Abstract
Fingolimod is an immunotherapeutic drug approved in certain countries as first-line therapy for relapsing–remitting multiple sclerosis (RRMS). The drug has been shown to alter the expression of several coding and non-coding genes. In the current study, we assessed the expression of miR-506-3p, miR-217, miR-381-3p, miR-1827, miR-449a and miR-655-3p in peripheral blood of patients with RRMS undergoing treatment with fingolimod compared with healthy controls. We also compared the expression of these miRNAs between fingolimod responders and non-responders to determine their relevance with regard to response to fingolimod. Expression of miR-381-3p was significantly higher in responders than in controls (RE difference = 3.903, P = 0.005), while expression of miR-655-3p was significantly lower in both responders and non-responders compared with controls (RE difference = −1.03, P = 0.014; RE difference = −1.41, P < 0.0001, respectively). No difference was found in the expression of other miRNAs between study subgroups. In addition, there was no significant difference in the expression of any miRNA between responders and non-responders. Although there were significant pairwise correlations between expression levels of all of the assessed miRNAs in controls, MS patients exhibited differences in correlation patterns. Expression of miR-381-3p was correlated with age in responders. However, expression of other miRNAs did not correlate with age in any study subgroup. The current study indicates a possible role for miR-655-3p and miR-381-3p in the pathogenesis of MS or possible effects of fingolimod on the expression of these miRNAs. Future studies are needed to verify these results in larger patient populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fingolimod is an immunotherapeutic drug which targets the sphingolipid-regulated signaling system (Brinkmann et al. 2002), and exerts effects on immune cells by binding with the sphingosine 1-P1 receptor. This binding leads to internalization and degradation of the sphingosine 1-P1 receptor and subsequent lymphocyte sequestration in peripheral lymphatic tissues. Thus, autoaggressive T cells cannot invade the central nervous system (CNS) (Chiba et al. 1998). As the first orally administered medicine for treatment of relapsing–remitting multiple sclerosis (RRMS), its efficacy in decreasing the rate of relapse has been proven in multiple clinical trials (Ayzenberg et al. 2016). Fingolimod is approved as a first-line treatment in the USA and certain other countries, and as a second-line treatment in other countries for patients who do not respond to first-line therapeutic approaches (Ayzenberg et al. 2016). This immunotherapeutic drug has been shown to modulate the expression of several coding and non-coding genes. Fiess et al. investigated transcriptomes of CD4+ T cells and found differential expression of 890 genes 3 months after initiation of fingolimod therapy. Notably, numerous microRNA (miRNA) precursors, including those for miR-216b, miR-142 and miR-548c, have been among the altered genes (Friess et al. 2017). Fenoglio et al. reported a slight decrease in serum levels of miR-15b, miR-23a and miR-223 after fingolimod treatment (Fenoglio et al. 2013). Marked changes in serum levels of these microRNAs (miRNAs) were found after 6 months of continuing fingolimod treatment (Fenoglio et al. 2016).
In the current study, we assessed the expression levels of miR-506-3p, miR-217, miR-381-3p, miR-1827, miR-449a and miR-655-3p in peripheral blood of RRMS patients undergoing treatment with fingolimod compared with healthy controls. Although other miRNAs have been shown to affect patient response to fingolimod (Nuzziello et al. 2020), we selected these miRNAs based on their roles in immune response in order to gain greater insight into the relationship between these miRNAs and the response to fingolimod. We also compared the expression of these miRNAs between fingolimod responders and non-responders to determine their relevance with regard to response to fingolimod. These miRNAs have a putative role in the regulation of immune responses, pathobiology of MS or effects of fingolimod. For instance, miRNA-506 can directly bind with its 3′-untranslated region (3’-UTR) of STAT3, thus inhibiting its expression (Su et al. 2019). STAT3 is involved in the process of fingolimod-mediated protection against ischemic white matter damage (Qin et al. 2017). miR-217 and miR-1827 are among the miRNAs found to modulate innate and adaptive immune responses through different mechanisms including modulation of STAT3 expression (Felix et al. 2019). miRNA-381 is a known negative regulator of toll-like receptor 4 (TLR4) signaling (Xu et al. 2015) and suppressor of antigen presentation by dendritic cells (Wen et al. 2016). miR-449a has a role in activation of CD4+ T cells during induction of alloimmune reactions through modulation of metabolic status (Huang et al. 2018). Finally, miR-655-3p is possibly involved in the suppression of immune responses by cyclooxygenase 2 (COX-2) (Majumder et al. 2018).
Materials and Methods
Enrolled individuals
The current investigation was conducted in 78 patients with MS and 79 healthy subjects. MS patients were assessed in Farshchian Hospital, Hamadan, Iran. Poser’s criteria (Fangerau et al. 2004) were used for assessment and diagnosis. All MS patients were being treated with 0.5 mg fingolimod administered orally once daily for at least 6 months. Subsequently, they were recognized as responders if there was no increase in the Expanded Disability Status Scale (EDSS) score, no relapse during the follow-up period (Eftekharian et al. 2019) and no evidence of disease activity based on the MRI data. Occurrence of at least one relapse during this period or a one-point increase in the EDSS score (lasting for 6 months) were regarded as indication of non-responsiveness. In total, 49 patients met the criteria for responsiveness, while 29 patients were regarded as non-responders. Controls were selected from age-matched healthy volunteers who had no history of inflammatory or chronic diseases or malignancy. The study protocol was approved by the local ethics committee of Hamadan University of Medical Sciences (IR.UMSHA.REC.1397.610). Written informed consent forms were obtained from all study participants.
Expression analysis
Three milliliters of peripheral blood were obtained from all enrolled individuals. These samples were used for RNA extraction using the Hybrid-R™ Blood RNA Kit (GeneAll, Seoul, South Korea). First-strand cDNA was synthesized from RNA samples using the TaqMan® MicroRNA Reverse Transcription Kit (ABI, Forest City, CA, USA). Expression of miRNAs was measured using the TaqMan® MicroRNA Assay (ABI, Forest City, CA, USA). The RNU6B gene was used as normalizer. Amplification was carried out in duplicate using the Corbett Rotor-Gene 6000 real-time PCR system.
Statistical analysis
The packages "quantreg" and "brms" in R 3.4.2 software were used for statistical assays. The relative expression (RE) of each miRNA in each sample was calculated using the Ln (Efficiency^-∆CT) formulation. Mean values of miRNA expression were compared between study subgroups using a Bayesian multilevel model using the probabilistic programming language Stan. Two-sided 95% credible intervals (95% CrI) were used to determine the significance of differences. The Spearman rank correlation coefficient was measured to evaluate correlations between miRNA expression and patient age.
Results
Demographic information for MS patients and controls
Demographic information for MS patients and controls is summarized in Table 1.
Relative expression of miRNAs in MS patients and controls
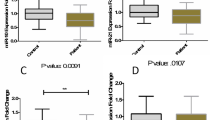
Figure 1 shows the relative expression of miRNAs in MS patients (responders and non-responders) and controls.
The expression of miR-381-3p was significantly higher in responders than in controls (RE difference = 3.903, P = 0.005), while expression of miR-655-3p was significantly lower in both responders and non-responders compared with controls (RE difference = −1.03, P = 0.014; RE difference = −1.41, P < 0.0001, respectively). The expression of other miRNAs did not differ between study subgroups. Table 2 shows the results of Bayesian multilevel modeling for comparison of the RE of miRNAs between MS patients and controls.
There was no significant difference in the expression of any miRNA between total responders and total non-responders. Moreover, no difference was found in the expression of any miRNA when expression of miRNAs in responder cases were compared with sex-matched non-responders (Table 3).
There was no correlation between the expression of any of the miRNAs and the age of study participants (Fig. 2).
Significant pairwise correlations were found between expression levels of all assessed miRNAs in controls, with the most robust correlation being detected between miR-1827 and miR-655-3p.
However, MS patients exhibited differing correlation patterns. Expression of miR-381-3p was correlated with the age of responders, while expression of miR-449a was correlated with the expression of miR-1827, miR-381-3p, miR-217 and miR-655-3p in responders. Expression of miR-655-3p was also correlated with the expression of miR-1827 in these patients (Figure 3).
Scatter matrix depicting correlation between expression of miRNAs in responders. The diagonal presents the distribution of expression values for each miRNA. The left diagonal has a scatterplot of the two variables above it and to its right; the right diagonal shows the correlation coefficient for the same variables, with the font size increasing for higher correlation
No significant correlation was detected between expression of any miRNA and age in non-responders. There were significant correlations between the following miRNA pairs: miR-506-3p/miR-381-3p, miR-381-3p/miR-217, miR-506-3p/miR-449a, miR-217/miR-449-3a, miR-217/miR-655-3p and miR-1827/miR-655-3p (Fig. 4).
Scatter matrix depicting correlation between expression of miRNAs in non-responders. The diagonal shows the distribution of expression values for each miRNA. The left diagonal has a scatterplot of the two variables above it and to its right; the right diagonal shows the correlation coefficient for the same variables, with the font size increasing for higher correlation
Discussion
In the present case–control study, we compared the expression of six immune-response-associated miRNAs between fingolimod-responder MS patients, fingolimod-non-responder MS patients and healthy subjects. We found higher levels of miR-381-3p expression in MS patients who responded to fingolimod compared with healthy controls. However, no difference in expression of this miRNA was found between non-responders and healthy controls. miR-381-3p binds with the 3’-UTR of the CD1c gene, thus suppressing its translation (Wen et al. 2016). CD1c is a transmembrane glycoprotein that participates in the presentation of lipid and glycolipid antigens of self or microbial sources to T cells (Posch and Hurley 2011). A previous study indicated that fingolimod was associated with a reduction in both the proliferation of CD1c+ dendritic cells and the release of proinflammatory cytokines from these cells (Luessi et al. 2015). Thus, the higher expression of miR-381-3p in fingolimod responders than in healthy controls might be associated with lower levels of CD1c in these patients compared with controls. Consequently, it is possible that the effect of fingolimod on CD1c expression is mediated through its effects on the expression of miR-381-3p. Meanwhile, miRNA-381 has been shown to be involved in the downregulation of TLR4 signaling (Xu et al. 2015). Our results are in accordance with the results of Andersson et al., who reported upregulation of TLR4 and its endogenous ligand in the CSF mononuclear cells of MS patients compared with healthy subjects (Andersson et al. 2008). However, Haegert et al. showed an unexpected correlation between increased TLR4 expression and a positive response of MS patients to fingolimod (Haegert et al. 2018). Thus, the correlation between miR-381-3p, TLR4 and response to fingolimod should be assessed in future studies.
We also detected lower levels of miR-655-3p in both responders and non-responders compared with controls. Downregulation of this miRNA has been associated with malignant phenotype in hepatocellular carcinoma and gastric cancer (Zhao et al. 2017; Wu et al. 2016; Alsaadoni et al. 2019). Moreover, this miRNA may be involved in the suppression of immune responses by COX-2 (Majumder et al. 2018). COX is involved in different pathogenic events in the evolution of MS, and COX-2 inhibitors have been effective in reversing these processes in animal models (Palumbo 2017). The downregulation of miR-655-3p observed in MS patients undergoing treatment with fingolimod suggests an effect of the drug on the attenuation of COX-2-mediated neuro-inflammatory processes. However, future functional studies are needed to verify this speculation.
Although there were significant pairwise correlations between expression levels of all assessed miRNAs in controls, MS patients exhibited differing correlation patterns. These differences might be due to the presence of disease-associated mechanisms, or alternatively may reflect the distinct effects of fingolimod in responders and non-responders.
Expression of miR-381-3p was correlated with age in fingolimod responders but not in the other study subgroups. The practical significance of this finding should be assessed in future studies. Expression of other miRNAs was not correlated with age in any study subgroup.
Taken together, the results of the current study indicate possible roles for miR-655-3p and miR-381-3p in the pathogenesis of MS or possible effects of fingolimod on the expression of these miRNAs. Future studies are needed to verify these results in larger patient populations including drug-naïve patients.
Our study has some limitations. First, there were no in vivo or in vitro data to support the meaningful impact of these alterations in the context of MS. Moreover, we did not assess expression of miRNAs in peripheral blood mononuclear cells or individual immune cell subsets. Whole blood miRNA measurements are not functionally meaningful for elucidation of key cellular and immune events that are responsible for driving disease pathology or responsiveness to therapy in MS. Finally, we did not assess expression of other miRNAs that have been reported to influence response to fingolimod.
References
Alsaadoni H, Çaykara B, Pençe S, Pençe HH, Bademler S (2019) The expression levels of miR-655-3p, miR127-5p, miR-369-3p, miR-544a in gastric cancer. Turkish Journal of Biochemistry 44:487–491
Andersson Å, Covacu R, Sunnemark D, Danilov AI, Dal Bianco A, Khademi M, Wallström E, Lobell A, Brundin L, Lassmann H (2008) Pivotal advance: HMGB1 expression in active lesions of human and experimental multiple sclerosis. Journal of Leukocyte Biology 84:1248–1255
Ayzenberg I, Hoepner R, Kleiter I (2016) Fingolimod for multiple sclerosis and emerging indications: appropriate patient selection, safety precautions, and special considerations. Therapeutics and Clinical Risk Management 12:261
Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, Bruns C, Prieschl E, Baumruker T, Hiestand P (2002) The immune modulator FTY720 targets sphingosine 1-phosphate receptors. Journal of Biological Chemistry 277:21453–21457
Chiba K, Yanagawa Y, Masubuchi Y, Kataoka H, Kawaguchi T, Ohtsuki M, Hoshino Y (1998) FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. I. FTY720 selectively decreases the number of circulating mature lymphocytes by acceleration of lymphocyte homing. The Journal of Immunology 160:5037–5044
Eftekharian MM, Komaki A, Mazdeh M, Arsang-Jang S, Taheri M, Ghafouri-Fard S (2019) Expression Profile of Selected MicroRNAs in the Peripheral Blood of Multiple Sclerosis Patients: a Multivariate Statistical Analysis with ROC Curve to Find New Biomarkers for Fingolimod. J Mol Neurosci 68:153–161
Fangerau T, Schimrigk S, Haupts M, Kaeder M, Ahle G, Brune N, Klinkenberg K, Kotterba S, Mohring M, Sindern E (2004) Diagnosis of multiple sclerosis: comparison of the Poser criteria and the new McDonald criteria. Acta Neurologica Scandinavica 109:385–389
Felix TF, Lapa RML, De Carvalho M, Bertoni N, Tokar T, Oliveira RA, Rodrigues MA, Hasimoto CN, Oliveira WK, Pelafsky L (2019) MicroRNA modulated networks of adaptive and innate immune response in pancreatic ductal adenocarcinoma. PloS one 14:e0217421
Fenoglio C, De Riz M, Pietroboni AM, Calvi A, Serpente M, Cioffi SM, Arcaro M, Oldoni E, Scarpini E, Galimberti D (2016) Effect of fingolimod treatment on circulating miR-15b, miR23a and miR-223 levels in patients with multiple sclerosis. Journal of Neuroimmunology 299:81–83
Fenoglio C, Ridolfi E, Cantoni C, De Riz M, Bonsi R, Serpente M, Villa C, Pietroboni AM, NAISMITH RT, Alvarez E (2013) Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Multiple Sclerosis Journal 19:1938–1942
Friess J, Hecker M, Roch L, Koczan D, Fitzner B, Angerer IC, Schröder I, Flechtner K, Thiesen H-J, Winkelmann A (2017) Fingolimod alters the transcriptome profile of circulating CD4+ cells in multiple sclerosis. Scientific Reports 7:42087
Haegert, D., Fitz-Gerald, L., Ghadiri, M., Bar-Or, A. & Antel, J. Analysis of a biomarker signature (TLR2, TLR4 and CCR1) by flow cytometry in patients with relapsing-remitting multiple sclerosis treated with fingolimod. Multiple Sclerosis Journal, 2018. Sage Publications Ltd 1 Olivers Yard, 55 City Road, London Ec1y 1SP, England, 307-308.
Huang, L., Li, F., Fu, Q., Yang, X., Deng, S. & Wei, L. Role of miR-449a in the Activation and Metabolism of CD4+ T Cells. Transplantation proceedings, 2018. Elsevier, 1519-1524.
Luessi F, Kraus S, Trinschek B, Lerch S, Ploen R, Paterka M, Roberg T, Poisa-Beiro L, Klotz L, Wiendl H (2015) FTY720 (fingolimod) treatment tips the balance towards less immunogenic antigen-presenting cells in patients with multiple sclerosis. Multiple Sclerosis Journal 21:1811–1822
Majumder M, Dunn L, Liu L, Hasan A, Vincent K, Brackstone M, Hess D, Lala PK (2018) COX-2 induces oncogenic micro RNA miR655 in human breast cancer. Scientific Reports 8:327
Nuzziello N, Ciaccia L, Liguori M (2020) Precision Medicine in Neurodegenerative Diseases: Some Promising Tips Coming from the microRNAs’ World. Cells 9:75
Palumbo, S. 2017. Pathogenesis and Progression of Multiple Sclerosis: The Role of Arachidonic Acid–Mediated Neuroinflammation.
Posch, P. & Hurley, C. 2011. Histocompatibility: HLA and other systems. Blood and Bone Marrow Pathology. Elsevier.
Qin C, Fan W-H, Liu Q, Shang K, Murugan M, Wu L-J, Wang W, Tian D-S (2017) Fingolimod protects against ischemic white matter damage by modulating microglia toward M2 polarization via STAT3 pathway. Stroke 48:3336–3346
Su Z, Ye X, Shang L (2019) MiR-506 Promotes natural killer cell cytotoxicity against human hepatocellular carcinoma cells by targeting STAT3. Yonsei medical journal 60:22–29
Wen Q, Zhou C, Xiong W, Su J, He J, Zhang S, Du X, Liu S, Wang J, Ma L (2016) MiR-381-3p regulates the antigen-presenting capability of dendritic cells and represses antituberculosis cellular immune responses by targeting CD1c. The Journal of Immunology 197:580–589
Wu G, Zheng K, Xia S, Wang Y, Meng X, Qin X, Cheng Y (2016) MicroRNA-655-3p functions as a tumor suppressor by regulating ADAM10 and β-catenin pathway in Hepatocellular Carcinoma. Journal of Experimental & Clinical Cancer Research 35:89
Xu, Z., Dong, D., Chen, X., Huang, H. & Wen, S. 2015. MicroRNA-381 negatively regulates TLR4 signaling in A549 cells in response to LPS stimulation. BioMed research international, 2015.
Zhao X, Liang B, JIANG K, Zhang H (2017) Down-regulation of miR-655-3p predicts worse clinical outcome in patients suffering from hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 21:748–752
Acknowledgements
The current study was supported by a grant from Hamadan University of Medical Sciences.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mazdeh, M., Kordestani, H., Komaki, A. et al. Assessment of expression profile of microRNAs in multiple sclerosis patients treated with fingolimod. J Mol Neurosci 70, 1274–1281 (2020). https://doi.org/10.1007/s12031-020-01537-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-020-01537-4