Abstract
Osmotin, a pathogenesis-related antifungal protein, is relevant in induced plant immunity and belongs to the thaumatin-like group of proteins (TLPs). This article describes comparative structural and functional analysis of the two osmotin isoforms cloned from Phytophthora-resistant wild Piper colubrinum. The two isoforms differ mainly by an internal deletion of 50 amino acid residues which separates them into two size categories (16.4 kDa—PcOSM1 and 21.5 kDa—PcOSM2) with pI values 5.6 and 8.3, respectively. Recombinant proteins were expressed in E. coli and antifungal activity assays of the purified proteins demonstrated significant inhibitory activity of the larger osmotin isoform (PcOSM2) on Phytophthora capsici and Fusarium oxysporum, and a markedly reduced antifungal potential of the smaller isoform (PcOSM1). Homology modelling of the proteins indicated structural alterations in their three-dimensional architecture. Tertiary structure of PcOSM2 conformed to the known structure of osmotin, with domain I comprising of 12 β-sheets, an α-helical domain II and a domain III composed of 2 β-sheets. PcOSM1 (smaller isoform) exhibited a distorted, indistinguishable domain III and loss of 4 β-sheets in domain I. Interestingly, an interdomain acidic cleft between domains I and II, containing an optimally placed endoglucanase catalytic pair composed of Glu–Asp residues, which is characteristic of antifungal PR5 proteins, was present in both isoforms. It is well accepted that the presence of an acidic cleft correlates with antifungal activity due to the presence of endoglucanase catalytic property, and hence the present observation of significantly reduced antifungal capacity of PcOSM1 despite the presence of a strong acidic cleft, is suggestive of the possible roles played by other structural features like domain I or/and III, in deciding the antifungal potential of osmotin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants defend themselves by deploying a plethora of defence-related molecules against the invading pathogens at each level of defence [1]. Plant pathogenesis-related (PR) proteins are such a group of effector molecules expressed under pathological conditions. These proteins are classified into different families based on functional features and serological relationships [2, 3]. PR 5 family proteins are a group of antimicrobial proteins with diverse functions in plants. Members of this family show similarity to thaumatin, a sweet tasting protein found in the arils of the plant Thaumatococcus danielli and hence are also called thaumatin-like proteins (TLPs) [3]. Members include the well-known antifungal proteins zeamatin and osmotin which show prominent antifungal activity in vitro and in vivo [4–8]. Transgenic over expression of PR 5 proteins in plants has resulted in delay in onset of disease symptoms, indicating their potential as candidate genes for genetic engineering of plants [7, 9, 10]. Experiments on isolated yeast (Saccharomyces cerevisiae) spheroplasts and other fungal cells have predicted plasma membrane as the target of protein activity [6, 11–13] and membrane permeabilization as the possible method of action [4, 6, 12]. Studies on yeast cells have also shown that cell wall components like carbohydrates and proteins could regulate osmotin sensitivity, as the presence of phosphomannans-enhanced osmotin activity while other cell wall-related proteins like PIR2 and SSD1 caused osmotin resistance [11, 14, 15]. Recent studies indicate that osmotin shares structural similarity to adiponectin, an antidiabetic, and antiatherosclerotic protein hormone in mammals that conditions sensing of energy status, fatty acid oxidation, and glucose transport upon interaction with adiponectin receptors [16, 17]. Both osmotin and adiponectin have lectin-like domain structure and their comparative functional studies in yeast cells and C2C12 myocytes have shown a receptor-mediated mechanism of action [16, 17]. Thus, osmotin, due to its functional similarity to adiponectin, has recently been highlighted as an agonist molecule with therapeutic potential [17].
The accepted model of TLPs comprises a three domain structure stabilized by disulphide bridges [18–22]. A notable feature is the presence of an acidic cleft between domains I and II, which possesses amino acid residues acting as catalytic pair capable of glucan hydrolysis. This is cited as a structural feature required for antifungal activity of TLP as it has been shown that several TLPs can bind to β-1,3-glucans and many of them can hydrolyse polymeric glucan molecules which are common constituents of fungal cell walls [23–26]. However, the glucanase activity exhibited by the TLPs is by far very low when compared to that of glucanases belonging to PR-2 class [25, 26]. There are also reports of antifungal TLPs which are devoid of glucanase activity, which leads to the inference that additional structural features could be involved in determining the antifungal activity of TLP [20, 21, 26].
In our earlier work, we identified two osmotin isoforms from a defence gene-enriched cDNA subtracted library of Phytophthora-resistant wild pepper—Piper colubrinum and carried out a comparative sequence analysis of the two forms. Accordingly, the smaller form PcOSM1, of molecular weight 16.4 kDa (EU271754.1) shows an internal deletion of 50 amino acids [27]. The present report focuses on expression of the recombinant isoforms in E. coli and their antifungal functional validation on the plant fungal pathogens Phytophthora capsici (causative organism of foot rot disease of cultivated black pepper Piper nigrum [28]) and Fusarium oxysporum. Protein functional studies of P. colubrinum osmotin isoforms was further extended to structural prediction using bioinformatics tools, in an attempt to provide possible insight into the hitherto unidentified roles played by additional structural features of TLPs in determining protein functionality.
Materials and Methods
Plant Material and Treatments
Healthy plants of P. colubrinum link, maintained in the greenhouse of Rajiv Gandhi Centre for Biotechnology, Trivandrum, India were used for this study. To analyse the relative effect of abiotic stress inducing compounds and different signalling compounds on the expression of P. colubrinum osmotin isoforms, fully expanded leaves were sprayed with ethephon (1 mg/ml)/1 mM SA/1 mM MeJ solutions in water. The leaves were inoculated with virulent PDA grown cultures of P. capsici to study the relative expression of the isoforms under biotic stress. The abaxial sides of leaves were needle-pricked and mycelial discs were placed on the wound. Needle-pricked leaves free of mycelial discs were used to study the effect of wounding on the expression level of the genes and also served as control for pathogen inoculation study. The treatments were carried out for a period of 24 h and the plants were kept covered using plastic bags until sampling.
Sequence Analysis
Homology searches were performed with BLAST to confirm sequence identity. The amino acid sequences of PR-5 proteins representing dicots and monocots were retrieved from NCBI Genbank and multiple sequence alignment was performed with PcOSM1 and PcOSM2 sequences using the online tool available at www.ebi.ac.uk and the programme GeneDoc (http://www.nrbsc.org/gfx/genedoc). The programmes Signal P (http://www.cbs.dtu.dk/services/SignalP/) and Target P (http://www.cbs.dtu.dk/services/TargetP/) were used to predict the cellular location of P. colubrinum osmotin isoforms.
Expression Analysis of P. colubrinum Osmotin Isoforms
Real-time PCR analyses were carried out to determine comparative tissue-specific transcript abundance of the osmotin isoforms and its accumulation in response to defence elicitors like SA, MeJ, ethylene and the pathogen P. capsici. Total RNA was isolated like from leaf, stem, root, spike and elicitor-treated leaves of P. colubrinum. Approximately, 1 μg of DNase (Sigma, St. Louis) treated RNA was used to prepare cDNA using MMLV-RT following manufacturer’s protocol (Promega, Madison, WI). Gene specific primers (Table 1) amplifying a 100 bp product were used for quantitative real-time PCR. The reaction was set up in a final volume of 20 μl containing 10 μl SYBR green PCR reagent (Applied Biosystems, CA), 1 μl of diluted cDNA and 300 nM each of the designed primers and the conditions were 50°C for 2 min initially followed by 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min in a real-time PCR machine (ABI 7500, Applied Biosystems, CA). Three replicates each from two biological samples were used for each analysis. The house keeping gene β-actin cloned from P. colubrinum was used for normalisation and the relative expression level of the genes was determined by the comparative C t method using the SDS software (Applied Biosystems, CA).
Statistical Analysis
Real-time analysis for transcript abundance of osmotin isoforms in each tissue as well as in response to each defence elicitor was performed in triplicate and each experiment was repeated twice (n = 6). Resultant mean values were analysed by Student’s ‘t’ test and values were considered significant at P < 0.05.
Prokaryotic Expression and In Vitro Antifungal Assay of Osmotin Isoforms
Osmotin isoforms cloned from P. colubrinum were expressed in recombinant E. coli BL21(DE3) pLyS (Invitrogen, Carlsbad) or JM109(DE3) pLyS (Promega, Madison, WI) using the expression vector pET 32a (Novagen, USA). The coding sequence for the mature protein of the isoforms (PcOSM1 and PcOSM2) was amplified using gene specific primers with sequences for histidine tag and restriction enzyme sites (Table 1). The amplicons were cloned into T/A cloning vector pGEMT—Easy (Promega, Madison, WI) and subcloned to pET 32 a (+) vector (Novagen, USA) at Nde I and Hind III sites. Recombinant constructs were confirmed by sequencing in an automated sequencer (3730 DNA analyser, Applied Biosystems, CA) using T7 promoter primer and introduced into the expression host. The recombinant protein was induced using 0.8–1.0 mM IPTG (Gibco BRL, CA). Expressed protein was purified by nickel affinity chromatography under denaturing conditions and eluted fractions were pooled and dialysed against 20 mM Tris HCl (pH 7.2) as described by Hu and Reddy [29]. Dialysed protein was concentrated (Amicon concentrators, Millipore) and used for in vitro antifungal assay after quantification by Bradford method [30]. The total bacterial protein from each sample was separated on 12% SDS-PAGE and transferred to nitrocellulose membrane (Hybond C, Amersham) for Western blot analysis using Mini Trans-Blot Electrophoretic Transfer cell (Biorad, CA). Recombinant proteins were probed with anti-his tag antibody (Sigma, MO). For in vitro antifungal assay, spores of P. capsici (1 × 103/100 μl) or conidia of F. oxysporum (1 × 103/100 μl) taken in a microplate in triplicate was incubated with different concentrations of purified recombinant protein for 48 h. Optical density of fungal growth in each well was measured at 595 nm to check growth inhibitory activity of the protein [31, 32].
Molecular Modelling of PcOSM1 and PcOSM2
The amino acid sequences of PcOSM1 and PcOSM2 were first subjected to PSI-BLAST [33] with three iterations for finding homologous template. Consequently, the refined crystal structure of antifungal banana fruit TLP at resolution 1.7 Å from the Brookhaven Protein Data Bank (PDB entry 1Z3Q) was retrieved and used as the template. Homology modelling was performed using the programme Modeller9v2 [34]. The three-dimensional structure prediction was carried out by alignment of target sequence with template structure. Models were built solely based on heavy atoms, and are likely to be a first approximation of the three-dimensional structure of antifungal banana fruit TLP. Modeller has an advantage over other such programs in allowing experimentally derived distance restraints to be used in conjunction with restraints from the homologous structures in deriving the model. A total of 50 models were produced and refined within Modeller. Conformational clustering of the final models, and identification of structures ‘representative’ of the ensemble of models, were performed using the program NMRCLUST [35]. Backbone conformation of the refined structure was evaluated by comparing the Z score of PcOSM1 and PcOSM2 and the template estimated using Prosa2003 [36]. Deviation of the modelled structure from the template was assessed by computing root-mean-square deviation (RMSD) values for positional differences between equivalent atoms following the superposition of Cα traces and backbone atoms of the model onto the template.
Docking of (1, 3)-β-d Glucan with PcOSM1 and PcOSM2
Docking of (1,3)-β-d glucan with PcOSM1/2 was performed with AUTODOCK4.1 [37]. The procedure involved creation of a 8 Å grid centred on acidic cleft. Ligand charges were computed using the Gasteiger method. The binding affinity of (1,3)-β-d glucan to the acidic cleft of PcOSM1/2 was studied using simulated annealing procedure of Accelrys Discovery Studio 2.0. Calculations were performed using the CHARMm Force-Fields PARM22 and the polysaccharide-specific CHEAT95 [38], and implemented in Discovery Studio. The simulation was performed for 500 ps with a time step of 0.001 and the temperature set to 300 K. The production type was set to NPT ensemble (constant temperature and pressure dynamics) with a radius of nonbonded interaction of 14.0 Å [39]. All simulation experiments were carried out using a DELL PowerEdge T300 Workstation with Quad Dual Intel Xeon 3.2 GHz processors.
Results
Sequence Analysis
Piper colubrinum osmotin isoforms share significant homology with corresponding sequences from other sources and show conserved residues of PR5 family members (Fig. 1). PcOSM1 and TLPs from monocots like Oryza sativa var. japonica and Secale cereale [40] differ from other plant members in having an internal deletion of amino acid residues. In monocots, the deletion is present near the C-terminal of the sequence, which differs from PcOSM1 which possesses a deletion proximal to N-terminus. Multiple alignments of the sequences reveal characteristic conserved residues of TLP in P. colubrinum osmotins. It is, however, noteworthy that while PcOSM2 retains the thaumatin signature sequence; it is partial in PcOSM1 (Fig. 1). An N-terminal signal peptide is predicted in both isoforms and their secretory nature is predicted by TARGET P. The pI values of the two isoforms suggest that PcOSM1 is an acidic protein (pI 5.6) and PcOSM2 is a basic protein (pI 8.3).
Sequence analysis. Multiple sequence alignment of amino acid sequences of Piper colubrinum osmotins (PcOSM1, PcOSM2) with TLPs from different taxa viz. Prunus avium (TLPPa, AAB38064), Malus domestica (TLPMa, CAC10270), Arabidopsis thaliana (OSMAt, AA61411), PR5 from Nicotiana tabacum (PR5Nt,CAA64620), PR-5d from tobacco [19], zeamatin (EU725369.1), thaumatin (THAU, AAA93095) Oryza sativa var. japonica (TLPOs, CAA48278.1), Secale cereale (TLPsec) [40] and Zea mays (NP_001147009.1). The red rectangle indicates the position of thaumatin signature sequence in PcOSM2 (GRGRCQTGDCGGVLQ) which is truncated to four residues (GVLQ) in PcOSM1 (Color figure online)
Expression Analysis of P. colubrinum Osmotin Isoforms
Real-time PCR analysis of transcript abundance (Fig. 2a) indicates that both PcOSM1and PcOSM2 have the highest expression level in inflorescence (spike). The expression levels of the two transcripts were comparable in leaf and root. The abundance of PcOSM1 transcript was greater than that of PcOSM2 in inflorescence. On the other hand, PcOSM2 transcript was more highly expressed than PcOSM1 in stem. It is therefore evident from real-time PCR analyses that transcript levels of the two P. colubrinum osmotin isoforms show significant differences in tissue-specific expression.
qRT-PCR analysis of P. colubrinum osmotin isoforms. a Tissue-specific expression analysis of PcOSM1 and PcOSM2. L—leaf, R—root, St—stem and Sp—spike. b Expression analysis of PcOSM1 and PcOSM2 in leaves treated with various defence elicitors. SA—salicylic acid, ET—ethylene, JA—jasmonic acid, Pc—Phytophthora capsici and W—Wounding. *Represents significance at P < 0.05; n = 6. Y-axis represents relative quantification values or fold expression change given by the Eq. 2−ΔΔCt
Piper colubrinum osmotin isoforms show varied levels of expression in response to stress signals and pathogen. Figure 2b summarises the gene expression levels in biotic or abiotic elicitor-treated P. colubrinum leaves. Among the elicitors, ethylene favoured the highest accumulation of both PcOSM1 and PcOSM2 transcripts. PcOSM2 was more abundant than PcOSM1 in MeJ, P. capsici and salicylic acid-treated leaves. PcOSM1 showed higher transcript accumulation in wounded leaves compared to PcOSM2. It is therefore evident from real-time PCR analyses that relative levels of the two P. colubrinum osmotin transcripts in leaves differ in response to various elicitor treatments.
Prokaryotic Expression and In Vitro Antifungal Assay of P. colubrinum Osmotin Isoforms
The isoforms were expressed in E. coli under IPTG induction. Figure 3 shows the gel analysis of purified pET-PcOSM1 and pET-PcOSM2 recombinant proteins. Both the recombinant forms were detected positive in Western blot analysis using anti-his tag antibody (Fig. 3c). The fungal inhibitory activity of purified recombinant proteins is shown in Fig. 4. Results show a significant difference in antifungal activity of the recombinant proteins. The larger form (PcOSM2) of P. colubrinum osmotin exhibits significant inhibitory activity against P. capsici and F. oxysporum (Fig. 4a, b).
Prokaryotic expression of P. colubrinum osmotin isoforms. a SDS-PAGE (12%) analysis of recombinant PcOSM1 (pET-PcOSM1) expression in E. coli. M—broad range protein marker (NEB, USA), 1—total protein of IPTG (0.8 mM)-induced bacteria harbouring recombinant construct, pET-PcOSM1, 2—purified recombinant PcOSM1 protein. b SDS-PAGE analysis of recombinant PcOSM2 (pET-PcOSM2) expression in E. coli. M—broad range protein marker (NEB,USA), 1—total protein of IPTG (0.8 mM)-induced bacteria harbouring recombinant construct, pET-PcOSM2, 2—purified recombinant PcOSM2 protein. c Western blot of histidine tagged recombinant P. colubrinum osmotin isoforms. The recombinant protein samples are probed with anti-His tag antibody and developed by alkaline phosphatase reaction. 1—pET-PcOSM2, M—broad range protein marker, 2—pET-PcOSM1
Antifungal activity of purified recombinant P. colubrinum osmotin isoforms. Effect of purified recombinant osmotins on germination and growth of Phytophthora capsici spore suspension (a) and Fusarium oxysporum conidial suspension (b). The experiment was repeated three times with triplicate samples which gave similar results. Shown are the results obtained from one experiment. Growth in each sample is expressed relative to growth in buffer-treated control and is the average of triplicate samples. Lanes 1—buffer control, 2–5—increasing concentrations of protein; 2—0.2 μM, 3—0.4 μM, 4—0.5 μM, 5—0.8 μM, 6—1 μM
Homology Modelling and Docking Studies
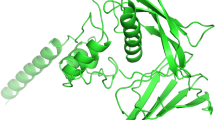
Homology models of PcOSM1 and PcOSM2 are shown in Fig. 5. PcOSM2 has the typical structure of TLPs with three distinct domains (I, II and III) but PcOSM1 shows significant deviation from the TLP structure. In the PcOSM1 model, domain I and II can be identified but the domain III is indistinguishable. Domain I of PcOSM1 has six β-sheets (10 in PcOSM2), but domain II containing α-helices are similar in both forms (Figs. 5, 6). The extent of structural deviation/alteration found in PcOSM1, from the typical TLP molecular structure can be inferred from Fig. 6. The loss of β-sheets in PcOSM1 causes a marked alteration in the surface of the protein. Homology models also show that the proteins have an overall globular architecture with a cleft region formed between domains I and II (Fig. 7). In both forms, the cleft region formed is acidic in nature. Residues found in and around the acidic cleft in PcOSM1 are 24Y, 25G, 27P, 29N, 31L, 33E, 44F, 46D, 48S, 50V, 51D, 92N, 94C, 95T, 98K, 102Y, 103C, 104C, 105N, 126Y, 128Y, 129P, 130K, 131D, 136T and in PcOSM2 the residues are 41G, 43R, 74Y, 75G, 77P, 79N, 81L, 83E, 96D, 98S, 100V, 101D, 142N, 114C, 145T, 148K, 152Y, 153C, 154C, 155N, 176Y, 178Y, 179P, 180K, 181D, 186T. Among these, negative charge contributing residues are 33E, 46D, 51D, 131D in PcOSM1 and 83E, 96D, 101D, and 181D in PcOSM2. The positions of the residues are relatively the same in the sequence as well as in the protein model of isoforms. 33E−51D residues in PcOSM1 and 83E−101D residues in PcOSM2 are situated in the cleft and extend across the cleft region (Fig. 5c, d). These residue pairs are most likely catalytic pairs for endoglucanase action as their distance and the position suit the definition of a catalytic pair. The distances calculated between the members of the pairs are 5.28 Å in PcOSM2 and 6.47 Å in PcOSM1 (Fig. 5c, d).
Homology models of TLPs. a Homology model of PcOSM1. Model shows the protein consisting of six β-sheets and four α-helices connected by various loops. b Homology model of PcOSM2. Model shows the protein comprising of 12 β-sheets and four α-helices. Models show differences in secondary structures. One common feature is that the proteins are cysteine rich and stabilized by disulphide bonds. PcOSM2 shows the typical thaumatin-like structure with three distinct domains, whereas PcOSM1 presents a distorted domain III. c, d shows the average distance between the acidic residues found in the cleft region of P. colubrinum osmotin isoforms. c, d Shows the catalytic pairs of PcOSM1 and PcOSM2 separated by 6.47 and 5.28 Å, respectively
Secondary structure formed by the ‘deletion’ residues. a Shows the overlapped image of the ‘ribbon models’ of osmotin isoforms. The portion coloured in pink is lacking in smaller osmotin isoform (PcOSM1). b Shows the extent of shift in the molecular surface of the protein caused by deletion. Green coloured portion in this figure corresponds to the internal deletion found in PcOSM1 (Color figure online)
Molecular surface architecture of P. colubrinum osmotin isoforms. Distribution of charges on the surface of the molecule is shown as red (acidic) and blue (basic). a Shows the PcOSM1 protein model harbouring a prominent acidic cleft between domains I and II similar to that found in PcOSM2 model (b) (Color figure online)
Docking experiments revealed residues that are able to interact by hydrogen bonding with glucan molecule (Fig. 8). In PcOSM1, the residues are 24Y, 25G, 26A, 27P, 31L, 33E, 35A, 50V, 51D, 86A, 91N, 94C, 95T, 96V, 98K, 103C, 104C. In PcOSM2, the residues are 41G, 42A, 43R, 63G, 64D, 74Y, 76A, 77P, 78P, 81L, 83E, 100V, 101D, 134L, 136A, 141N, 144C, 145T, 146V, 148K, 179P. The interaction energies of the molecules are −287.3 kcalmol−1 for PcOSM1 and −253.93 kcalmol−1 for PcOSM2. The homology modelling and docking studies reveal the presence of catalytic pairs and the possibility of involvement of the two osmotin isoforms in the cleavage of carbohydrate molecules like glucans.
Discussion
Transcript analysis of P. colubrinum osmotin isoforms shows a general over expression against various biotic and abiotic stimuli. It is evident that transcripts of smaller isoform, PcOSM1 show high expression level similar to PcOSM2 in response to pathogen, wounding, jasmonic acid and ethylene. This suggests a role for smaller forms of TLPs in plant–pathogen or plant–environment interaction. These smaller proteins may not have a direct inhibitory activity on the pathogen as evidenced by the absence of antifungal potential in vitro, but their possible indirect role in impeding pathogen progress in plants cannot be ruled out.
The deduced amino acid sequences of PcOSM1 and 2 are nearly identical except at the site of deletion [27]. The functional difference between the two osmotin isoforms in this study can possibly be attributed to the internal deletion of 50 amino acids in the smaller form. The internal deletion separates them into two size categories. Both forms of the protein possess an N-terminal signal peptide sequence indicating their extracellular location [27]. Structural information obtained from earlier models of antifungal TLPs—zeamatin, thaumatin, PR-5d, Osmotin and NP24-I provide a general model of TLPs having three distinct domains stabilized by disulphide bonds. An inter-domain acidic cleft has often been highlighted as a structural feature which is able to bind or hydrolyse β-1, 3-glucans, the constituents of fungal cell walls [19, 21, 25, 26]. In the homology models of P. colubrinum osmotin isoforms, an acidic cleft is recognised between domains I and II. The most striking feature of PcOSM1 was the deletion of amino acid residues in the sequences and the distortion of domain III and four β-sheets in its domain I. Though the deletion caused distortion in the overall structure of PcOSM1, it is interesting to note that it has retained an inter-domain acidic cleft containing glucan-interacting residues. A closer analysis of the cleft region of osmotin isoforms shows that the major residues in the cleft are intact, including optimally placed catalytic pair, enabling them to retain glucan hydrolytic property. PcOSM2 possesses a catalytic pair comprising 83E and 101D residues with a distance of 5.28 Å between them. In PcOSM1, the distance between the carboxylic oxygen atoms of the catalytic pair 33E and 51D is 6.47 Å. This data suggests that the glucan hydrolysing catalytic pair residues are at agreeable distance to effect a glucan hydrolysis action [21, 26] suggestive of a glucan-catalytic function for both forms.
In spite of the presence of the structural features necessary for antifungal action, PcOSM1 does not show any detectable inhibitory activity on the fungal species studied (P. capsici, F. oxysporum), compared to PcOSM2, which exhibits a strong antifungal potential. This observation reinforces earlier findings that the presence of acidic cleft alone may not be sufficient for the antifungal activity of TLPs, as the glucanase activity of TLP members to cause an inhibitory action against fungi is much lower compared to that shown by members of PR-2 family [21, 26].
It is possible that the structural alteration brought about by the internal deletion of amino acid residues in PcOSM1 results in the marked difference in fungal inhibitory activity between the two isoforms, as even a subtle change in structure can bring about significant difference in protein action. Comparison with PcOSM2 shows that the deletion residues comprise of six β-strands, four of which are constituents of domain I and the two remaining β-strands constitute a distinct domain III in PcOSM2. The deletion also causes a shift in the surface charge potential which probably determines the interaction of PcOSM1 with other membrane-associated structures, a feature thought to be essential for antifungal activity of proteins [41–43].
Adiponectin is a protein hormone produced and secreted by fat cells (adipocytes), involved in the regulation of metabolism of lipids and glucose in mammals [16]. It was shown earlier in yeast that osmotin (24 kDa protein from Nicotiana tabacum) binds to a receptor which is a homologue of mammalian adiponectin receptor. It also shares functional similarity with adiponectin in triggering an AMP kinase pathway in C2C12 myocytes [16, 17]. In this study, both the osmotin isoforms are predicted as globular proteins and domains I and III of osmotin may significantly contribute to the similarity to adiponectin molecule. In the light of a previous observation that the action of TLPs could resemble K1 Killer toxin of yeast which has distinct binding and effector domains [44, 45], it is reasonable to infer from this study that the deleted residues in PcOSM1 constitute or form a part of an effector domain which is responsible for the architectural similarity to the globular adiponectin molecule. The present comparative structural study of osmotin isoforms from P. colubrinum has identified putative structural features that could determine its activity. Analysis of these natural variants of the TLP family will provide additional insights into the evolutionary trend operating in the family where the different members are functionally optimised for diverse roles including defence. Though the osmotin isoforms in P. colubrinum differ in their inhibitory capacity on phytopathogenic fungi, the possibility of one form facilitating the action of the other, thus effecting a synergestic inhibitory activity on the fungal pathogen cannot be ruled out. This study warrants further in planta functional analysis of the isoforms which will rule out the possibility of protein misfolding associated with bacterial expression system, a point to be considered before establishing their functional divergence.
In conclusion, osmotin isoforms identified in P. colubrinum differ significantly in their antifungal activity. Despite the presence of a prominent acidic cleft, the smaller form of the protein is unable to exhibit detectable antifungal activity. Structural analysis showed that the internal deletion of PcOSMI caused significant structural alteration involving the loss of a proper domain III and four β-sheets from domain I. It brought changes in the surface charge potential of the protein as well. The absence of antifungal potential could be attributed to the loss of structural features caused by internal deletion highlighting the importance of domain III and domain I besides the presence of an acidic cleft in determining the antifungal activity of osmotins.
References
Agrios, G. N. (2005). Plant pathology. Burlington: Elsevier Academic press.
Van Loon, L. C., Pierpoint, W. S., Boller, T., & Conejero, V. (1994). Recommendations for naming plant pathogenesis-related proteins. Plant Molecular Biology Reporter, 12, 245–264.
Velazhahan, R., Datta, S. K., & Muthukrishnan, S. (1999). The PR-5 family: Thaumatin-like proteins in plants. In S. K. Datta & S. Muthukrishnan (Eds.), Pathogenesis-related proteins in plants (pp. 107–129). Boca Raton: CRC press.
Roberts, W. K., & Selitrennikoff, C. P. (1990). Zeamatin, an antifungal protein from maize with membrane permeabilizing activity. Journal of General Microbiology, 136, 1771–1778.
Woloshuk, C. P., Meulenhoff, J. S., Sela-Buurlage, M., van den Elzen, P. J. M., & Cornelissen, B. J. C. (1991). Pathogen-induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell, 3, 619–628.
Abad, L. R., D’Urzo, M. P., Liu, D., Narasimhan, M. L., Reuveni, M., Zhu, J. K., et al. (1996). Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilisation. Plant Science, 118, 11–23.
Liu, D., Rhodes, D., D’Urzo, M. P., Xu, Y., Narasimhan, M. L., Hasegawa, P. M., et al. (1996). In vivo and in vitro activity of truncated osmotin that is secreted into the extracellular matrix. Plant Science, 121, 123–131.
Jami, S. K., Anuradha, T. S., Guruprasad, L., & Kirti, P. B. (2007). Molecular, biochemical and structural characterization of osmotin-like protein from black nightshade (Solanum nigrum). Journal of Plant Physiology, 164, 238–252.
Liu, D., Raghothama, K. G., Hasegawa, P. M., & Bressan, R. A. (1994). Osmotin over expression in potato delays development of disease symptoms. Proceedings of the National Academy of Science of the United States of America, 91, 1888–1892.
Datta, K., Velazhahan, R., Oliva, N., Ona, I., Mew, T., & Khush, G. S. (1999). Over-expression of the cloned rice thaumatinlike protein (PR-5) gene in transgenic rice plants enhances environmently friendly resistance to Rhizoctonia solani causing sheath blight disease. Theoretical and Applied Genetics, 98, 1138–1145.
Yun, D. J., Zhao, Y., Pardo, J. M., Narasimhan, M. L., Damsz, B., & Lee, H. (1997). Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proceedings of the National Academy of Science of the United States of America, 94, 7082–7087.
Anžlovar, S., Dalla Serra, M., Dermastia, M., & Menestrina, G. (1998). Membrane permeabilizing activity of pathogenesis-related protein linusitin from flax seed. Molecular Plant-Microbe Interactions, 7, 610–617.
Anžlovar, S., & Dermastia, M. (2003). The comparative analysis of osmotins and osmotin-like PR-5 proteins. Plant Biology, 5, 116–124.
Ibeas, J. I., Lee, H., Damsz, B., Prasad, D. T., Pardo, J. M., Hasegawa, P. M., et al. (2000). Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant Journal, 23, 375–383.
Narasimhan, M. L., Lee, H., Damsz, B., Singh, N. K., Ibeas, J. L., Mat-sumoto, T. K., et al. (2003). Overexpression of a cell wall glycoprotein in Fusarium oxysporum increases virulence and resistance to a plant PR-5 protein. Plant Journal, 36, 390–400.
Kadowaki, T., & Yamauchi, T. (2005). Adiponectin and adiponectin receptors. Endocrine Reviews, 26, 439–451.
Narasimhan, M. L., Coca, M. A., Jin, J., Yamauchi, T., Ito, Y., Kadowaki, T., et al. (2005). Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Molecular Cell, 17, 171–180.
Batalia, M. A., Monzingo, A. F., Ernst, S., & Robertus, J. D. (1996). The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family. Nature Structural Biology, 3, 19–23.
Koiwa, H., Kato, H., Nakatsu, T., Oda, J., Yamada, Y., & Sato, F. (1999). Crystal structure of tobacco PR-5d protein at 1.8 Å resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. Journal of Molecular Biology, 286, 1137–1145.
Min, K., Ha, S. C., Hasegawa, P. M., Bressan, R. A., Yun, D., & Kim, K. K. (2004). Crystal structure of osmotin, a plant antifungal protein. Proteins: Structure, Function, and Bioinformatics, 54, 170–173.
Leone, P., Menu-Bouaouiche, L., Peumans, W. J., Payan, F., Barre, A., Roussel, A., et al. (2006). Resolution of the structure of the allergenic and antifungal banana fruit thaumatin like protein at 1.7-A°. Biochimie, 88, 45–52.
Ghosh, R., & Chakrabarti, C. (2008). Crystal structure analysis of NP24-I: A thaumatin-like protein. Planta, 228, 883–890.
Trudel, J., Grenier, J., Potvin, C., & Asselin, A. (1998). Several thaumatin-like proteins bind to β-1, 3-glucans. Plant Physiology, 118, 1431–1438.
Grenier, J., Potvin, C., Trudel, J., & Asselin, A. (1999). Some thaumatin-like proteins hydrolyse polymeric β-1, 3-glucans. Plant Journal, 19, 473–480.
Osmond, R. I., Hrmova, M., Fontaine, F., Imberty, A., & Fincher, G. B. (2001). Binding interactions between barley thaumatin-like proteins and (1, 3)-β-D-glucans. Kinetics, specificity, structural analysis and biological implications. European Journal of Biochemistry, 268, 4190–4199.
Menu-Bouaouiche, L., Vriet, C., Peumans, W. J., Barre, A., Van Damme, E. J., & Rougé, P. (2003). A molecular basis for the endo-beta 1, 3-glucanase activity of the thaumatin-like proteins from edible fruits. Biochimie, 85, 123–131.
Mani, T., & Manjula, S. (2010). Cloning and characterization of two osmotin isoforms from Piper colubrinum. Biologia Plantarum, 54, 377–380.
Vanaja, T., Neema, V. P., Mammootty, K. P., & Rajeshkumar, R. (2008). Development of a promising interspecific hybrid in black pepper (Piper nigrum L.) for Phytophthora foot rot resistance. Euphytica, 161, 437–445.
Hu, X., & Reddy, A. S. N. (1997). Cloning and expression of a PR5-like protein from Arabidopsis: Inhibition of fungal growth by bacterially expressed protein. Plant Molecular Biology, 349, 49–59.
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry, 723, 41–74.
Broekaert, W. F., Terras, F. R. G., Cammue, B. P. A., & Vanderleyden, J. (1990). An automated quantitative assay for fungal growth inhibition. FEMS Microbiology Letters, 69, 55–60.
Salzman, R. A., Koiwa, H., Ibeas, J. I., Pardo, J. M., Hasegawa, P. M., & Bressan, R. A. (2004). Inorganic cations mediate plant PR5 protein antifungal activity through fungal Mnn1-and Mnn4-regulated cell surface glycans. Molecular Plant-Microbe Interactions, 17, 780–788.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acid Research, 25, 3389–33402.
Sali, A., & Blundell, T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology, 234, 779–815.
Kelley, L. A., Gardner, S. P., & Sutcliffe, M. J. (1996). An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Engineering, 9, 1063–1065.
Sippl, M. J. (1993). Recognition of errors in three-dimensional structures of proteins. Proteins, 17, 355–362.
Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., et al. (1998). Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. Journal of Computational Chemistry, 19, 1639–1662.
Kouwizjer, M. L. C. E., & Grootenhuis, P. D. J. (1995). Parametrization and application of CHEAT95, an extended atom force field for hydrated oligosaccharides. Journal of Physical Chemistry, 99, 13426–13436.
Aswati Nair, R., Kiran, A. G., Sivakumar, K. C., & Thomas, G. (2010). Molecular characterization of an oomycete-responsive PR-5 protein gene from Zingiber zerumbet. Plant Molecular Biology Reporter, 28, 128–135.
Chan, Y. W., Tung, W. L., Griffith, M., & Chow, K. C. (1999). Cloning of a cDNA encoding the thaumatin-like protein of winter rye (Secale cereale L. Musketeer) and its functional characterisation. Journal of Experimental Botany, 50, 627–1628.
Thevissen, K., Osborn, R. W., Acland, D. P., & Broekaert, W. F. (1997). Specific, high affinity binding sites for an antifungal plant defensin on Neurospora crassa hyphae and microsomal membranes. Journal of Biological Chemistry, 272, 32176–32181.
Thevissen, K., Osborn, R. W., Acland, D. P., & Broekaert, W. F. (2000). Specific binding sites for an antifungal plant defensin from Dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Molecular Plant-Microbe Interactions, 13, 54–61.
Veronese, P., Ruiz, M. T., Coca, M. A., Hernandez-Lopez, A., Lee, H., Ibeas, J. I., et al. (2003). In defense against pathogens both plant sentinels and foot soldiers need to know the enemy. Plant Physiology, 131, 1580–1590.
Hutchins, K., & Bussey, H. (1983). Cell wall receptor for yeast killer toxin: Involvement of (1–6)-β-d-glucan. Journal of Bacteriology, 1549, 161–169.
Bussey, H. (1991). K1 killer toxin, a pore-forming protein from yeast. Molecular Microbiology, 5, 2339–23431.
Acknowledgments
Authors would like to thank Dr. N. Anith, Kerala Agricultural University, Vellayani, Trivandrum for the fungal strains. T.M. would like to acknowledge Council for Scientific and Industrial Research, New Delhi, Government of India, for CSIR-Junior Research Fellowship, and MS gratefully acknowledges the Department of Biotechnology, Government of India, for financial support in the form of research grant.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mani, T., Sivakumar, K.C. & Manjula, S. Expression and Functional Analysis of Two Osmotin (PR5) Isoforms with Differential Antifungal Activity from Piper colubrinum: Prediction of Structure–Function Relationship by Bioinformatics Approach. Mol Biotechnol 52, 251–261 (2012). https://doi.org/10.1007/s12033-011-9489-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9489-0