Abstract
High mortality rate and poor survival in melanoma are associated with efficient metastatic colonization. The underlying mechanisms remain elusive. Elucidating the role of exosomes in mediating the interactions between cancer cells and the metastatic microenvironment has been focused on cancer cell derived exosomes in modulating the functions of stromal cells. Whether cancer stem cells (CSCs) can modify the metastatic properties of non-CSC cells, and whether exosomal crosstalk plays a role have not been demonstrated prior to this report. In this study, a paired M14 melanoma derivative cell line, i.e., melanoma parental cell (MPC) and its CSC derivative cell line melanoma stem cell (MSC) were employed. We demonstrated that exosomal crosstalk betwen MSCs and non-CSC MPCs is a new mechanism that underlies melanoma metastasis. Low metastatic melanoma cells (MPCs) can acquire the “metastatic power” from highly metastatic melanoma CSCs (MSCs). We illustrated an uncharacterized microRNA, miR-4535 in mediating such exosomal crosstalk. MSCs deliver its exosomal miR-4535 to the targeted MPCs. Upon entering MPCs, miR-4535 augments metastatic colonization of MPCs by inactivating the autophagy pathway.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metastasis is the leading cause of relapse and death [1]. Prognosis of metastatic melanoma is poor and the mean survival is less than 1 year [2]. Efficient metastatic colonization at the distant organ is a feature of metastatic melanoma. However, the underlying mechanisms remain to be fully elucidated.
Cancer stem cells (CSCs) are a group of tumor cells with self-renewal capability and play an important role in tumor metastasis [3]. Current studies suggest that the presence of CSCs is the main reason for the failure of radiotherapy and chemotherapy [4, 5]. The influence of CSCs on tumor progression is multifaceted [6, 7]. Proliferative colonization of the extravasated cancer cells at the distant organ is rate-limiting for metastasis [8,9,10] and requires the formation of a new metastatic microenvironment (MME). Establishment of MME is the result of communication and synergistic action between CSCs, tumor cells and stromal cells [11,12,13,14].
Exosomes emerge as a new class of mediators for such interaction [15,16,17,18]. Exosomes are double-membrane vesicles with particle size between 30 and 140 nm [19, 20]. Exosomes contain a variety of substances, including DNA, proteins, non-coding RNAs, liposomes, mitochondrial DNA, metabolites and so on [21, 22]. Exosomes are heterogeneous in size, content, species specificity, tissue specificity and tumor specificity [23]. Exosomes mediates cell-to-cell communication by transferring biologically active substances from donor cells to the recipient cells. Exosomes from the donor cells can modify the biological functions, or cellular properties of the target cells, and hence may influence the course of cancer initiation, progression or development of resistance to therapies [24, 25]. On the other hand, exosomes may also transport anti-tumor substances, such as microRNAs that have tumor suppressor function [26].
The research on exosomes in cancer progression has gained increasing attention [27, 28]. In the tumor microenvironment, exosomes secreted by cancer cells, stromal cells, and cancer stem cells can remodel the tumor microenvironment. Whether exosomes promote or inhibit tumor progression depends on the source of exosomes and the context of tumor development [29]. Most of the studies are focused on cancer cell-derived exosomes in modulating the functions of stromal cells [30,31,32,33]. Some studies have shown that in the tumor microenvironment, endothelial cells derived from tumors can change the immune capacity of the body and tumor angiogenesis ability through exosomes [34,35,36].There are increasing evidence showing the ability of stromal cell derived exosomes in modifying the behavior of cancer cells. There are few studies that demonstrate the exosomal crosstalk between cancer cells [37,38,39,40]. In melanoma research, the role of exosomal crosstalk in melanoma oncogenesis and progression has been illustrated [41,42,43]. At metastatic colonization stage, whether the CSCs can alter the metastatic properties of non-CSCs through exosomal crosstalk, a new mechanism of metastatic colonization, has not been demonstrated prior to this report.
In this study, a paired M14 melanoma derivative cell line, i.e., MPC and its CSC derivative cell line MSC were employed. We show that exosomal crosstalk between MSCs and non-CSC MPC cells is a new mechanism that underlies melanoma metastasis. Low metastatic melanoma cells (MPCs) can acquire the “metastatic power” from highly metastatic melanoma CSCs (MSCs). We illustrated an uncharacterized microRNA, miR-4535 in mediating such exosomal crosstalk. MSC deliver its exosomal miR-4535 to the targeted MPC cells. Upon entering MPC cells, miR-4535 augments metastatic colonization of MPC by inactivating the autophagy pathway.
Results
Exosomes Secreted by MSCs Can Promote the Invasiveness of the Low-Metastatic MPC Cells In Vitro
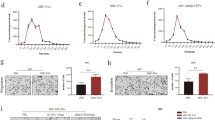
In this study, we used a paired derivative cell line of M14, the MPCs and MSCs that we generated and characterized [44], M14-OL cells exhibit oligometastatic phenotype in vivo upon tail-vein injection, forming limited number of metastatic foci on the mouse lungs(defined as oligometastatic-OL) [45]. Melanoma Stem Cells (OL-SCs) were isolated and purified from OL cells, and the stem cell properties were characterized. Here, for convenience, we define OL cell line as MPC (melanoma parental cell line) and OL-SCs cell line as MSC (melanoma stem cell line). Exosomes were isolated and purified from the culture supernatants of MPC and MSCs cells (Materials and Methods). The quality and the purity of the obtained exosomes were analyzed by: (1) Western blot for the expression of exosomal marker proteins CD81, CD63, Alix and ER marker Calnexin (Fig. 1A); by transmission electron microscopy for visualization of their round or elliptical membranous vesicle morphology (Fig. 1B); by Nanoparticle Tracking Analysis (NTA) for their size range (30 ~ 140 nm, with an average around 100 nm, Fig. 1C).
Exosomes of MSCs can promote migration and invasion of MPCs in vitro. A. Expression of exosome marker proteins CD81, CD63 and Alix, and negative control Calnexin in exosomes of MPCs and MSCs. B. TEM images of exosomes of MPCs and MSCs, the bar is 100 nm. C(i). NTA map of exosomes of MPCs. C(ii). NTA map of exosomes of MSCs. D(i)(ii). Migration assay and quantification of migrated cell numbers of MPCs treated with exosomes of MSC. The scale bar is 64 μm, ** refers to p < 0.01. E(i)(ii). Invasion assay and quantification of invaded cell numbers MPCs treated with exosomes of MSC. The scale bar is 64 μm, ** refers to P < 0.01
At the site of metastasis, whether CSCs can enhance the invasiveness of extravasated non-CSC cancer cells has not been characterized. We thus investigated the effects of MSC-exo on the metastatic function of MPC cells in vitro. MPC cells incubated with 40μg MSC-exo showed significant increases in invasiveness, measured by the migration and invasion assays (Fig. 1D-E) compared to the control cells incubated with PBS (MPC-PBS).
RAB27 family regulates the secretion of exosomes and RAB27a/b knockdown can inhibit the secretion of exosomes [46]. To confirm that the enhancement effects we observed with the addition of MSC-exo were caused by the MSC secreted exosomes, we inhibited exosome secretion function of MSC cells by Rab27a siRNA silencing (Materials and Methods). Effective inhibition of Rab27a expression was confirmed by qRT-PCR (Fig. 2A), WB (Fig. 2B) and NTA analysis (Fig. 2C), respectively. As anticipated, inhibition of Rab27a largely prevented the increase of the number of migratory (Fig. 2D) and invaded (Fig. 2E) MPC cells seen after addition of MSC-exo to MPC cells, respectively. These results show that MSC-exo can enhance the invasiveness of MPC cells in vitro.
The influence of MSC exosomes to MPCs after knockdown of RAB27a. A. The knockdown efficiency of RAB27 measured by QPCR, *** refers to P < 0.001. B. RAB27 knockdown efficiency measured by WB. C(i). NTA map of exosomes in MSC-shNC group. C(ii). NTA map of exosomes in MSC-shRAB27 group. D(i). The migration ability of MPCs co-cultured with exosomes of MSC-shNC and MSC-shRAB27 cells. The scale bar is 64 μm. D(ii). Quantification of migrated cells, *** refers to P < 0.001 and * refers to P < 0.05. E(i). The invasion ability of MPCs co-cultured with exosomes of MSC-shNC and MSC-shRAB27 cells. The scale bar is 64 μm. E(ii). Quantification of invaded cells, ** refers to P < 0.01, * refers to P < 0.05
We hypothesize that “MSC cells can enhance the metastatic capability of MPC cells by transferring biologically active factors to MPC cells via exosomes.”
MSC Exosome miR-4535 Promotes the Metastatic Colonization Ability of MPC Cells In Vitro and In Vivo
Increasing studies have shown that the involvement of exosomal microRNAs can significantly impact tumor metastasis [30, 31, 47,48,49,50]. We thus focused our hypothesis and mechanistic investigation on exosomal miRNAs. We collected the exosomes of MSCs and MPCs for microRNA sequencing and analysis (Materials and Methods, Fig. 3A). We found that eight microRNAs were highly expressed in the exosomes of MSCs (Fig. 3B). We selected seven microRNAs that exhibit |log2(FC)| >2 (FC: fold-change) and P < 0.001 for biological validation (Fig. 3C). We performed RT-qPCR in cells and exosomes of MPCs and MSCs, and found that five microRNAs were consistently and highly expressed in MSCs and their exosomes (Fig. 5D). Among the five microRNAs, miRNA-4535 is novel and its function is unknown. We thus selected miR-4535 for further mechanistic investigation.
Exosomal microRNA 4535 is a novel microRNA that promotes migration and invasion of MPCs. A. Heat map of microRNA sequencing. B. Volcano Plot shows the distribution of differentially expressed microRNAs. C. microRNAs with high expression in exosomes of MSCs. D(i). The expression levels of differentially expressed microRNAs in exosomes of MPCs and MSCs. *** refers to P < 0.001, ** refers to P < 0.01. D(ii). The expression levels of differentially expressed microRNAs in MPCs and MSCs. *** refers to P < 0.001, ** refers to P < 0.01, * refers to p < 0.05. E. Overexpression efficiency of miRNA-4535 by microRNA mimics. *** refers to p < 0.001. F(i). Migration ability of MPCs after miRNA-4535 was overexpressed by microRNA mimics. The scale bar is 153 μm. F(ii). Quantification of migrated cells, ***refers to P < 0.001. G(i). Invasion ability of MPCs after miRNA-4535 was overexpressed by microRNA mimics. The scale bar is 153 μm. G(ii). Quantification of invaded cells, ***refers to P < 0.001. H. Proliferation curve of MPCs after miRNA-4535 was overexpressed by microRNA mimics, p > 0.05
To verify whether miR-4535 expression status affects the invasiveness of MPC, microRNA mimics was used to overexpress miR-4535 in MPCs (Fig. 3E). while mimic treatment significantly enhanced the migration and invasion of MPC cells in vitro (Fig. 3F and G), it had no significant effect on MPC proliferation (Fig. 3H). In order to prove that exosomal miR-4535 can enter the MPC cells, we increased miR-4535 content in the exosomes of MSC cells directly by electro-perforation method (Supplementary Fig. 1A). After co-culturing of miR-4535 overexpressed MSC exosomes with MPC cells, the expression of miR-4535 in MPC cells was increased (Supplementary Fig. 1B), and the migration and invasion ability of MPC cells was enhanced (Supplementary Fig. 1C and 1D). To further prove the specificity of miR-4535 in MSC exosomes in affecting the migration and invasion abilities of MPC cells, we overexpressed miR-4535 in MSC cells with mimics. The level of miR-4535 in MSC exosomes was also increased accordingly (Supplementary Fig. 1E). Upon co-culturing with miR-4535 overexpressing MSC exosomes, the expression of miR-4535 in MPC cells was increased (Supplementary Fig. 1F), and the migration and invasion ability of MPC cells was also enhanced (Supplementary Fig. 1G and 1H). These observations collectively show that MSC-derived miR-4535 can be packaged into MSC-exo and transported to MPC cells. MSC-exo miR-4535 upon entering MPC cells can lead to increased miR-4535 expression and consequently enhanced invasiveness of MPC cells in vitro.
We next inhibited miR-4535 expression in MSCs using miR-4535 specific inhibitor. After confirming the inhibitory efficiency both in MSC cells and MSC exosomes (Fig. 4A and Supplementary Fig. 1I), exosomes of non-specific control (NC) and miR-4535 inhibitor treated MSCs were collected and co-cultured with MPCs for 24 h, respectively. Inhibitor-treated MSC-exo failed to enhance the migration and invasion of MPCs (Figs. 4B-4C). This set of observations indicate that MSC-exo augmentation of MPC invasiveness in vitro requires exosomal miR-4535 transferred to MPCs.
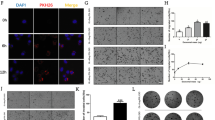
Exosomal microRNA 4535 of MSCs can promote metastasis of melanoma. A. Inhibition efficiency of miRNA-4535 inhibited by microRNA inhibitor, ** refers to p < 0.01. B(i). Migration ability of MPCs after inhibiting the expression of miRNA-4535. The scale bar is 64 μm. B(ii). Quantification of migrated cells, ***refers to P < 0.001. C(i). Invasion ability of MPCs after inhibiting the expression of miRNA-4535. The scale is 64 μm. C(ii). Quantification of migrated cells, **refers to P < 0.01. D. Overexpression efficiency of miRNA-4535 overexpressed using lentivirus, **refers to P < 0.01. E. Co-localization of Cy5 labelled miRNA-4535 and PKH26 labelled exosomes in MPCs at different times. The scale bar is 100 μm. F. Lung metastases after tail vein injection of MPCs with miRNA-4535 overexpressed (MPC-OE-mir-4535) into NOD/SCID mice, the scale bar is 1 cm. G(i). HE staining of metastatic foci of the lungs, scale is 5 mm. G(ii). Quantification of metastasis foci, ** refers to p < 0.01
We next confirmed that miR-4535 can be transferred from MSC cells to MPC cells by MSC-exo. MSCs were transfected with Cy5 labeled miR-4535 mimics [51]. Exosomes were extracted and labelled with PKH26. PKH26-labeled MSC-exo were added to MPC cell culture and confocal microscopy imaging was conducted to visualize the time course (6, 9 and 12 h) of MSC-exo (PKH26-labeled, red) uptake by MPC cells and exosomal miR-4535 (Cy5-labeled, green) release within MPC cells. Shown in Fig. 4E, at 9 h, PKH26-labeled MSC-exo entered MPC cells and accumulated mostly in the cytosol (Fig. 4E, 3rd lane, red). In addition, Cy5-labeled miR-4535 mimics were found inside MPC cells (Fig. 4E, 2nd lane, green). Co-localization of Cy5- miR-4535 mimics with PKH26-MSC-exo (Fig. 4E, 4th lane, yellow) indicates that miRNA-4535 can be transferred from MSC cells to MPC cells via the route of exosomes.
To investigate the effect of miR-4535 overexpression on MPCs metastasis in vivo, MPC cells stably overexpressing miR-4535 were generated via lentiviral infection (Materials and Methods) (Fig. 4D). 5*105 MPC-OE-NC and MPC-OE-miR-4535 cells were injected into NOD/SCID mice via the tail vein (Materials and Methods). All mice were sacrificed at day 30 post tumor cell injection. Since OE-NC and OE-miR-4535 cells were RFP-labeled, macroscopic metastatic foci formed at the lung surface can be visualize under external fluorescence imaging using Sellstrom Z87 fluorescence goggles and an LDP 532 nm bright green flashlight. As shown in Fig. 4F, mice receiving MPC-OE-miR-4535 cells produced significantly more macroscopic lung foci than that of MPC-OE-NC cells (representative n = 3). H&E analysis was conducted and significantly more metastatic foci were present in the lungs that received MPC-OE-miR-4535 cells (Fig. 4G). These observations demonstrate that miR-4535 can enhance metastatic colonization efficiency of MPC cells in vivo.
Collectively, we have demonstrated so far that miR-4535, an uncharacterized miRNA, transferred from MSC-derived exosomes to MPC cells, can effectively augment the clonogenic colonization capability of MPC cells to give arise to more extensive macroscopic metastases. Thus, miR-4535 is oncogenic in the context of melanoma metastatic colonization.
MSC Exosomal miR-4535 Promotes Melanoma Metastasis by Inhibiting the Autophagy Pathway of MPCs
To explore the molecular mechanisms that mediate the oncogenic activities of exosomal miR-4535, TargetScan Human (http://www.targetscan.org/) and BGI Gene Database were analyzed for deriving miR-4535 predicted gene targets and for pathway enrichment of the predicted gene targets. Most of the predicted target genes of miR-4535 were enriched in the “autophagy pathway”, suggesting that miR-4535 may regulate autophagy (Fig. 5A). Our prior study showed that activation of autophagy inhibits melanoma metastasis [52].
MicroRNA 4535 promotes melanoma metastasis via suppressing autophagy of MPCs. A. KEGG Pathway enrichment of miRNA-4535 target genes. B. Accumulation of autophagosomes was detected by immunofluorescence after MPC cells overexpressed miRNA-4535. The bar is 10 μm. C. WB analysis of autophagy marker proteins LC3 and P62 after miRNA-4535 were overexpressed in MPC cells. D(i). Autophagosomes were observed by transmission electron microscopy after miRNA-4535 were overexpressed in MPC cells. The bar of the left picture is 2 μm and of the right picture is 1 μm. D(ii). Statistical difference of autophagosome in D(i). ***refers to P < 0.001. E(i). The migration ability of MPC cells overexpressing miRNA-4535 that were treated with autophagy activator rapamycin. The bar is 64 μm. E(ii). The statistical difference of the number of migrating cells in E(i). ** refers to p < 0.01 and * refers to p < 0,05. F(i). The invasion ability of MPC cells overexpressing miRNA-4535 that were treated with autophagy activator. The bar is 64 μm. F(ii). Statistics of the difference in the number of invasive cells in F(i). ***refers to P < 0.001 and ** refers to p < 0.01. G. WB analysis of autophagy marker proteins LC3 and P62 after miRNA-4535 were overexpressed in MPC cells and the autophagy were activated by rapamycin. H. Accumulation of autophagosomes was detected by immunofluorescence after MPC cells overexpressed miRNA-4535 in which autophagy was activated by rapamycin. The bar is 10 μm
We hypothesized that “miRNA-4535 affects melanoma metastasis by affecting autophagy of MPCs.” To validate this hypothesis, we analyzed autophagy status when miR-4535 expression was altered by gene manipulation. We found that overexpression of miR-4535 by treatment of MPC cells with miR-4535 mimics resulted in the inhibition of autophagic activity in MPC cells, supported by: [1] increased mcherry-LC3 punta by immunofluoresence (Fig. 5B), accumulation of LC3 and P62 by WB (Fig. 5C) [53], and accumulation of autolysosomes by transmission electron microscopy [54] (Fig. 5D).
Our prior study has shown that activation of autophagy in MPC cells hinders MPC cell invasion both in vitro and in vivo [52]. Thus, inhibition of autophagy upon miR-4535 overexpression in MPC cells should result in augmented invasiveness. To confirm that autophagy pathway mediates the pro-metastatic activity of miR-4535 in MPC cells, miR-4535 overexpression was achieved by miR-4535 mimics and the autophagic activity was increased by Rapamycin (RaPa) treatment. Activation of autophagy by RaPa (Fig. 5E-F) in MPC-OE-miR-4535-mimics cells prevented the increase of cell migration and invasion in vitro caused by miR-4535 overexpression (Fig. 5G-H). These observations indicate that MSC exosomal miR-4535, upon taking up by MPC cells, can promote metastatic colonization by targeting the autophagy pathway.
To explore the molecular mechanism underlying miR-4535 regulation of autophagy, we performed bioinformatics analysis and prioritized ATG13 as a potential gene target of miR-4535 (Supplementary Fig. 2A). We conducted double luciferase assay (Supplementary Fig. 2B). The results showed that there was no targeting relationship between miR-4535 and ATG13 (Supplementary Fig. 2C). This result suggests that targeting autophagy pathway by miR-4535 may not be achieved by direct targeting of autophagy genes.
Collectively, these results indicated that melanoma stem cells (MSCs) can enhance the metastatic capability of low metastatic non-CSC MPC cells by transferring of their exosomal miR-4535 to the MPC cells which in turn targets the autophagy pathway.
Discussion
Patients with metastatic melanoma have poor survival and very limited treatment options [55]. Effective management of melanoma requires improved understanding of the molecular mechanisms underlying its metastatic progression. Proliferative colonization is the rate-limiting step of metastasis [8,9,10], that requires the formation of a new microenvironment.
The role of exosomes of tumor cells in the establishment of metastatic microenvironment has gained increasing attention in cancer research. The exosomal crosstalk mediates the transfer of miRNA, mRNA and proteins between the donor cells and the neighboring recipient cells, thus can modify the biological properties of the recipient cells [56]. Few published studies in melanoma research have demonstrated that exosomes can mediate the interactions between melanoma cancer cells and stromal cells, as well as between cancer cells and normal melanocytes, implicating an important role of exosomes in melanoma oncogenesis and progression. However, research on the functional role of exosomes of CSCs is limited in scope and in depth.
Few recent studies show that exosomes of CSCs are regulators of tumor microenvironment, capable of changing the fate of non-CSC target cells [40]. Non-coding RNAs in the exosomes of CSCs can promote metastatic progression in renal cancer [38], lung cancer [57] and in glioma [58]. At the mechanistic level, CSC exosomes can: (1) promote EMT [59] and development of drug resistance [60] of the target non-CSCs; modify the function of immune cells [61,62,63]; promote the stemness of non-CSC tumor cells [64, 65]. However, the effect of CSC exosomes on melanoma metastasis has not been reported prior to this study.
The present study has made the following novel findings that have elucidated a new CSC-exosome-based mechanism that underlies melanoma progression:
First, CSCs Exosomes Can Enhance the Metastatic Colonization Capability of Non-CSC Melanoma Cells
CSCs can alter tumor microenvironment by directly releasing a variety of biologically active material, or indirectly by packaging such material in vesicles such as exosomes. The role of non-coding RNAs in mediating the interactions between CSCs and tumor microenvironment has been reported [66]. Previous research on the role of exosomes in metastasis has been mainly focused on cancer cell exosomes in modifying the functions of stromal cells in the tumor microenvironment [67]. At the stage of metastatic colonization, a rate-limiting stage for metastasis, whether exosomes of CSCs can modify the metastatic properties of the neighboring non-CSCs has not been demonstrated prior to this study.
In-depth mechanistic investigation of melanoma metastasis has been limited by the lack of clinically relevant metastatic melanoma cellular and in vivo models. Here, we used a paired derivative cell line of M14, the MPC and MSC that we generated and characterized, Tail-vein injection of MPC cells produces limited number of metastatic foci on the mouse lungs in vivo (defined as oligometastatic – OL). MSC represents the CSC component of the MPC as we characterized. Exosomal profiling of microRNAs has identified a new molecular feature and entity that can differentiate CSCs and non-CSCs. Using this paired cell lines, we show that melanoma CSCs can promote metastasis by enhancing the metastatic colonization capability of non-CSCs through exosomal transfer.
Second, Exosomal miR-4535, a Novel microRNA from Melanoma CSCs Augments the Invasiveness of Non-CSC cancer Cells
Through profiling, we have identified and prioritized miRNA-4535 for mechanistic investigation. The novelty of this study is that the function of miR-4535 has not been reported prior to this study. Only Yoshikawa et al. reported the potential of using miR-4535 for predicting the probability of fetal infection [68]. Here, we show that miR-4535, transferred from MSC exosomes to MPC cells, can effectively augment the clonogenic colonization capability of MPC cells in vitro and in vivo.
Exosome-based biomarker development has gained increasing attention. Exosomal biomarkers have been identified for various types of cancer, such as breast cancer [69], lung cancer [70], and gastrointestinal cancer [71]. Whether exosomal miR-4535 can serve as a potential prognostic marker for melanoma and for other types of cancer awaits future investigations and validation using clinical samples.
It is reported that CSC exosomes can promote the stemness of parental tumor cells. It has been found that glioblastoma stem cell exosomes can transport Notch1 into parental glioblastoma cells, resulting in the acquiring of stemness of parental glioma cells [64]. Another study has shown that papillary thyroid cancer stem cell exosomes lncRNA DOCK9-AS2 can induce stemness of parental papillary thyroid cancer cells by activating the WNT /β-catenin signaling pathway [65]. Since miR-4535 is an uncharacterized microRNA, we also explored whether melanoma CSCs can modify the stemness features of non-CSCs via exosomal interactions. miR-4535 overexpression in MPC cells failed to increase the stemness of MPC cells, as measured by the 6-well spheroid formation assay and the 96-well single-cell cloning assay (data not shown). This preliminary observation suggests that overexpression of miR-4535 alone is not sufficient to transform MPCs into MSCs. While inhibition of miR-4535 expression in MSC cells using specific inhibitors resulted in significantly decreased spheroid formation efficiency (Supplementary Fig. 3A), single cell cloning efficiency was not significantly changed (Supplementary Fig. 3B). In addition, we examined the expression of stemness genes in MSCs after miRNA-4535 was inhibited. We found that the expression of stemness genes sox2, klf4, bmi1 and aldh1 decreased (Supplementary Fig. 3C). These results indicate that miR-4535 can affect the stemness of MSCs. In-depth and mechanistic understanding on the role of miR-4535 in MSC self-renewal is beyond the scope of this study but merits future investigations.
Third, the Oncogenic Activity of MSC Exosomal miR-4535 Is Achieved through Inhibition of the Autophagy Pathway in MPC Cells
miR-4535 is an uncharacterized microRNA. To investigate the molecular mechanisms underlying the pro-metastatic activity of exosomal miR-4535, we performed bioinformatics analysis of predicted gene targets of miR-4535. Pathway enrichment analysis has identified “autophagy pathway” to be most pronouncedly associated with differential miR-4535 expression. Both the anti- and pro-metastatic roles of autophagy have been reported and appear to be context and stage-dependent [72]. At the colonization stage of metastasis, on the one hand, autophagy prevents proliferation and colonization of the newly extravasated tumor cells by keeping them in the dormancy stage [73]. On other hand, once micro-metastases are established, autophagy promotes macro-metastases by helping tumor cells adapt to the stressful foreign microenvironment [74, 75]. Our previous research shows that activation of autophagy through the “SEC23A-S1008-BECLIN1-autophagy axis” inhibits melanoma metastasis at the step of metastatic colonization [52]. In this study, we show that miR-4535 promotes metastasis by targeting autophagy pathway. However, the results of our experiments shows that the predicted target gene ATG13 did not bind directly to miR-4535. MicroRNA 4535 may affect other upstream signaling pathways which regulate autophagy and thus affect autophagy. The specific mechanism underlying miR-4535 regulation of autophagy requires future investigations.
In conclusion, we show for the first time that exosomal crosstalk between the melanoma cancer stem cells (MSCs) and the melanoma cancer cells (MPCs) are a new mechanism that underlies melanoma metastasis and heterogeneity. Low metastatic melanoma cells (MPC) can acquire the “metastatic power” from highly metastatic melanoma CSCs (MSC). We illustrated an uncharacterized microRNA, miR-4535 in mediating such exosomal crosstalk. MSC deliver its exosomal miR-4535 to the targeted MPC cells. Upon entering MPC cells, miR-4535 augments metastatic colonization of MPC by inactivating the autophagy pathway.
Materials and Methods
Cell Line and Cell Culture
M14 melanoma cell line was kindly provided by Dr. Robert Hoffman (University of California San Diego). As we previously described, M14-OL, a M14 derivative cell line was derived from three-rounds of in vivo passage, isolation and purification, and forms limited number of metastatic foci on the mouse lungs upon orthotopic injection (defined as oligometastatic–OL). In the present study, we define OL cell line as MPC (melanoma parental cell line). Melanoma Stem Cells (MSCs) were isolated and purified from MPC cells as we described. MPC cells were maintained in DMEM high glucose supplemented (Hyclone, America, CODE: SH30285.02) with 10% fetal bovine serum (FBS) (Gibco, America, CODE: 16140071), and 1% penicillin-streptomycin (Hyclone, America, CODE: SV30010). MSC cells were cultured in DMEM/F12-based normal stem-cell media (Hyclone, America, CODE: SH30023.01) supplemented with 2% B27 (Gibco, America, CODE: 17504044) and 1% penicillin-streptomycin.
Exosome Isolation and Characterization
MPC and MSC cells were cultured in 10 cm diameter petri dishes (Thermo Fisher Scientific, America, CODE: 150464). Each dish contains 15 mL culture medium, and 10 dishes were prepared at one time for collecting conditioned culture medium. Culture medium was collected after the cells reached 90%. The collected culture medium was first concentrated using a centrifugal concentrator with an aperture of 100 kDa MWCO (Millipore, America, CODE: UFC9100). Thereafter, exosomes were extracted by ultracentrifugation. Exosomes were extracted by differential and ultracentrifugation. Cells were harvested when MPC and MSC cultures reached 90% confluency. After centrifugation at 800 g for 5 min, the supernatants were collected and centrifuged at 2000 g at 4 °C for 10 min. For differential ultracentrifugation, the supernatants obtained from low-speed centrifugation were centrifuged at 10000 g at 4 ̊C for 10 min and then passed through a 0.22 μm filter (Millipore, America, CODE: MPGL04GH2). Thereafter, the supernatants were centrifuged at 100000 g at 4 ̊C for additional 70 min. The resultant pellets were resuspended in 200-500 μl PBS (Hyclone, America, CODE: SH30256.01) and stored at −80 ̊C for further analysis.
Exosomal protein content was determined by Micro BCA Protein Assay (CWBIO, China, CODE: CW20115). Exosome characterization was conducted by: (1) transmission electron microscopy (TEM, JEM-1400PLUS, JEOL) for morphology; [53] nanoparticle Tracking Analysis (NTA) for size distribution and concentration; [53] Western blot(WB)for the expression of exosomal markers.
Western Blot
Cells lysates were boiled at 100 °C for 5 min. 20 μg of each protein sample was separated by electrophoresis with 12% polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, America, CODE: 1620177). The membranes were incubated with appropriate primary and secondary antibodies according to the manufacturer’s instruction. Primary antibodies of Alix (CODE: 12422–1-AP), CD63 (CODE: 25682–1-AP), CD81 (CODE: 66866–1-Ig), Rab27a (CODE: 17817–1-AP), calnexin (CODE: 10427–1-AP) and the loading controls (GAPDH (CODE: 10494–1-AP) and β-Actin (CODE: 66009–1-Ig)) were purchased from Proteintech Group. LC3 antibody was purchased from Abcam, America, CODE: ab51520. P62 antibody was purchased from Sigma-Aldrich, America, CODE: P0067.
Transwell Migration and Invasion Assays
Transwell inserts (8 μm pore size, BD Falcon, America, CODE: 353097) were used to perform invasion assay with Matrigel (100 μl, 1:10 dilution in serum free medium, BD Biosciences, America, CODE: 356234) and migration assay without Matrigel. In the migration and invasion experiment, 800ul of high glucose DMEM containing 10% FBS was added in the lower chamber while 300ul of high glucose DMEM without FBS was added in the upper chamber. The migration assay used 30,000 cells and 50,000 cells were used in the invasion test. After 24 h culture, Cells were removed from the upper surface of the porous membrane with a cotton swab and cells migrated and invaded to the lower surface of the membrane were fixed with 70% ethanol for 30 min, followed by staining with crystal violet (Solarbio, China, CODE: G1062) for 10 min. The stained cells were counted under light microscopy. 9 random fields from three replicate Transwells were counted. The number of migrated and invaded cells was presented as the number of cells counted per field of the porous membrane.
Quantitative Polymerase Chain Reaction (QPCR)
Total RNA from cells was extracted using Trizol (Invitrogen, America, CODE: 15596026) and Total RNA from exosomes was extracted using miRNeasy Serum/Plasma Kit (QIAGEN, Germany, CODE: 1071073), respectively. Reverse transcription was performed using PrimeScript RT Master Mix (Takara, Japan, CODE: RR036A) and Mir-X™ miRNA First Strand Synthesis kit (Takara, Japan, CODE: 638313). QPCR was performed using a SYBR Green Real-time PCR Master Mix kit (Takara, Japan, CODE: RR420A), with the following program: pre-incubation at 95 °C for 30s, followed by 39 cycles at 95 °C for 5 s and 60 °C for 30s, respectively. The relative mRNA levels were analyzed using the 2 − ΔΔCt method. The relative expression level of miRNAs was calculated through normalization to U6 internal controls, and mRNAs were normalized with GAPDH. The sequences of primers are listed in Table 1.
CCK-8 Cell Proliferation Assay
The proliferation of cells was examined using CCK-8 kit (Solarbio, China, CODE: CA1210). First, 2 × 103 melanoma cells were planted in 96-well plates, The absorbance of each well was measured with an enzyme-linked immunosorbent assay reader at 450 nm. The proliferation curve was drawn after all tests were completed in 5 days.
Animal Experiments
Ten NOD/SCID male mice aged about 8 weeks and weighing about 20 g were used in this animal experiment. The mice were obtained from the Animal Experiment Center of Chongqing Medical University. And the license number of experimental animal is SYXK2018–0003. 200ul of liquid containing 5 × 105 MPC-OE-NC and MPC-OE-miR-4535 cells were injected via the tail vein. The weight of the mice was measured every other day. The mouse was sacrificed after 30 days of tail vein injection. Sellstrom Z87 fluorescence goggles and an LDP 532 nm bright green flashlight were used for the examination of the metastatic foci. Metastatic nodules in the lungs were counted at the time of sacrifice and confirmed by H&E staining.
Lentivirus Infection and Oligonucleotide Transfection
The lentivirus particles of N.C., miR-4535 overexpression and RAB27a/b silencing plasmids were purchased from Shanghai GenePharma Company. Cells were infected with a multiplicity of infection (MOI) of 50 according to the manufacturer’s protocol. miR-4535 mimics or inhibitor and control miRNAs were chemically synthesized by GenePharma. Cells were transfected with the siRNA or miRNA mimics by Lipofectamine 2000 (Thermo Fisher Scientific, America, CODE: 11668500) according to manufacturer’s instructions. RT-qPCR and western blotting assays were performed to confirm the efficiency of silencing or overexpression after 48 h of transfection.
Detection of Cy5-Labelled miR-4535 Transfer
MSCs was transfected with Cy5 labeled miR-4535 mimics, and extracted its exosomes by the above method of separation and purification of exosomes. MSC exosomes were stained with PKH26(Sigma, America, CODE: MINI26-1KT) according to the manufacturer’s instructions. After MPCs were cultured on 24-well plate cell slides for 24 h and respectively co-cultured with stained exosomes for 6, 9 or 12 h. Cells were fixed with ice ethanol for 30 min, washed with PBS and stained the nucleus with DAPI for 10 min. The images were taken with a confocal fluorescence microscope.
Sequencing and Analysis
The sequencing of microRNA and mRNA was completed by BGI, and the bioinformatic analysis was performed by the online analysis system of BGI, Dr.Tom (https://biosys.bgi.com/).
Induction of Autophagy
We used rapamycin, a recognized autophagy inducer, to induce autophagy in MPC cells. Rapamycin was purchased from Solarbio, China, CODE: R8140. MPC cells was treated with Rapamycin for 24 h at the concentration of 20 μM.
Assessment of Autophagy Activity
Autophagy state was analyzed by three assays: first, autophagy flux analysis was performed by measuring LC3B puncta as we previously reported. Adenovirus expressing mCherry-GFP-LC3B fusion protein (Ad-mCherry-GFP-LC3B) was purchased from Beyotime, China, CODE: C3011. Cells were let grow to 50%–70% confluence in 6-well plates at the time of Ad-mCherry-GFP-LC3B transfection. The presence of mCherry-LC3B puncta indicated the autolysosomes in red fluorescent images. Second, WB analysis of P62 and LC3. Third, Transmission electron microscopy analysis of autophagosome [54]. TEM was conducted by JEM- 1400Plus transmission electron microscope (JEOL, Japan).
Data Analysis
All experiments were performed at least three times for statistical analysis. Quantitative results were shown as mean ± SEM (standard error of the mean). Proliferation curves were drawn using Graphpadprism 9, and all statistical analysis were performed by Student’s independent t-test of variance using GraphPad Prism software. *P < 0.05, **P < 0.01 and ***P < 0.001 were considered statistically significant. The numbers of migrated cells and invaded cells were counted by Image J software. Statistics of metastases in animal experiment were also completed by Image J software.
Data Availability
The data used to support the findings of this study are available from every author upon request.
References
Gensbittel, V., et al. (2021). Mechanical adaptability of tumor cells in metastasis. Developmental Cell, 56(2), 164–179.
Schadendorf, D., et al. (2018). Melanoma. The Lancet, 392(10151), 971–984.
Batlle, E., & Clevers, H. (2017). Cancer stem cells revisited. Nature Medicine, 23(10), 1124–1134.
Regan, J. L., et al. (2017). Non-canonical hedgehog signaling is a positive regulator of the WNT pathway and is required for the survival of Colon Cancer stem cells. Cell Reports, 21(10), 2813–2828.
Lee, S. Y., et al. (2017). Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Molecular Cancer, 16(1), 1–25.
Hu, J., et al. (2017). A CD44v(+) subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death & Disease, 8(3), 1–9.
Lawson, D. A., et al. (2015). Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature, 526(7571), 131–135.
Luzzi, K. J., et al. (1998). Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Multistep Nature of Metastatic Inefficiency, 153, 865–873.
Chen, D., et al. (2016). The neuropeptides FLP-2 and PDF-1 act in concert to arouse Caenorhabditis elegans locomotion. Genetics, 204(3), 1151–1159.
Barnes, D. G., et al. (2013). Embedding and publishing interactive, 3-dimensional, scientific figures in portable document format (PDF) files. PLoS One, 8(9), 1559–1564.
Lenos, K. J., et al. (2018). Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nature Cell Biology, 20(10), 1193–1202.
Li, F., et al. (2019). Retinoblastoma inactivation induces a Protumoral microenvironment via enhanced CCL2 secretion. Cancer Research, 79(15), 3903–3915.
Wilson, J. L., et al. (2020). Inverse data-driven modeling and multiomics analysis reveals Phgdh as a metabolic checkpoint of macrophage polarization and proliferation. Cell Reports, 30(5), 1542–1552.
Plaks, V., et al. (2015). The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell, 16(3), 225–238.
Wortzel, I., et al. (2019). Exosome-mediated metastasis: Communication from a distance. Developmental Cell, 49(3), 347–360.
Devhare, P. B., & Ray, R. B. (2018). Extracellular vesicles: Novel mediator for cell to cell communications in liver pathogenesis. Molecular Aspects of Medicine, 60, 115–122.
Rauner, G., et al. (2018). High expression of CD200 and CD200R1 distinguishes stem and progenitor cell populations within mammary repopulating units. Stem Cell Reports, 11(1), 288–302.
Mathieu, M., et al. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nature Cell Biology, 21(1), 9–17.
Bonsergent, E., et al. (2021). Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nature Communications, 12(1), 1–11.
Ratajczak, M. Z., & Ratajczak, J. (2020). Extracellular microvesicles/exosomes: Discovery, disbelief, acceptance, and the future? Leukemia, 34(12), 3126–3135.
Pegtel, D. M., & Gould, S. J. (2019). Exosomes. Annual Review of Biochemistry, 88, 487–514.
Spilak, A., et al. (2021). Implications and pitfalls for cancer diagnostics exploiting extracellular vesicles. Advanced Drug Delivery Reviews, 175, 1–18.
Kalluri, R., & LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. science, 367, 1–15.
Galasso, M., et al. (2012). MicroRNA expression signatures in solid malignancies. The Cancer Journal, 18, 238–243.
Xie, Y., et al. (2019). The role of exosomal noncoding RNAs in cancer. Molecular Cancer, 18(1), 1–10.
Pomatto, M. A. C., et al. (2019). Improved loading of plasma-derived extracellular vesicles to encapsulate antitumor miRNAs. Mol Ther Methods Clin Dev, 13, 133–144.
Sun, Z., et al. (2018). Effect of exosomal miRNA on cancer biology and clinical applications. Molecular Cancer, 17(1), 1–19.
Zhang, L., & Yu, D. (2019). Exosomes in cancer development, metastasis, and immunity. Biochimica Et Biophysica Acta. Reviews on Cancer, 1871(2), 455–468.
Cavallari, C., et al. (2020). Extracellular vesicles in the tumour microenvironment: Eclectic supervisors. International Journal of Molecular Sciences, 21(18), 1–21.
Zeng, Z., et al. (2018). Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nature Communications, 9(1), 1–14.
Fang, T., et al. (2018). Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nature Communications, 9(1), 1–13.
Yu, L., et al. (2021). Exosomes derived from osteogenic tumor activate osteoclast differentiation and concurrently inhibit osteogenesis by transferring COL1A1-targeting miRNA-92a-1-5p. J Extracell Vesicles, 10(3), 1–27.
Zhao, S., et al. (2020). Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. Journal of Hematology & Oncology, 13(1), 1–19.
Lopatina, T., et al. (2020). Targeting IL-3Ralpha on tumor-derived endothelial cells blunts metastatic spread of triple-negative breast cancer via extracellular vesicle reprogramming. Oncogenesis, 9(10), 1–14.
Lopatina, T., et al. (2020). Extracellular vesicles released by tumor endothelial cells spread immunosuppressive and transforming signals through various recipient cells. Frontiers in Cell and Development Biology, 8, 1–14.
Lombardo, G., et al. (2018). IL-3R-alpha blockade inhibits tumor endothelial cell-derived extracellular vesicle (EV)-mediated vessel formation by targeting the beta-catenin pathway. Oncogene, 37(9), 1175–1191.
Yang, B., et al. (2020). High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene, 39(42), 6529–6543.
Wang, L., et al. (2019). CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: Role of remote MiR-19b-3p. Molecular Cancer, 18(1), 1–15.
Wang, Z. F., et al. (2019). Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma. Journal of Experimental & Clinical Cancer Research, 38(1), 1–15.
Lopez de Andres, J., et al. (2020). Cancer stem cell secretome in the tumor microenvironment: A key point for an effective personalized cancer treatment. Journal of Hematology & Oncology, 13(1), 1–22.
Somasundaram, R., & Herlyn, M. (2012). Melanoma exosomes: messengers of metastasis. Nature Medicine, 18(6), 853–854.
Kim, J., et al. (2018). Replication study: Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Elife, 7, 1–17.
Vignard, V., et al. (2020). MicroRNAs in tumor exosomes drive immune escape in melanoma. Cancer Immunology Research, 8(2), 255–267.
Wang, J., et al. (2017). Comparison of tumor biology of two distinct cell sub-populations in lung cancer stem cells. Oncotarget, 8(57), 96852–96864.
Sun, Z., et al. (2018). Sec23a mediates miR-200c augmented oligometastatic to polymetastatic progression. EBioMedicine, 37, 47–55.
Song, L., et al. (2019). KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nature Communications, 10(1), 1–13.
Makarova, J., et al. (2021). Extracellular miRNAs and cell-cell communication: Problems and prospects. Trends in Biochemical Sciences, 46(8), 640–651.
Wu, K., et al. (2021). Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nature Communications, 12(1), 1–18.
Fang, J. H., et al. (2018). Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology, 68(4), 1459–1475.
Liu, C., et al. (2017). MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nature Communications, 8, 1–14.
Barkan, D., et al. (2008). Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Research, 68(15), 6241–6250.
Sun, Z., et al. (2020). S100A8 transported by SEC23A inhibits metastatic colonization via autocrine activation of autophagy. Cell Death & Disease, 11(8), 1–13.
Ono, M., et al. (2014). Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Science Signaling, 7(332), 1–10.
Temoche-Diaz, M. M., et al. (2019). Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife, 8, 1–34.
Grzywa, T. M., et al. (2017). Intratumor and Intertumor heterogeneity in melanoma. Translational Oncology, 10(6), 956–975.
Zhang, X., et al. (2015). Exosomes in cancer: Small particle, big player. Journal of Hematology & Oncology, 8, 1–13.
Wang, L., et al. (2020). Lung CSC-derived exosomal miR-210-3p contributes to a pro-metastatic phenotype in lung cancer by targeting FGFRL1. Journal of Cellular and Molecular Medicine, 24(11), 6324–6339.
Li, J., et al. (2021). Hypoxic glioma stem cell-derived exosomes containing Linc01060 promote progression of glioma by regulating the MZF1/c-Myc/HIF1alpha Axis. Cancer Research, 81(1), 114–128.
Hardin, H., et al. (2018). Thyroid cancer stem-like cell exosomes: Regulation of EMT via transfer of lncRNAs. Laboratory Investigation, 98(9), 1133–1142.
Yang, Z., et al. (2020). Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cellular Oncology (Dordrecht), 43(1), 123–136.
Hwang, W. L., et al. (2019). Tumor stem-like cell-derived exosomal RNAs prime neutrophils for facilitating tumorigenesis of colon cancer. Journal of Hematology & Oncology, 12(1), 1–17.
Naseri, M., et al. (2021). Dendritic cells loaded with exosomes derived from cancer stem cell-enriched spheroids as a potential immunotherapeutic option. Journal of Cellular and Molecular Medicine, 25(7), 3312–3326.
Yuan, Y., et al. (2021). Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8(+) T cells. Cancer Letters, 500, 98–106.
Sun, Z., et al. (2020). Glioblastoma stem cell-derived exosomes enhance Stemness and Tumorigenicity of glioma cells by transferring Notch1 protein. Cellular and Molecular Neurobiology, 40(5), 767–784.
Dai, W., et al. (2020). Exosomal lncRNA DOCK9-AS2 derived from cancer stem cell-like cells activated Wnt/beta-catenin pathway to aggravate stemness, proliferation, migration, and invasion in papillary thyroid carcinoma. Cell Death & Disease, 11(9), 1–17.
Cha, S. Y., et al. (2017). Clinical impact of microRNAs associated with Cancer stem cells as a prognostic factor in ovarian carcinoma. Journal of Cancer, 8(17), 3538–3547.
Conigliaro, A., et al. (2015). CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Molecular Cancer, 14, 1–11.
Yoshikawa, K., et al. (2021). Diagnostic predictability of miR-4535 and miR-1915-5p expression in amniotic fluid for foetal morbidity of infection. Placenta, 114, 68–75.
Hannafon, B. N., et al. (2016). Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Research, 18(1), 1–14.
Hydbring, P., et al. (2018). Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer, 124, 45–52.
Lee, C. H., et al. (2018). Discovery of a diagnostic biomarker for colon cancer through proteomic profiling of small extracellular vesicles. BMC Cancer, 18(1), 1–11.
Kenific, C. M., et al. (2010). Autophagy and metastasis: Another double-edged sword. Current Opinion in Cell Biology, 22(2), 241–245.
Lu, Z., et al. (2008). The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. The Journal of Clinical Investigation, 118(12), 3917–3929.
Mowers, E. E., et al. (2017). Autophagy in cancer metastasis. Oncogene, 36(12), 1619–1630.
Su, Z., et al. (2015). Apoptosis, autophagy, necroptosis, and cancer metastasis. Molecular Cancer, 14, 1–14.
Acknowledgments
I would like to thank all the students in the lab for their help and the members of the research group for their efforts. I would also like to thank my advisors for their guidance. In addition, thanks for the funding supports of the National Natural Science Fund (Grant No. 82073277 and 82173247), the Science and Technology Project Affiliated to the Education Department of Chongqing (Grant No. KJQN202100404), Natural Science Fund of Chongqing (Grant No. cstc2019jcyj-msxmX0868) and Science and Technology Project of Chongqing Yuzhong District (Grant No. 20200110).
Funding
This work was supported by the National Natural Science Fund (Grant No. 82073277 and 82173247), the Science and Technology Project Affiliated to the Education Department of Chongqing (Grant No. KJQN202100404), Natural Science Fund of Chongqing (Grant No. cstc2019jcyj-msxmX0868) and Science and Technology Project of Chongqing Yuzhong District (Grant No. 20200110).
Author information
Authors and Affiliations
Contributions
Doudou Liu and Xiaoshuang Li completed the molecular experiment, Bin Zeng and Qiting Zhao completed the animal experiment, Hao Chen was responsible for exosome extraction, Yuhan Zhang and Yuting Chen completed the analysis of sequencing data. Doudou Liu wrote the article, Jianyu Wang and H. Rosie Xing were responsible for the overall design of this study and the revision and improvement of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Ethical Approval
All animal work was conducted in accordance with an approved protocol and carried out in accordance with the institutional animal welfare guidelines of the Chongqing Medical University. The license number of experimental animal is SYXK2018–0003.
Consent to Participate
All authors agreed to participate in the study of the subject.
Consent for Publication
All authors agree to revise the paper for publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Supplementary Figure 1
A. Efficiency of transfection of miR-4535 mimics into MSC exosomes by electroporation, **refers to p < 0.01. B. Expression of miR-4535 in MPC cells was increased after MPC cells co-cultured with MSC exosomes that overexpressed miR-4535, *refers to p < 0.05. C-D. Overexpression of miR-4535 in MSC exosomes promoted the migration (C) and invasion (D) of MPC cells, the bar is 210 μm, **refers to p < 0.01. E. The level of miR-4535 in MSC exosomes was increased when miR-4535 was overexpressed in MSC cells, ***refers to p < 0.001. F. After overexpression of miR-4535 in MSC cells, MSC exosomes were co-cultured with MPC, and the expression of miR-4535 in MPC was also increased, *refers to p < 0.05. G-H. After miR-4535 overexpression in MSC cells, MSC exosomes promoted the migration (G) and invasion (H) of MPC cells, the bar is 210 μm, ***refers to p < 0.001. I. After inhibiting the expression of miR-4535 in MSC cells with the inhibitor, the level of miR-4535 in exosomes was also decreased, ***refers to p < 0.001. (PNG 2269 kb)

Supplementary Figure 2
A. Predicted binding site and score of miRNA-4535 and ATG13 in TargetScan. B. Predicted binding sites and mutation sites of miRNA-4535 and ATG13. C. Relative luciferase activities were analyzed in 293 T cells co-transfected with ATG13 3 ′ -UTR (wild type or mutant) reporter plasmid, and microRNA 4535 mimic or NC mimics. (PNG 475 kb)

Supplementary Figure 3
A. Assessment of the stemness of MSC cells by the spheroid formation ability of 1000 cells in a 6-well plate. The scale bar is 300 μm. B. Assessment of the stemness of MSC cells by single cell cloning rate in 96-well plates after the expression of miRNA 4535 was inhibited in MSC cells. C. The expression of stemness genes after the expression of miRNA 4535 was inhibited in MSC cells. ***refers to P < 0.001 and **refers to p < 0.01. (PNG 1728 kb)
Rights and permissions
About this article
Cite this article
Liu, D., Li, X., Zeng, B. et al. Exosomal microRNA-4535 of Melanoma Stem Cells Promotes Metastasis by Inhibiting Autophagy Pathway. Stem Cell Rev and Rep 19, 155–169 (2023). https://doi.org/10.1007/s12015-022-10358-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-022-10358-4