos and elevated levels of PTEN, caspase-3, and Bax. Co-treatment of oe-HES1 and MW167 could reverse the changes regarding the upregulation of Notch1, HES1, Bcl-2, c-MYC, and c-fos as well as downregulation of PTEN, caspase-3, and Bax caused by oe-HES1. Taken together, HES1 can cause activation of the Notch1 signaling pathway in CLL cells.
HES1 activates the Notch1 pathway in CLL cells. A, mRNA expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in the MEC1 cell line treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167 as detected by RT-qPCR. B, mRNA expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in the HG3 cell line treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167 as detected by RT-qPCR. C, Protein expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in the MEC1 cell line treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167 as detected by Western blot assay. D, Protein expression of HES1, Notch1, Bcl-2, c-Myc, c-Fos, PTEN, caspase-3, and Bax in the HG3 cell line treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167 as detected by Western blot assay. *p < 0.05 vs. the MEC1 or HG3 cell line treated with sh-NC. #p < 0.05 vs. the MEC1 or HG3 cell line treated with Vector. &p < 0.05 vs. the MEC1 or HG3 cell line treated with DMSO. @p < 0.05 vs. the MEC1 or HG3 cell line treated with oe-HES1. Treatment concentration of MW167 was 50 µM. The experiment was repeated three times independently
HES1 Activated the Notch1 Signaling Pathway to Promote CLL Cell Proliferation and Cell Cycle Progression But to Suppress CLL Cell Apoptosis
We then focused on the effect of HES1 on the biological functions of MEC1 and HG3 cell lines. As reflected by MTT assay, after MEC1 and HG3 cells were cultured for 24 h, no significant difference was detected in their proliferation following varied treatment. However, after 48 and 72 h of culture, cell proliferation was reduced in the presence of sh-HES1 but it was notably enhanced upon oe-HES1. In addition, treatment with MW167 attenuated cell proliferation. Lower cell proliferation was observed in the presence of oe-HES1 + MW167 than oe-HES1 alone (Fig. 5). The above results indicate that overexpression of HES1 can activate the Notch1 signaling pathway and thus promote the proliferation of CLL cells.
HES1 activates the Notch1 signaling pathway to stimulate the proliferation of CLL cells. A Proliferation of MEC1 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167 detected by MTT assay. B Proliferation of HG3 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167 detected by MTT assay. *p < 0.05 vs. the MEC1 or HG3 cell line treated with sh-NC. #p < 0.05 vs. the MEC1 or HG3 cell line treated with Vector. &p < 0.05 vs. the MEC1 or HG3 cell line treated with DMSO. @p < 0.05 vs. the MEC1 or HG3 cell line treated with oe-HES1. Treatment concentration of MW167 was 50 µM. The experiment was repeated three times independently
Moreover, flow cytometric data revealed increased proportion of MEC1 and HG3 cells arrested at the G1 phase yet decreased proportion of MEC1 and HG3 cells arrested at the S phase upon silencing of HES1, the effect of which was abolished by overexpression of HES1. The proportion of cells arrested at the G1 phase was increased, while that of cells arrested at the S phase was reduced in response to MW167. Notably, co-treatment with oe-HES1 and MW167 counteracted the above effects induced by individual treatment with oe-HES1 (Fig. 6). Cumulatively, overexpression of HES1 could increase cells arrested at the S phase by activating the Notch1 signaling pathway.
HES1 activates the Notch1 signaling pathway to arrest more cells at the S phase. A Detection on cell cycle distribution of MEC1 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. B Detection on cell cycle distribution of HG3 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. C Histogram for cell cycle distribution of MEC1 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. D Histogram for cell cycle distribution of HG3 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. *p < 0.05 vs. the MEC1 or HG3 cell line treated with sh-NC. #p < 0.05 vs. the MEC1 or HG3 cell line treated with Vector. &p < 0.05 vs. the MEC1 or HG3 cell line treated with DMSO. @p < 0.05 vs. the MEC1 or HG3 cell line treated with oe-HES1. Treatment concentration of MW167 was 50 µM. The experiment was repeated three times independently
Subsequently, flow cytometric data suggested that MEC1 and HG3 cell apoptosis were promoted by silencing of HES1 or treatment with MW167 but it was reduced by overexpression of HES1. Co-treatment with oe-HES1 and MW167 could neutralize the effects triggered by oe-HES1 (Fig. 7). The above results indicate that overexpression of HES1 can activate the Notch1 signaling pathway and thus repress the apoptosis of CLL cells.
HES1 activates the Notch1 signaling pathway to prevent the apoptosis of CLL cells. A Plots for apoptosis of MEC1 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. B Plots for apoptosis of HG3 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. C Apoptotic rates of MEC1 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. D Apoptotic rates of HG3 cells treated with sh-HES1, oe-HES1, MW167, or oe-HES1 + MW167. *p < 0.05 vs. the MEC1 or HG3 cell line treated with sh-NC. #p < 0.05 vs. the MEC1 or HG3 cell line treated with Vector. &p < 0.05 vs. the MEC1 or HG3 cell line treated with DMSO. @p < 0.05 vs. the MEC1 or HG3 cell line treated with oe-HES1. Treatment concentration of MW167 was 50 µM. The experiment was repeated three times independently
CLL is a malignancy that entails the slow proliferation of B-cell lymphocyte in the bone marrow and blood [26]. It has become a global problem accounting for approximately 25% of total leukemia cases, which renders the necessity of development of novel targets for CLL treatment [27]. In this study, we explored the role of Notch/HES1/PTEN signaling in CLL and found that HES1 could activate the Notch1 pathway and downregulate the expression of PTEN to promote CLL cell progression.
Initial results of this study demonstrated upregulation of HES1 and Notch1 expression, yet low expression of PTEN in B lymphocytes in CLL patients. At present, a large number of previous studies have highlighted the upregulated expression of HES1 in various types of leukemia. For example, significant upregulation of HES1 was found in the most primitive CD34 + Thy + subset of CD34 + cells in chronic myeloid leukemia [28]. High HES1 expression was detected in patients with chronic myelogenous leukemia in blast crisis [29]. It was previously demonstrated that the Notch pathway, interplaying with the NF-κB pathway, could be activated in CLL in an unmutated state [30]. In concert with our finding, the inactivated Notch1 signaling pathway was found to aid bepridil to exert anti-leukemic activity in a CLL xenotransplant model, by diminishing the percentage of leukemic cells infiltrating the spleen [31]. Similarly, a lack of PTEN expression was detected by Kapoor et al. in ibrutinib-resistant CLL cell lines [32]. In addition, Bailon et al. unfolded reduced PTEN expression in CLL cells, which played a part in stimulating development of the disease [33]. All the aforementioned reports indeed support our finding regarding the participation of Notch1/HES1/PTEN in CLL.
Mechanistically, we revealed that overexpression of HES1 could decrease the expression of PTEN by binding to the promoter region of PTEN, while treatment with the Notch1 signaling pathway inhibitor, MW167, could lead to an opposite result. This result demonstrated that Notch1 downregulates PTEN by upregulating HES1 expression in CLL. The Notch-HES1-cylindromatosis-NF-κB axis played a key role in maintaining T-cell acute lymphoblastic leukemia, and cylindromatosis could aid to weaken the ability of Notch1 to induce the leukemia by knocking down HES1 targeted by Notch or upregulating cylindromatosis targeted by HES1 [34]. Moreover, the Notch1-CCN3 signaling pathway in chronic myeloid leukemia K562 cells led to decreased expression of the Notch1 target genes c-Myc and HES1, which could diminish formation of K562/CCN3 colonies and thus induce the disease pathogenesis [35]. These previous reports revealed the upregulation of HES1 by Notch1 in leukemia. So far, the interaction among Notch-HES1-PTEN has been highlighted in multiple cancers. For instance, the modulation of PTEN expression by Notch via HES1 was revealed; silencing of Notch or HES1 could upregulate PTEN expression in human glioblastomas and thus both the Notch and HES1 pathways could improve therapeutic efficacy [36]. Additionally, Guo et al. reported the correlation between silencing of Notch1 and inhibited HES1 as well as activated PTEN/p53 pathway in intrahepatic cholangiocarcinoma cells [37]. Of note, it was also reported by Hales et al. that HES1 could inhibit PTEN in a transcriptional manner and thus activate AKT, which could be neutralized by Notch1 inhibition using γ-secretase inhibitors in T-cell acute lymphoblastic leukemia [38]. Moreover, Palomero et al. demonstrated the interplay between Notch1 and HES1 to inactivate PTEN in T-cell acute lymphoblastic leukemia [19]. In our study, we revealed the HES1-mediated Notch/PTEN signaling in CLL.
Furthermore, we found that silencing of HES1 or inactivated Notch1 signaling pathway inhibited the proliferation while facilitating apoptosis of CLL cells. HES1 is well known for its capacity of regulating cell proliferation and differentiation [39]. Consistently, downregulated HES1 by miR-182-5p can contain the development of myeloid in chronic myeloid leukemia leukemic cells [40]. In line with this finding, interfered Notch signaling could diminish peripheral lymphocytes in lymph nodes in a rat model [41]. A prior study once identified that Notch1, with the ability to target gene regulators of B-cell proliferation, was capable of transactivating Myc by binding to B-cell-specific factors, which thus serves as an oncogene in the progression of CLL [42]. Similarly, nuclear PTEN expression could suppress growth arrest while promoting apoptosis induction of CLL cells, thereby acting as a promising anti-tumor factor in CLL [43]. Moreover, downregulated PTEN by overexpressed miR-22 was found responsible for proliferative B cells in CLL [44]. In this study, we demonstrated that silencing of HES1 by downregulated Notch1 could activate the expression of PTEN, thereby curbing the growth of CLL cells.
Conclusion
In summary, we demonstrated that HES1 promoted the activity of Notch1 to downregulate expression of PTEN, thereby promoting proliferation and suppressing apoptosis of CLL cells (Fig. 8). Despite of this, clinical trials are warranted for validation of this finding.
References
Savvopoulos, S., Misener, R., Panoskaltsis, N., Pistikopoulos, E. N., & Mantalaris, A. (2016). A personalized framework for dynamic modeling of disease trajectories in chronic lymphocytic leukemia. IEEE Transactions on Biomedical Engineering, 63, 2396–2404.
Wiestner, A. (2012). Emerging role of kinase-targeted strategies in chronic lymphocytic leukemia. Blood, 120, 4684–4691.
Hus, I., Salomon-Perzynski, A., & Robak, T. (2020). The up-to-date role of biologics for the treatment of chronic lymphocytic leukemia. Expert Opinion on Biological Therapy, 20, 799–812.
Stauder, R., Eichhorst, B., Hamaker, M. E., Kaplanov, K., Morrison, V. A., Osterborg, A., Poddubnaya, I., Woyach, J. A., Shanafelt, T., Smolej, L., Ysebaert, L., & Goede, V. (2017). Management of chronic lymphocytic leukemia (CLL) in the elderly: A position paper from an international Society of Geriatric Oncology (SIOG) Task Force. Annals of Oncology, 28, 218–227.
Eichhorst, B., Cramer, P., & Hallek, M. (2016). Initial therapy of chronic lymphocytic leukemia. Seminars in Oncology, 43, 241–250.
Lee, J. C., & Lamanna, N. (2020). Is there a role for chemotherapy in the era of targeted therapies? Current Hematologic Malignancy Reports, 15, 72–82.
Wu, Y., Gong, L., Xu, J., Mou, Y., Xu, X., & Qian, Z. (2017). The clinicopathological significance of HES1 promoter hypomethylation in patients with colorectal cancer. Oncotargets and Therapy, 10, 5827–5834.
Nakahara, F., Sakata-Yanagimoto, M., Komeno, Y., Kato, N., Uchida, T., Haraguchi, K., Kumano, K., Harada, Y., Harada, H., Kitaura, J., Ogawa, S., Kurokawa, M., Kitamura, T., & Chiba, S. (2010). Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood, 115, 2872–2881.
Monahan, P., Rybak, S., & Raetzman, L. T. (2009). The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology, 150, 4386–4394.
Ferrante, F., Giaimo, B. D., Bartkuhn, M., Zimmermann, T., Close, V., Mertens, D., Nist, A., Stiewe, T., Meier-Soelch, J., Kracht, M., Just, S., Kloble, P., Oswald, F., & Borggrefe, T. (2020). HDAC3 functions as a positive regulator in Notch signal transduction. Nucleic Acids Research, 48, 3496–3512.
Close, V., Close, W., Kugler, S. J., Reichenzeller, M., Yosifov, D. Y., Bloehdorn, J., Pan, L., Tausch, E., Westhoff, M. A., Dohner, H., Stilgenbauer, S., Oswald, F., & Mertens, D. (2019). FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood, 133, 830–839.
Gianfelici, V. (2012). Activation of the NOTCH1 pathway in chronic lymphocytic leukemia. Haematologica, 97, 328–330.
Zou, Z. J., Fan, L., Wang, L., Xu, J., Zhang, R., Tian, T., Li, J. Y., & Xu, W. (2015). miR-26a and miR-214 down-regulate expression of the PTEN gene in chronic lymphocytic leukemia, but not PTEN mutation or promoter methylation. Oncotarget, 6, 1276–1285.
Zou, Z. J., Zhang, R., Fan, L., Wang, L., Fang, C., Zhang, L. N., Yang, S., Li, Y. Y., Li, J. Y., & Xu, W. (2013). Low expression level of phosphatase and tensin homolog deleted on chromosome ten predicts poor prognosis in chronic lymphocytic leukemia. Leukaemia & Lymphoma, 54, 1159–1164.
Del Papa, B., Baldoni, S., Dorillo, E., De Falco, F., Rompietti, C., Cecchini, D., Cantelmi, M. G., Sorcini, D., Nogarotto, M., Adamo, F. M., Mezzasoma, F., Silva Barcelos, E. C., Albi, E., Iacucci Ostini, R., Di Tommaso, A., Marra, A., Montanaro, G., Martelli, M. P., Falzetti, F., … Sportoletti, P. (2019). Decreased NOTCH1 activation correlates with response to ibrutinib in chronic lymphocytic leukemia. Clinical Cancer Research, 25, 7540–7553.
Rosati, E., Sabatini, R., Rampino, G., Tabilio, A., Di Ianni, M., Fettucciari, K., Bartoli, A., Coaccioli, S., Screpanti, I., & Marconi, P. (2009). Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood, 113, 856–865.
Weng, A. P., Millholland, J. M., Yashiro-Ohtani, Y., Arcangeli, M. L., Lau, A., Wai, C., Del Bianco, C., Rodriguez, C. G., Sai, H., Tobias, J., Li, Y., Wolfe, M. S., Shachaf, C., Felsher, D., Blacklow, S. C., Pear, W. S., & Aster, J. C. (2006). c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & Development., 20, 2096–2109.
Tottone, L., Zhdanovskaya, N., Carmona Pestana, A., Zampieri, M., Simeoni, F., Lazzari, S., Ruocco, V., Pelullo, M., Caiafa, P., Felli, M. P., Checquolo, S., Bellavia, D., Talora, C., Screpanti, I., & Palermo, R. (2019). Histone modifications drive aberrant notch3 expression/activity and growth in T-ALL. Frontiers in Oncology, 9, 198.
Palomero, T., Dominguez, M., & Ferrando, A. A. (2008). The role of the PTEN/AKT pathway in NOTCH1-induced leukemia. Cell Cycle, 7, 965–970.
Hwang, K. K., Chen, X., Kozink, D. M., Gustilo, M., Marshall, D. J., Whitesides, J. F., Liao, H. X., Catera, R., Chu, C. C., & Yan, X. J. (2012). Enhanced outgrowth of EBV-transformed chronic lymphocytic leukemia B cells mediated by coculture with macrophage feeder cells. Blood, 119(7), e35-44.
Yan, P., Xue, Z., Li, D., Ni, S., Wang, C., Jin, X., Zhou, D., Li, X., Zhao, X., & Chen, X. (2021). Dysregulated CRTC1-BDNF signaling pathway in the hippocampus contributes to Abeta oligomer-induced long-term synaptic plasticity and memory impairment. Experimental Neurology, 345, 113812.
Nelson, J. D., Denisenko, O., Sova, P., & Bomsztyk, K. (2006). Fast chromatin immunoprecipitation assay. Nucleic Acids Research, 34(1), e2. https://doi.org/10.1093/nar/gnj004
Wirth, M., Doescher, J., Jira, D., Meier, M. A., Piontek, G., Reiter, R., Schlegel, J., & Pickhard, A. (2016). HES1 mRNA expression is associated with survival in sinonasal squamous cell carcinoma. Oral Surgery, Oral Medicine, Oral Pathology, and Oral Radiology, 122, 491–499.
Ahadi, M., Andrici, J., Sioson, L., Sheen, A., Clarkson, A., & Gill, A. J. (2016). Loss of Hes1 expression is associated with poor prognosis in colorectal adenocarcinoma. Human Pathology, 57, 91–97.
Liu, Z., Fu, Q., Fu, H., Wang, Z., Xu, L., An, H., Li, Y., & Xu, J. (2016). A three-molecule score based on Notch pathway predicts poor prognosis in non-metastasis clear cell renal cell carcinoma. Oncotarget, 7, 68559–68570.
Fonseka, L. N., & Tirado, C. A. (2015). C-myc involvement in chronic lymphocytic leukemia (CLL): A molecular and cytogenetic update. Journal of the Association of Genetic Technologists, 41, 176–183.
Bhattacharya, M., Sharma, A. R., Sharma, G., Patra, B. C., Lee, S. S., & Chakraborty, C. (2020). Interaction between miRNAs and signaling cascades of Wnt pathway in chronic lymphocytic leukemia. Journal of Cellular Biochemistry, 121, 4654–4666.
Aljedai, A., Buckle, A. M., Hiwarkar, P., & Syed, F. (2015). Potential role of Notch signalling in CD34+ chronic myeloid leukaemia cells: Cross-talk between Notch and BCR-ABL. PLoS ONE, 10, e0123016.
Nakahara, F., Kitaura, J., Uchida, T., Nishida, C., Togami, K., Inoue, D., Matsukawa, T., Kagiyama, Y., Enomoto, Y., Kawabata, K. C., Chen-Yi, L., Komeno, Y., Izawa, K., Oki, T., Nagae, G., Harada, Y., Harada, H., Otsu, M., Aburatani, H., … Kitamura, T. (2014). Hes1 promotes blast crisis in chronic myelogenous leukemia through MMP-9 upregulation in leukemic cells. Blood, 123, 3932–3942.
Baldoni, S., Sportoletti, P., Del Papa, B., Aureli, P., Dorillo, E., Rosati, E., Ciurnelli, R., Marconi, P., Falzetti, F., & Di Ianni, M. (2013). NOTCH and NF-kappaB interplay in chronic lymphocytic leukemia is independent of genetic lesion. International Journal of Hematology, 98, 153–157.
Baldoni, S., Del Papa, B., Dorillo, E., Aureli, P., De Falco, F., Rompietti, C., Sorcini, D., Varasano, E., Cecchini, D., Zei, T., Di Tommaso, A., Rosati, E., Alexe, G., Roti, G., Stegmaier, K., Di Ianni, M., Falzetti, F., & Sportoletti, P. (2018). Bepridil exhibits anti-leukemic activity associated with NOTCH1 pathway inhibition in chronic lymphocytic leukemia. International Journal of Cancer, 143, 958–970.
Kapoor, I., Li, Y., Sharma, A., Zhu, H., Bodo, J., Xu, W., Hsi, E. D., Hill, B. T., & Almasan, A. (2019). Resistance to BTK inhibition by ibrutinib can be overcome by preventing FOXO3a nuclear export and PI3K/AKT activation in B-cell lymphoid malignancies. Cell Death & Disease, 10, 924.
Bailon, E., Ugarte-Berzal, E., Amigo-Jimenez, I., Van den Steen, P., Opdenakker, G., Garcia-Marco, J. A., & Garcia-Pardo, A. (2014). Overexpression of progelatinase B/proMMP-9 affects migration regulatory pathways and impairs chronic lymphocytic leukemia cell homing to bone marrow and spleen. Journal of Leukocyte Biology, 96, 185–199.
D’Altri, T., Gonzalez, J., Aifantis, I., Espinosa, L., & Bigas, A. (2011). Hes1 expression and CYLD repression are essential events downstream of Notch1 in T-cell leukemia. Cell Cycle, 10, 1031–1036.
Suresh, S., McCallum, L., Crawford, L. J., Lu, W. H., Sharpe, D. J., & Irvine, A. E. (2013). The matricellular protein CCN3 regulates NOTCH1 signalling in chronic myeloid leukaemia. The Journal of Pathology, 231, 378–387.
Saito, N., Hirai, N., Aoki, K., Suzuki, R., Fujita, S., Nakayama, H., Hayashi, M., Ito, K., Sakurai, T., & Iwabuchi, S. (2019). The oncogene addiction switch from NOTCH to PI3K requires simultaneous targeting of NOTCH and PI3K pathway inhibition in glioblastoma. Cancers, 11, 121.
Guo, J., Fu, W., Xiang, M., Zhang, Y., Zhou, K., Xu, C. R., Li, L., Kuang, D., & Ye, F. (2019). Notch1 drives the formation and proliferation of intrahepatic cholangiocarcinoma. Current Medical Science, 39, 929–937.
Hales, E. C., Orr, S. M., Larson Gedman, A., Taub, J. W., & Matherly, L. H. (2013). Notch1 receptor regulates AKT protein activation loop (Thr308) dephosphorylation through modulation of the PP2A phosphatase in phosphatase and tensin homolog (PTEN)-null T-cell acute lymphoblastic leukemia cells. Journal of Biological Chemistry, 288, 22836–22848.
Gao, F., Huang, W., Zhang, Y., Tang, S., Zheng, L., Ma, F., Wang, Y., Tang, H., & Li, X. (2015). Hes1 promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3beta pathway in human colon cancer. Oncotarget, 6, 38667–38680.
Arya, D., Sachithanandan, S. P., Ross, C., Palakodeti, D., Li, S., & Krishna, S. (2017). MiRNA182 regulates percentage of myeloid and erythroid cells in chronic myeloid leukemia. Cell Death Disease, 8, e2547.
Simutis, F. J., Sanderson, T. P., Pilcher, G. D., & Graziano, M. J. (2018). Nonclinical safety assessment of the gamma-secretase inhibitor avagacestat. Toxicological Sciences, 163, 525–542.
Fabbri, G., Holmes, A. B., Viganotti, M., Scuoppo, C., Belver, L., Herranz, D., Yan, X. J., Kieso, Y., Rossi, D., Gaidano, G., Chiorazzi, N., Ferrando, A. A., & Dalla-Favera, R. (2017). Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the USA, 114, E2911–E2919.
Carra, G., Panuzzo, C., Torti, D., Parvis, G., Crivellaro, S., Familiari, U., Volante, M., Morena, D., Lingua, M. F., Brancaccio, M., Guerrasio, A., Pandolfi, P. P., Saglio, G., Taulli, R., & Morotti, A. (2017). Therapeutic inhibition of USP7-PTEN network in chronic lymphocytic leukemia: A strategy to overcome TP53 mutated/deleted clones. Oncotarget, 8, 35508–35522.
Palacios, F., Prieto, D., Abreu, C., Ruiz, S., Morande, P., Fernandez-Calero, T., Libisch, G., Landoni, A. I., & Oppezzo, P. (2015). Dissecting chronic lymphocytic leukemia microenvironment signals in patients with unmutated disease: MicroRNA-22 regulates phosphatase and tensin homolog/AKT/FOXO1 pathway in proliferative leukemic cells. Leukaemia & Lymphoma, 56, 1560–1565.
Funding
This study was funded by the General items of Health Department of Zhejiang Province (2021KY1078), the Zhejiang Provincial Health Science and Technology Plan in 2022 (Clinical Research Application Project) (2022KY350), and the Wenzhou Science and Technology Bureau (Y20210155).
Author information
Authors and Affiliations
Contributions
CZ and WZ conceived and designed research. WY performed experiments. BL interpreted results of experiments. SW analyzed data. YC prepared figures. QZ performed the bioinformatics analysis and drafted paper. JG, ZZ, and YH edited and revised the manuscript. All authors read and approved final version of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2022_476_MOESM1_ESM.jpg
Supplementary file1 (DOCX 1499 KB). Supplementary Figure 1. Bioinformatics prediction of HES1-related signaling pathways in CLL. A, HES1-related signaling pathways predicted by the Wikipathways website. B, Protein–protein interaction network of the top 30 differentially expressed genes in the GSE26725 dataset analyzed on STRING.
12033_2022_476_MOESM2_ESM.jpg
Supplementary file2 (DOCX 429 KB). Supplementary Figure 2. The highest expression of HES1 in the MEC1 cell line and the lowest in the HG3 cell line. A, HES1 mRNA expression in CLL-AAT, MEC2, 183-E95, MEC1, and HG3 cell lines determined by RT-qPCR. B, NOTCH1 and PTEN mRNA expression in MEC1 and HG3 cell lines determined by RT-qPCR. *p < 0.05 vs. CLL-AAT cells. #p < 0.05 vs. MEC1 cell line. The experiment was repeated 3 times independently.
Rights and permissions
About this article
Cite this article
Zhang, Q., Zhu, Z., Guan, J. et al. Hes1 Controls Proliferation and Apoptosis in Chronic Lymphoblastic Leukemia Cells by Modulating PTEN Expression. Mol Biotechnol 64, 1419–1430 (2022). https://doi.org/10.1007/s12033-022-00476-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00476-2