Abstract
Triple negative breast cancer (TNBC) is the most aggressive subtype of breast cancer. More and more studies have shown that the tumor immune microenvironment (TME) of TNBC is closely related to its poor prognosis and early metastasis. We try to explain how tumor-associate macrophages (TAMs), an important component of the TME, function in the matrix of TNBC. Therefore, we induced THP-1 cells to become M1-TAMs and M2-TAMs, investigated their influence on breast cancer cells. 82 TNBC paraffin samples were made into tissue microarrays. The expression of macrophages makers were measured by immunohistochemistry. Scratch assay, Transwell assay, CCK-8 cell proliferation assay were performed in the co-culture system of breast cancer cells lines and macrophages to observe the invasion and proliferation ability of breast cancer cell lines. Western Blot (WB) was performed to detect the expression of E-cadherin (CDH1) and N-cadherin (CDH2). M2-TAMs were more numerous than M1-TAMs in the matrix of TNBC cancer nests and associated with poor prognosis. M2-TAMs promoted the invasion, migration, and proliferation of TNBC cells. M1-TAMs had inhibitory effects. In MCF-7 cells, WB showed a decrease in CDH1 and an increase in CDH2. In MDA-MB-231 cells and BT549 cells, CDH2 expression was reduced and CDH1 expression was increased. All of the above results were statistically significant, p < 0.001. M2-TAMs were more numerous in TNBC and associated with poor prognosis. M2-TAMs promoted the invasion, migration, and proliferation of breast cancer cells. The mechanism may be related to the epithelial-mesenchymal transition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
TNBC is the most aggressive subtype of breast cancer, which is defined as no expression of estrogen receptor (ER), progesterone receptor (PR), and no amplification or overexpression of HER2. Early recurrence, easy transfer, and most importantly, lack of therapeutic targets are major problems in TNBC [1]. Multiple mechanisms are associated with poor prognosis of TNBC, include the TME. But patients with TNBC have little benefits [2]. This may be due to the lack of understanding the TME in TNBC.
TME plays an important role in the study of TNBC immunogenicity [3], tumor-associated macrophages (TAMs) are the most inflammatory immune cells in TME, accounting for about 30% [4]. TAMs, have influence on tumor progression and distant metastasis by regulate tumor cells invasion, migration, and angiogenesis [5]. TAMs can be divided into many subtypes, of which the classical classification method is divided into the classical activated M1 subtype and the alternately activated M2 subtype, according to their receptor, cytokine, and chemokine expression and their effect function [6]. In brief, M1-TAMs express a large amount of inducible nitric oxide synthase (iNOS) and tumor necrosis factor (TNF)-α, and exert anti-tumor activity by promoting pro-inflammatory effect and immune response [7]. M2-TAMs express argininase1 (Arg1), high levels of cytokines, growth factors, and proteases, play a role in promoting cancer [8].
Studies have shown that M2-TAMs can promote tumor metastasis by promoting the entry of tumor cells into blood vessels through the paracrine circuit composed of macrophage colony-stimulating factor-1 (M-CSF) of tumor cells and EGF of macrophages and their receptors [9]. Other studies have shown that TAMs can express a variety of cytokines that stimulate the proliferation and survival of tumor cells, such as epithelial growth factor (EGF), platelet-derived growth factor TGF-β1, hepatocyte growth factor (HGF), and epithelial growth ligand of the EGFR family [10]. In addition, other studies have shown that M2-TAMs promote angiogenesis, forms stem cell characteristics, and promotes distant metastasis [11]. In comparison, most studies on M1-TAMs suggest that they recognize and kill tumors cells directly by secreting some tumor killing molecules including ROS and NO [12]. Tumor cells can also be killed through the antibody dependent cell-mediated cytotoxicity (ADCC) mechanism [13].
In this study, we intend to analyze the differences between M1-TAMs and M2-TAMs in the matrix of TNBC cancer nests by detecting the differences in the expression levels of CD163, CD206, CD80, CD86, IL-10, and IL-6 in tissue microarrays. Co-culture system of breast cancer cells (MCF-7, MDA-MB-231, BT549) and TAMs were established. Cell Scratching assay, Transwell invasion and migration assay, CCK-8 proliferation assay, and WB were used to explore the effect of M2 on invasion, proliferation, and preliminary mechanism in TNBC.
Materials and methods
Patients and tissue samples
82 patients with TNBC, who underwent surgery for stage I to III tumors from January 2018 to December 2020 at Department of Pathology, The First Affiliated Hospital of USTC, were included in this study. Patients with ER, PR, HER2 all negative breast cancer were included, ductal carcinomas in situ (DCIS), stage IV tumors, and other type invasive BC were excluded. Table 1 shows the clinical and pathological characteristics of the patients enrolled in this study. This study was conducted in accordance with ethical principles, including the Helsinki declaration, and was approved by the Institutional Review Board of The First Affiliated Hospital of USTC. The requirement for written informed consent was waived owing to the retrospective nature of the study.
Tissue microarrays and IHC
All patients received surgical intervention for primary breast tumors. All specimens were formalin fixed, paraffin embedded, and cut into 4 μm thick sections for hematoxylin and eosin (H&E) staining. Histological grades were assigned based on the Nottingham system, using surgical specimens. Tissue microarrays were made by Anhui Xinle Biotechnology Co., LTD. The expression of the CD163, CD206, CD80, CD86, IL-10, and IL-6 was examined by IHC in tissue microarray. The following primary antibodies were used: anti-CD163 (ready-to-use, 2 m-0428, ZSBG-BIO, China); anti-CD206 (dilution of 1:50, ab64693, Abcam, Cambridge, UK); anti-CD80 (dilution of 1:20, GeneTex Inc., Irvine, CA, USA); anti-CD86 (dilution of 1:50, GeneTex Inc., Irvine, CA, USA); anti-IL-10(dilution of 1:100, FNab04211, FineTest, China); anti-IL-6 (dilution of 1:200, FNab04282, FineTest, China). Immunostaining was performed on a Roche Benchmark XT automated staining system (Roche/Ventana) according to the instructions. Procedure: Three-micrometer sections were dewaxed in EZprep concentration buffer at 75 °C for 4 min. Epitope repair was performed in cell conditioning solution at 100 °C for 64/76 min. Antibodies were both incubated at 37 °C for 32 min. Then, goat anti-mouse/anti-rabbit IgG/IgM secondary antibody coupled with horseradish peroxidase (HRP) was added for 8 min followed by DAB visualization and finally hematoxylin staining. CD163, CD206, CD80, CD86 were localized in the cytoplasm and/or membrane of macrophages, IL-10 and IL-6 were localized in the nucleus. Yellow–brown or tan particles were defined as positive. The stained cells were calculated using Image J (1.8.0.112; version 2.1.4.7, National Institutes of Health, Maryland, USA) to get the quantitative results.
Cell culture and reagents
Human monocyte cell line THP-1, Human breast carcinoma cell lines MCF-7, MDA-MB-231, BT549 were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. THP-1, MCF-7, MDA-MB-231, BT549 cells were maintained in RPMI-1640 medium, supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin (Thermo Fisher Scientific, USA). Other than that, THP-1 were maintained with 0.05 nM β mercaptoethanol (M301574, Aladdin, China). All cells were cultured at 37 °C and 5% CO2.
Induction and identification of macrophages
9 × 106 THP-1 cells were inoculated in a 100 mm culture dish, and different cytokines were added into the culture flask periodically (Table 2). PMA (100 ng/mL, HY-18739, MCE, China), IFN-γ(20 ng/mL, HY-p7025, MCE, China), LPS (100 ng/mL, L6529, Sigma, USA), IL-4 (20 ng/mL, C610006, BBI, China). Cells were collected for qRT-PCR detection. The markers of M1-TAMs (HLA-DR、TNF-α、IL-6、IL-1β) and M2-TAMs (CD206、IL-10、MRC-1、DC-SIGN、SOCS1、Arg-1、TGF-β) were analyzed in THP-1 cells, M1 THP-1 cells, and M2 THP-1 cells using the synthesized primers (Sangon Bio, China). (Online Resource 1).
Quantitative real-time PCR
The RNA extraction procedure for paraffin tissues was performed according to the instructions of FFPE RNA Kit (R6954, OMEGA, USA). Total RNA was extracted from THP-1 cells, M1 THP-1 cells, and M2 THP-1 cells using TRIzol reagent and 2 μg RNA was reverse transcribed to cDNA using reverse transcriptase (Vazyme). Real-time quantitative PCR (qRT-PCR) was done using the SYBR Green I Real-Time Detection kit (CWBio) on the CFX96 Detection System (Bio-Rad). Gene expression was normalized to GAPDH expression. Relative gene expression was normalized to macrophage expression.
Cell scratch assay
Marked on the back of 6-well plate, about every 1.0 cm, drew at least 5 lines. Each hole with 5 × 105 cells, added 2 ml medium, incubated overnight. Then, drew 2 or 3 straight lines with tips, washed the plate with PBS three times, incubated for 24 h with serum-free culture medium, took pictures at 0 h, 24 h.
Transwell migration assay
Plate cells (5 × 105) in the upper chamber of 8 μm Transwell (Corning, NY), and allowed to migrate for 8 h at 37 °C. Induced cell ability to migration by placing 1640 THP-1 cells, M1 THP-1 cells and M2 THP-1 cells in the lower chamber of Transwell inserts. Counted the number of cells that migrated across the filters in 4 high-power fields per insert, and average values afterward. For each migration condition, three identical replicates should be performed.
Transwell invasion assay
Thawed the stock 10 ml Matrigel (BD, Biosciences) overnight at 4 °C by placing inside a refrigerator or cold room. Once thawed, always keep Matrigel chilled on wet ice. Diluted to 1/8, put 100 μl on the upper chamber, incubated the plates at 37 °C for at least 30 min before starting the invasion assay. The rest of the steps were to reference Transwell Migration Assay.
The CCK-8 cell proliferation assay
Prepared cell suspension, counted 5 × 104 cells /ml, mixed evenly, took about 100 ul, inoculated it into a 96-well plate. The same samples were repeated 5 times and put into 37 °C incubators. After cell adherence, 10 μl CCK8 was added, and continued culture for 4 h. The absorbance was measured at 450 nm.
Western blots
Total cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to Polyvinylidene difluoride membranes (Millipore Corporation, Billerica, MA, USA); the immune complexes were detected by enhanced chemiluminescence (Cell Signaling Technology, Beverly, MA). Antibodies were used: rabbit monoclonal anti-human E-cadherin (dilution of 1:1000, Abcam, Cambridge, UK); mouse monoclonal anti-human N-cadherin (dilution of 1:1000, Abcam, Cambridge, UK); rabbit monoclonal anti-human CD86 (dilution of 1:1000, Abcam, Cambridge, UK); rabbit monoclonal anti-human iNOS (dilution of 1:500, Abcam, Cambridge, UK); rabbit monoclonal anti-human CD206 (dilution of 1:1000, Abcam, Cambridge, UK); rabbit monoclonal anti-human Arg-1 (dilution of 1:1000, Cell Signaling Technology, Boston, USA); β-actin (dilution of 1:5000, Abcam, Cambridge, UK). Blots were quantitated by densitometry and normalized using β-actin to correct for differences in protein loading. For densitometry analyses, protein bands on the blot were measured by use of Image J.
Statistical analysis
All experiments were repeated three times, and data were expressed as mean ± standard deviation. The data were tested for significant differences by paired two-tailed t test, after a Bartlett test had shown that variances were homogenous using GraphPad Prism 6.01 software (GraphPad Software Inc, La Jolla, CA, USA). The difference was considered statistically significant at p < 0.05.
Results
M2-TAMs were more numerous than M1-TAMs in the matrix of TNBC cancer nests and associated with poor prognosis
CD206+/CD163+ TAMs and CD86+/CD80+ TAMs were closely correlated with tumor size (p < 0.001), Nottingham Classification (p < 0.001), lymph node metastasis (p < 0.001), and Ki-67 index (p < 0.01) of TNBC patients (Fig. 1). CD206+/CD163+ TAMs expression was positively correlated (Fig. 1a), while CD86+/CD80+ TAMs expression was negatively correlated (Fig. 1b). IHC results showed that CD206 was expressed more than CD86 in the matrix of TNBC carcinoma nests (Fig. 2a). Manual counting, the number of M2-TAMs is also more than M1-TAMs in the same field (Fig. 2b). The results were statistically different, p < 0.001. The expression of CD206 and IL-10 was also higher than that of CD86 and IL-6 (Fig. 3c). We also found that CD206+/CD163+ cells (M2-TAMs) were more than CD86+/CD80+ cells (M1-TAMs) in the same filed (Fig. 3d). The results were statistically different, p < 0.001.
Correlation of various clinicopathological features in TNBC with the expression of M2-TAMs and M1-TAMs. a CD206+/CD163+ TAMs expression was positively correlated with lymph node metastasis, tumor size, Nottingham classification, and ki-67 index. b CD86+/CD80+ TAMs expression was negatively correlated with lymph node metastasis, tumor size, Nottingham classification, and ki-67 index
Expression of M2-TAMs and M1-TAMs in the matrix of TNBC cancer nests. a Tissue microarray and IHC for CD206 and CD86. In the same HE field, there were significantly more CD206+ M2-TAMs (red triangles) than CD86+ M1-TAMs (red pentagons) around the cancer cells (red arrows). Scale bar: 50 μm. b Independent counts of M2-TAMs and M1-TAMs separately. Bars represent the average ± standard deviation of three independent counts (***p < 0.001)
M2-TAMs were more numerous than M1-TAMs in the matrix of TNBC cancer nests. a Surface markers (CD163, CD206) and expression of specific cytokine (IL-10) in M2-TAMs. Upper Scale bar: 200 μm. Lower Scale bar: 50 μm. b Surface markers (CD80, CD86) and expression of specific cytokines (IL-6) in M1-TAMs. Upper Scale bar: 200 μm. Lower Scale bar: 50 μm. c The CD206 expression were stronger than CD86 expression, and IL-10 expression were higher than IL-6. d There were more M2-TAMs (CD206+ and CD163+) than M1-TAMs (CD86+ and CD80+). Positive rates of expression were measured with Image J software. Bars represent the average ± standard deviation of three independent counts (***p < 0.001)
TAMs were induced and identification in vitro
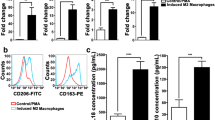
Co-culture system for breast cancer cells and TAMs (Fig. 4a). Observe the changes in THP-1 cell morphology after induction. THP-1 cells were full in shape and had good refractive index, a single cell grew in suspension. M1 THP-1 macrophages showed diversity in morphology, and some cells were in long spindle shape. M2 THP-1 macrophages were polygons, increased in volume, the refractive index became worse, and the cytoplasm increased and became rough (Fig. 4b). The qRT-PCR results showed that the markers of M1 macrophages (HLA-DR, TNF-α, IL-6, IL-1β) were increased in M1 THP-1 macrophages. The markers of M2 macrophages (CD206, IL-10, MRC-1, DC-SIGN, SOCS1, Arg-1, TGF-β) were increased in M2 THP-1 macrophages (Fig. 4c, d). The WB results showed that the markers of M1 macrophages (CD86, iNOS) were increased in M1 THP-1 macrophages. The markers of M2 macrophages (CD206, Arg-1) were increased in M2 THP-1 macrophages (Fig. 4e, f).
Induction and identification of macrophage. a Diagram of the co-culture of macrophages and breast cancer cells. b Morphological changes of THP-1 cells after induction of PMA and different cytokines. Upper panel: 0.1% crystalline violet staining, Scale bar: 200 μm. Lower panel: Scale bar: 50 μm. c The relative mRNA expression of M2 THP-1 macrophages was analyzed by qRT-PCR in M0 THP-1 macrophages (THP-1 cells induced by PMA) with or without IL-4 treatment for 48 h (n = 3). (***p < 0.001). d The relative mRNA expression of M1 THP-1 macrophages was analyzed by qRT-PCR in M0 THP-1 macrophages (THP-1 cells induced by PMA) with or without LPS, IFN-γ treatment for 48 h (n = 3). (***p < 0.001). e, f Western blot for CD86, iNOS, CD206, Arg-1 in M0 THP-1 macrophages, M1 THP-1 macrophages, and M2 THP-1 macrophages. (***p < 0.001)
M2 THP-1 macrophages enhance the migration and invasion of breast cancer cells
In the Scratch assay, the migratory ability of MCF-7 cells co-cultured with M2 THP-1 macrophages was significantly enhanced, and co-cultured with M1 THP-1 macrophages was not significantly changed (Fig. 5a). MDA-MB-231 cells and BT549 cells co-cultured with M2 THP-1 macrophages had significantly enhanced migratory ability, while co-cultured with M1 THP-1 macrophages had significantly reduced (Fig. 5b, c). In Transwell migration and invasion assays, the migration and invasion abilities were enhanced for all these cell lines co-cultured with M2 and diminished for those co-cultured with M1 (Fig. 5d–f).
Changes in biological behavior of breast cancer cell lines MCF-7, MDA-MB-231, and BT549 after co-culture with M1 THP-1 macrophages / M2 THP-1 macrophages. a, b, c Scratching assay of MCF-7, MDA-MB-231, BT549 cells after co-culture with M1 THP-1 macrophages / M2 THP-1 macrophages. Pictures were taken at 0 h and 24 h. Scale bar: 200 μm. d, e, f Transwell migration and invasion assay of MCF-7, MDA-MB-231, BT549 cells after co-culture with M1 THP-1 macrophages / M2 THP-1 macrophages. Upper panel: migration assay. Lower panel invasion assay. d, e Scale bar: 200 μm. f Scale bar: 100 μm. g MCF-7, MDA-MB-231, and BT549 cells have enhanced healing ability after co-culture with M2 THP-1 macrophages. MDA-MB-231 and BT549 have diminished healing ability after co-culture with M1 THP-1 macrophages. h, i MCF-7, MDA-MB-231, and BT549 cells have enhanced migration and invasion ability after co-culture with M2 THP-1 macrophages. Migration and invasion ability were diminished after co-culture with M1 THP-1 macrophages. j MCF-7, MDA-MB-231, and BT549 cells had enhanced proliferation ability after co-culture with M2 THP-1 macrophages. Weakened proliferation ability after co-culture with M1 THP-1 macrophages. The control group was breast cancer cells without co-culture with TAMs. Bars represent the average ± standard deviation of three independent counts (*p < 0.05; **p < 0.01; ***p < 0.001)
M2 THP-1 macrophages promote proliferation of breast cancer cells
In CCK-8 proliferation assays, the proliferation abilities of MCF-7, MDA-MB-231, BT549 cells co-cultured with M2 THP-1 macrophages was enhanced, whereas the proliferation abilities of those cell lines co-cultured with M1 THP-1 macrophages was diminished (Fig. 5j).
M2 THP-1 macrophages promotes EMT of breast cancer cells
The expression of CDH1, which was originally highly expressed, was significantly reduced after co-culture with M2 THP-1 macrophages, and CDH2 was expressed instead in MCF-7 cells (Fig. 6a, b). In MDA-MB-231 and BT549 cells, CDH1 was expressed to a certain extent, while CDH2 was weakened, after co-culture with M1 THP-1 macrophages. Compared with M2 THP-1 macrophages showed strong expression in both CDH1 and CDH2 (Fig. 6c–f).
Expression levels of EMT-associated proteins CDH1 and CDH2 in MCF-7, MDA-MB-231, and BT549 cells after co-culture with M1 THP-1 macrophages / M2 THP-1 macrophages. a, b, c Western blot for E-cadherin and N-cadherin in MCF-7、MDA-MB-231、BT549 cells after treatment with M1 THP-1 macrophages, M2 THP-1 macrophages for 48 h. β-actin was used as a loading control. d MCF-7 cells co-cultured with M2 THP-1 macrophages showed reduced expression of CDH1 and increased expression of CDH2. After co-culture with M1 THP-1 macrophages, the expression of CDH1 was decreased and the expression of CDH2 was increased. e MDA-MB-231 cells co-cultured with M2 THP-1 macrophages showed increased expression of CDH1and no significant changes in CDH2 expression levels. After co-culture with M1 THP-1 macrophages, the expression of CDH1 was significantly increased and CDH2 was decreased. f BT549 cells co-cultured with M2 THP-1 macrophages showed elevated expression of CDH1 and decreased expression of CDH2. After co-culture with M1 THP-1 macrophages, the expression of CDH1 was increased and the expression of CDH2 was decreased. The CDH1/β-actin and CDH2/β-actin ratio was quantified and was shown as fold changes compared with control. The control group was breast cancer cells without co-culture with TAMs. Bars represent the average ± standard deviation of three independent counts (*p < 0.05; **p < 0.01; ***p < 0.001)
Discussion
TNBC is recognized as a subtype of breast cancer with poor prognosis. Multiple factors are associated with poor prognosis, among which tumor-associated macrophages (TAMs) have an irreplaceable position [4]。The polarization of TAMs is continuously changing state. Most studies divide TAMs into two polarization tops, M1-TAMs and M2-TAMs [14]。The results of this study showed that the number of M2-TAMs in TNBC was positively correlated with tumor size, Nottingham Classification, and lymph node metastasis, suggesting a poor prognosis. It has been shown that a large number of CD206+ M2-TAMs is associated with high proliferation and low differentiation of tumors and these cells are more frequently found in TNBC [15]. It was also investigated to assess the status of TAMs in the tumor microenvironment using the number of M2-TAMs/the number of M1-TAMs, the TAMs balance fraction (MBF). Confirmed that in breast cancer, high MBF was associated with poor prognosis and could promote tumor metastasis [16, 17]. Macrophages can be differentiated into subtypes of macrophages in different directions after stimulated by cytokines in vitro. In this paper, a model of THP-1 mononuclear cell line differentiation into macrophages induced by PMA was used [18]. Pathological histological observations showed that M2-TAMs were predominant in the matrix of TNBC cancer nests. In vitro, experiments revealed that M2-TAMs promoted the migration, invasion, and proliferation of breast cancer cells, supporting the pro-tumor progressive role of M2-TAMs. It was shown that M2-TAMs were involved in the whole metastatic process of breast cancer, including local invasion, promotion of tumor cells into blood vessels, and extravasation from distant sites [19]. It has been shown that the mechanism is that they go through the production of matrix metalloproteinases (MMPs), cysteine histone proteases and serine proteases that destroy the cell matrix and subsequently, and then invade tumor cells into the surrounding tissue [20]. TAMs also secrete acidic substances, including chemokine ligands (e.g., CCL18) and epidermal growth factor (EGF), all of which have tumorigenic effects [21]. This study found that breast cancer cells may promote the polarization of M1 phenotype to M2 phenotype in co-culture mode. Some studies have reported that the polarization status of TAMs is influenced by several factors in breast cancer [22], as the lactate metabolism [23]. It was also confirmed that changes in adenosine deaminase 2 (ADA2) activity may contribute to the differentiation of TAMs to the M2 phenotype in TNBC [24]. In addition, M1-TAMs inhibited the migration, invasion, and proliferation of breast cancer cells in this study and had some inhibitory effect on tumor progression. Most studies have shown that M1-TAMs show anti-tumor capacity mainly through the release of pro-inflammatory cytokines such as TNF and interleukin (IL)-2, as well as reactive nitrogen and oxygen intermediates [22, 25]. It has also demonstrated that the AMPK-NF-κB signaling pathway is involved in regulating the polarization of macrophage into the anti-tumor M1 phenotype [26].
In recent years, there have been numerous studies on the mechanism of action of TAMs. The results of this study showed that after co-culture with M1-TAMs, the expression of CDH1 and CDH2 was reversed in MDA-MB-231 cells and BT549 cells, suggesting a reversal of the epithelial-mesenchymal transition (EMT) process. After co-culture with M2-TAMs, the process of EMT was promoted in MCF-7 cells. Investigations showed that the majority of breast cancer patients (90%) died due to the invasion and metastasis, which associated with EMT [27]. Weng et al. showed that the oncogene Multiple Copies in T-cell Malignancy 1 (MCT-1/MCTS1) stimulated the secretion of interleukin-6 (IL-6), promoted monocyte THP-1 polarization to the M2 phenotype, stimulated the development of EMT, and increased the invasiveness of TNBC cells [28]. It was also shown that miR-200c, a potent EMT suppressor, upregulated cytokines, such as granulocyte–macrophage colony-stimulating factor (GM-CSF), promoted the polarization of TAMs to M1 phenotype [29]. In addition, we observed that some breast cancer cells aggregated into spheres after co-culture with M2-TAMs. The study confirmed that exosomes secreted by mesenchymal stem cells (MSCs) drive accelerated breast cancer progression by inducing monocytes to differentiate into M2 phenotype macrophages [30]. MSCs were also shown to differentiate into S100A4-secreting cancer-associated fibroblasts (CAFs), and S100A4 stimulates cells to secrete CCL2, which polarizes macrophages into M2-TAMs, promoting their metastatic potential through a bidirectional interaction between MSCs/CAFs and M2-TAMs [31]. Jia et al. used M2-conditioned medium to culture mesenchymal stem cells (cMSCs) and found that their ability to promote tumor growth was greatly enhanced in vivo. Moreover, IL-6 secreted by cMSCs could polarize infiltrating TAMs to the M2 phenotype [32]. Therefore, targeting TAMs is an extremely promising therapeutic strategy. Either to reverse the EMT process or to reduce the stemness of TNBC cells.
Conclusion
In conclusion, the predominance of M2-TAMs in the matrix of TNBC cancer nests was associated with poor prognosis of TNBC. M2-TAMs promote breast cancer cell invasion and proliferation by a mechanism related to EMT. M1-TAMs, on the other hand, have been shown to have some anti-tumor effects, inhibited breast cancer cell invasion and proliferation in vitro, associated with the EMT mechanism.
References
Vagia E, Mahalingam D, Cristofanilli M. The landscape of targeted therapies in TNBC. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12040916.
Keenan TE, Tolaney SM. Role of immunotherapy in triple-negative breast cancer. J Natl Compr Canc Netw. 2020;18(4):479–89. https://doi.org/10.6004/jnccn.2020.7554.
Deepak K, Vempati R, Nagaraju GP, Dasari VR, S N, Rao DN, et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. https://doi.org/10.1016/j.phrs.2020.104683.
Niu M, Valdes S, Naguib YW, Hursting SD, Cui Z. Tumor-associated macrophage-mediated targeted therapy of triple-negative breast cancer. Mol Pharm. 2016;13(6):1833–42. https://doi.org/10.1021/acs.molpharmaceut.5b00987.
Lundgren C, Bendahl PO, Ekholm M, Fernö M, Forsare C, Krüger U, et al. Tumour-infiltrating lymphocytes as a prognostic and tamoxifen predictive marker in premenopausal breast cancer: data from a randomised trial with long-term follow-up. Breast Cancer Res. 2020;22(1):140. https://doi.org/10.1186/s13058-020-01364-w.
Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs classically and M2(LPS-) vs. alternatively activated macrophages. Front Immunol. 2019;10:1084. https://doi.org/10.3389/fimmu.2019.01084.
Paul S, Chhatar S, Mishra A, Lal G. Natural killer T cell activation increases iNOS(+)CD206(-) M1 macrophage and controls the growth of solid tumor. J Immunother Cancer. 2019;7(1):208. https://doi.org/10.1186/s40425-019-0697-7.
Arlauckas SP, Garren SB, Garris CS, Kohler RH, Oh J, Pittet MJ, et al. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics. 2018;8(21):5842–54. https://doi.org/10.7150/thno.26888.
Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun. 2018;9(1):21. https://doi.org/10.1038/s41467-017-02481-5.
Onal S, Turker-Burhan M, Bati-Ayaz G, Yanik H, Pesen-Okvur D. Breast cancer cells and macrophages in a paracrine-juxtacrine loop. Biomaterials. 2021;267:120412. https://doi.org/10.1016/j.biomaterials.2020.120412.
Yang Y, Hao E, Pan X, Tan D, Du Z, Xie J, et al. Gomisin M2 from Baizuan suppresses breast cancer stem cell proliferation in a zebrafish xenograft model. Aging (Albany NY). 2019;11(19):8347–61. https://doi.org/10.18632/aging.102323.
Cotzomi-Ortega I, Nieto-Yañez O, Juárez-Avelar I, Rojas-Sanchez G, Montes-Alvarado JB, Reyes-Leyva J, et al. Autophagy inhibition in breast cancer cells induces ROS-mediated MIF expression and M1 macrophage polarization. Cell Signal. 2021;86:110075. https://doi.org/10.1016/j.cellsig.2021.110075.
Pellizzari G, Hoskin C, Crescioli S, Mele S, Gotovina J, Chiaruttini G, et al. IgE re-programs alternatively-activated human macrophages towards pro-inflammatory anti-tumoural states. EBioMedicine. 2019;43:67–81. https://doi.org/10.1016/j.ebiom.2019.03.080.
Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. https://doi.org/10.3389/fimmu.2020.583084.
Qiu SQ, Waaijer S, Zwager MC, de Vries E, van der Vegt B, Schröder CP. Tumor-associated macrophages in breast cancer: innocent bystander or important player. Cancer Treat Rev. 2018;70:178–89. https://doi.org/10.1016/j.ctrv.2018.08.010.
Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12(:306. doi:https://doi.org/10.1186/1471-2407-12-306.
Wang C, Cao M, Jiang X, Yao Y, Liu Z, Luo D. Macrophage balance fraction determines the degree of immunosuppression and metastatic ability of breast cancer. Int Immunopharmacol. 2021;97:107682. https://doi.org/10.1016/j.intimp.2021.107682.
Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol. 2014;23(1):37–45. https://doi.org/10.1016/j.intimp.2014.08.002.
Lin L, Chen YS, Yao YD, Chen JQ, Chen JN, Huang SY, et al. CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer. Oncotarget. 2015;6(33):34758–73. https://doi.org/10.18632/oncotarget.5325.
Pan Y, Wang W, Huang S, Ni W, Wei Z, Cao Y, et al. Beta-elemene inhibits breast cancer metastasis through blocking pyruvate kinase M2 dimerization and nuclear translocation. J Cell Mol Med. 2019;23(10):6846–58. https://doi.org/10.1111/jcmm.14568.
Zhu L, Fu X, Chen X, Han X, Dong P. M2 macrophages induce EMT through the TGF-β/Smad2 signaling pathway. Cell Biol Int. 2017;41(9):960–8. https://doi.org/10.1002/cbin.10788.
Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. https://doi.org/10.12703/P6-13.
Dias AS, Almeida CR, Helguero LA, Duarte IF. Metabolic crosstalk in the breast cancer microenvironment. Eur J Cancer. 2019;121:154–71. https://doi.org/10.1016/j.ejca.2019.09.002.
Kutryb-Zajac B, Harasim G, Jedrzejewska A, Krol O, Braczko A, Jablonska P, et al. Macrophage-derived adenosine deaminase 2 correlates with M2 macrophage phenotype in triple negative breast cancer. Int J Mol Sci. 2021. https://doi.org/10.3390/ijms22073764.
Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–96. https://doi.org/10.1038/ni.1937.
Chiang CF, Chao TT, Su YF, Hsu CC, Chien CY, Chiu KC, et al. Metformin-treated cancer cells modulate macrophage polarization through AMPK-NF-κB signaling. Oncotarget. 2017;8(13):20706–18. https://doi.org/10.18632/oncotarget.14982.
Felipe Lima J, Nofech-Mozes S, Bayani J, Bartlett JM. EMT in breast carcinoma—a review. J Clin Med. 2016. https://doi.org/10.3390/jcm5070065.
Weng YS, Tseng HY, Chen YA, Shen PC, Al Haq AT, Chen LM, et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol Cancer. 2019;18(1):42. https://doi.org/10.1186/s12943-019-0988-0.
Williams MM, Christenson JL, Neill KI, Hafeez SA, Ihle CL, et al. MicroRNA-200c restoration reveals a cytokine profile to enhance M1 macrophage polarization in breast cancer. NPJ Breast Cancer. 2021;7(1):64. https://doi.org/10.1038/s41523-021-00273-1.
Biswas S, Mandal G, Roy Chowdhury S, Purohit S, Payne KK, Anadon C, et al. Exosomes produced by mesenchymal stem cells drive differentiation of myeloid cells into immunosuppressive M2-polarized macrophages in breast cancer. J Immunol. 2019;203(12):3447–60. https://doi.org/10.4049/jimmunol.1900692.
Ryan D, Koziol J, ElShamy WM. Targeting AXL and RAGE to prevent geminin overexpression-induced triple-negative breast cancer metastasis. Sci Rep. 2019;9(1):19150. https://doi.org/10.1038/s41598-019-55702-w.
Jia XH, Feng GW, Wang ZL, Du Y, Shen C, Hui H, et al. Activation of mesenchymal stem cells by macrophages promotes tumor progression through immune suppressive effects. Oncotarget. 2016;7(15):20934–44. https://doi.org/10.18632/oncotarget.8064.
Acknowledgements
Thanks to all the peer reviewers and editors for their opinions and suggestions.
Funding
The present study was supported by the Innovative Research Group Project of the National Natural Science Foundation of China (No. 81872055).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Z., Wu, J., Wang, L. et al. Tumor-associated macrophages of the M1/M2 phenotype are involved in the regulation of malignant biological behavior of breast cancer cells through the EMT pathway. Med Oncol 39, 83 (2022). https://doi.org/10.1007/s12032-022-01670-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01670-7