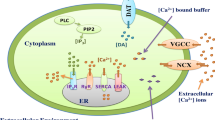
Abstract
The bioenergetic system of calcium ([Ca2+]), inositol 1, 4, 5-trisphophate (IP3) and nitric oxide (NO) regulate the diverse mechanisms in neurons. The dysregulation in any or all of the calcium, IP3 and nitric oxide dynamics may cause neurotoxicity and cell death. Few studies are noted in the literature on the interactions of two systems like [Ca2+] with IP3 and [Ca2+] with nitric oxide in neuron cells, which gives limited insights into regulatory and dysregulatory processes in neuron cells. But, no study is available on the cross talk in dynamics of three systems [Ca2+], IP3 and NO in neurons. Thus, the cross talk in the system dynamics of [Ca2+], IP3 and NO regulation processes in neurons have been studied using mathematical model. The two-way feedback process between [Ca2+] and IP3 and two-way feedback process between [Ca2+] and NO through cyclic guanosine monophosphate (cGMP) with plasmalemmal [Ca2+]-ATPase (PMCA) have been incorporated in the proposed model. This coupling handles the indirect two-way feedback process between IP3 and nitric oxide in neuronal cells automatically. The numerical outcomes were acquired by employing the finite element method (FEM) with the Crank-Nicholson scheme (CNS). The present model incorporating the sodium-calcium exchanger (NCX) and voltage-gated calcium channel (VGCC) provides novel insights into the various regulatory and dysregulatory processes due to buffer, IP3-receptor, ryanodine receptor, cGMP kinetics through PMCA channel, etc. and their impacts on the interactive spatiotemporal system dynamics of [Ca2+], IP3 and NO in neurons. It is concluded that the behavior of different crucial mechanisms is quite different for interactions of two systems of [Ca2+] and NO and the interactions of three systems of [Ca2+], IP3 and nitric oxide in neuronal cell due to mutual regulatory adjustments. The association of several neurological disorders with the alterations in calcium, IP3 and NO has been explored in neurons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Various agents have emerged as indispensable signal carriers for the proper functioning of cellular life. Several crucial mechanisms including transport of different ions and molecules, buffer approximation, different inflow and outflow processes, etc. constitute the bioenergetic dynamical systems for several signaling ions and molecules like calcium, IP3 and NO in neuronal cells. The interactions of calcium with other signaling molecules like IP3 and nitric oxide regulate a variety of cellular mechanisms. The IP3 mobilizes the intracellular calcium ions and controls numerous processes including metabolism, secretion, etc. in cells. The NO with several secondary messengers including [Ca2+], proteins, O2, etc. influence the regulatory mechanisms in different cells. Nitric oxide is produced from L-arginine via NO enzymes synthase [1] and serves as a biological messenger, cytotoxic agent and regulator in a variety of central nervous system tissues.
The association of [Ca2+] buffer and the reduction in free calcium during transient calcium entry was reported in neuronal cells [2]. The different elementary [Ca2+] signaling events through several channels like IP3-receptor (IP3R), ryanodine receptor (RyR), etc. were explored for intracellular [Ca2+] signaling [3]. The IP3-induced [Ca2+] fluctuations have been influenced by [Ca2+] buffering mechanisms [4], and this buffer approximation was validated close to the source location of calcium ions in cells [5]. Also, the behavior of [Ca2+]-induced calcium waves along with spatial dimension has been analyzed in excitable systems [6]. The higher concentration of slow buffer leads to fluctuations in calcium signaling [7], which is crucial to generate information of neuronal encoding and processing [8]. The alterations in neuronal calcium signaling contribute in different neurological disorders including Alzheimer’s [9]. Several research workers have investigated the [Ca2+] regulation in numerous cells including myocytes [10,11,12], fibroblast [13,14,15], astrocytes [16, 17], T-lymphocyte [18, 19], neurons [20,21,22,23,24,25,26], hepatocyte [27,28,29], acinar cells [30, 31], β-cell [32] and Oocytes [33, 34], etc. utilizing analytical and numerical methods in recent years. Incorporating biophysical components of [Ca2+] signaling like calcium channel, sodium channel, potassium channel, etc. the study on the behavior of [Ca2+] dynamics has been conducted in neurons [21], which was extended by utilizing the modified Bessel function for a one-dimensional cylindrical-shaped neurons [23]. The analytical work on the reaction-diffusion model of [Ca2+] dynamics was explored by employing Laplace and Fourier transformation with Atangana–Baleanu–Caputo derivatives in neuronal cells [35]. The neuronal calcium dynamics in association with NCX, endoplasmic reticulum (ER), buffer, VGCC, etc. were discussed concerning Alzheimer’s disorder [36].
The association of hydrolysis of PIP2 and an elevation in intracellular [Ca2+] concentration has been noticed in human cells [37]. The different cellular processes like learning, memory, fertilization, cell development, etc. are regulated by the IP3-induced calcium release through IP3-receptor [38, 39]. The foundation for intricate calcium regulatory patterns is provided by the intracellular [Ca2+] release from ER via RyR and IP3R in cells [40]. The messenger action ranges of calcium and IP3 signaling mechanisms was examined by the measurements of their diffusion coefficients in cells [41]. To describe the calcium activation and inhibition of IP3R in ER, the nine variables kinetic model has been constructed [42], which was minimized into two variables kinetic system [43]. The role of [Ca2+] signaling and IP3 concentration in the chaotic oscillations was reported in bursting neuron cells [44]. The study on the association of the elevation in IP3-induced calcium release and Alzheimer’s disorder was explored in neuronal cells [45]. The interdependent calcium and IP3 dynamics has been studied in diverse cells including myocytes [11, 12], hepatocyte [46], etc. In neurons, Few studies are available on the interdependence of [Ca2+] and IP3 signaling systems regulating the NO, β-amyloid and ATP formations [47,48,49].
The [Ca2+]-dependent and [Ca2+]-independent NO generation and their biological importance were discussed in neuronal cells [50]. The neuronal NO formation is simulated via [Ca2+] influx and induced by a glutamate receptor [51, 52]. The neuronal nitric oxide generation is regulated by different calcium channels including VGCC [53], R-type [Ca2+] channel [54], [Ca2+]-dependent K+ channel [55], etc. NO exhibits cytoprotective properties at low concentrations and cytotoxic effects at high concentration levels in neurons [56]. To maintain adequate nitric oxide concentration and prevent the neurons from toxicity, NO needs tight regulation by calcium signaling in cells [57]. The formation of potent vasodilators including NO controls the high concentration of calcium in neurons [58]. The elevated [Ca2+] concentration is reported during Ischemia [59], which may produce high amounts of nitric oxide in cells [60]. A computational model for synaptic activities associated with the variations in cerebral blood volume was explored in neuron cells [61]. The elevated NO levels cause an elevation in cGMP concentration, which further causes reduction in the cytosolic [Ca2+] levels through PMCA channel, and these reduced calcium levels lead to the decrease in the [Ca2+]-dependent NO generation in cells [62, 63]. The interaction of neuronal calcium signaling with nitric oxide dynamics was reported incorporating one-way feedback process between [Ca2+] and NO in neuronal cells with different neuronal illnesses like Parkinson’s, etc. [64]. The regulation of ATP via the mechanisms of the interdependent calcium and IP3 systems has been reported in fibroblast cells [65].
The neuron cell forms a complex media involving reaction-diffusion of bioenergetic dynamical systems of calcium, IP3 and NO, etc. The study of independent dynamical systems of calcium, IP3 and nitric oxide signaling offer very limited information about cellular functioning of neuronal cells. Few studies are reported on the cooperation of two system dynamics namely calcium dynamics with either IP3 or nitric oxide in nerve cells that cast light on the mutual regulation and dysregulation of different cellular activities. The IP3 concentration was taken as constant in the past to study mutual regulation of calcium and NO in neuronal cells [64]. But in fact the dynamics of IP3 also influences the calcium signaling in neuronal cells and thus have consequent impacts on NO dynamics. No research is noted on the spatiotemporal cross talk of three systems namely [Ca2+], IP3, and NO in neurons. Here, a new model of system dynamics of calcium, IP3 and NO is proposed to explore the insights of their mutual spatiotemporal regulation in neuronal cells. Therefore, the study of interdependent dynamics of calcium, IP3 and NO will give insights into the impacts of feedback control mechanics on their mutual regulation. The two-way feedback process between calcium and IP3 systems, and two-way feedback between [Ca2+] and NO through NO/cGMP pathways and PMCA channel were incorporated in the proposed model. A FEM with the Crank-Nicholson technique is utilized to obtain the numerical findings of interactive systems dynamics of [Ca2+], IP3 and NO in the presence of NCX and VGCC channels, and the effects of different parameters including buffer, IP3-receptor, ryanodine channel, NO/cGMP kinetics with PMCA channel, etc. on the cross talk of [Ca2+], IP3, and nitric oxide has been analyzed in neuron cells. The crucial information about the behavior of different mechanisms under the interactions of three dynamical systems of [Ca2+], IP3 and NO has been explored in neuronal cells.
Mathematical Formulation
Incorporating buffer concentration, VGCC, NCX, RyR and PMCA channel in the Wagner et al. [66] model, the [Ca2+] regulation with IP3 in neuron cells can be depicted as,
Here, [Ca2+]∞ and [B]∞ are respectively representing the steady state concentration levels of [Ca2+] and buffer. DCa and K+ are depicting the [Ca2+] diffusion coefficient and buffer association rate respectively. t and x respectively depict the time and distance parameters.
In Eq. (1), the expressions of the distinct inflow and outflow terms are shown as follows [66]:
Where, fluxes for IP3R, RyR, SERCA Pump and Leak are denoted respectively by JIPR, JRyR, JSERCA and JLEAK. Also, VRyR, VLEAK and VIPR are representing sequentially the flux rates for RyR, Leak and IP3R. KSERCA and VSERCA represent the Michaelis constant and flux rate concerning SERCA pump.
The variable m and h from Eq. (2) are expressed as:
Here, KIP3, KAc and Kinh are denoting correspondingly the dissociation parameters of binding site of IP3 and [Ca2+] activations and [Ca2+] inhibition.
JVGCC represents the VGCC flux and framed by employing the following Goldmann-Hodgkin-Kartz (GHK) current equations [67],
Where, the intracellular and extracellular [Ca2+] levels are expressed sequentially by [Ca2+]i and [Ca2+]0. PCa and ZCa respectively demonstrate the permeability and valency of [Ca2+]. The Faradays constant and membrane potential are correspondingly represented by F and Vm. The absolute temperature and the real gas constant are sequentially depicted by T and R. The following equation converts Eq. (8) into molar/sec,
The GHK equation for current yields the current density as a voltage function. The NCX involves the exchange of one [Ca2+] ion for three [Na+] ions and regulates the calcium concentrations as depicted below [68,69,70]:
Where, Nai and Na0 are correspondingly expressing the intracellular and extracellular Na+ levels.
The IP3 dynamics with [Ca2+] proposed by Wagner et al. [66] is used and the IP3 regulation with [Ca2+] can be depicted as,
Here, Di represents the IP3 transport coefficient. The [Ca2+]-dependent IP3 generation flux can be denoted as [66],
Where, the IP3 production flux is expressed by JProduction. The Michaelis constant for [Ca2+] activation and IP3 formation rate are respectively KProduction and VProduction.
The IP3 elimination fluxes by JKinase and Jphosphatase as depicted below [71],
Where, V1 and V2 are the rate constants concerning low and high [Ca2+] (3-kinase) and Vph is a rate constant for phosphatase. λ is an controllable variable [66] used for the estimation of elimination rate. The expression for [Ca2+]ER is depicted as follows,
The nitric oxide dynamics with [Ca2+] is reported by Gibson et al. [61] and expressed below as,
Where, the nitric oxide diffusion coefficient is depicted by DNO. The NO formation flux is given by as follows [61]:
Where, VNO and KNO are the rate constants.
Where, the NO degradation flux and NO degradation rate constant are respectively depicted by Jdegradation and Kdeg.
The NO/cGMP kinetics is expressed below as follows [72]:
Where, the flux of cGMP generation is depicted as a function of NO distribution and g0, g1, a0, a1, VcGMP, XcGMP and KcGMP are constant and their values are acquired in the model by fitting NO/cGMP pathways to experiment data [72].
The expression of plasmalemmal [Ca2+]-ATPase (PMCA) in the presence of cGMP can be represented as follows:
Here, \({{\rm{V}}}_{{\rm{cGMP}}}^{{\rm{PMCA}}}\,{\rm{and}}\,{{\rm{K}}}_{{\rm{cGMP}}}^{{\rm{PMCA}}}\) are respectively the cGMP levels at half-activation of PMCA, maximum current in cGMP concentration’s absence.
Initial conditions
The reported initial conditions for [Ca2+] [73], IP3 [74], and NO and cGMP [75] are sequentially depicted as follows:
Boundary conditions
For [Ca2+], the reported boundary conditions as expressed below [73],
Where, σ is calcium source influx term.
At opposite end from the source site, the [Ca2+] reaches concentration levels of 0.1 µm [73].
The following are the derivations of the boundary concentrations for IP3 regulation [74],
The deduced boundary conditions for NO regulation are depicted as follows [76]:
Appendix illustrates the description of the FEM with the Crank-Nicholson scheme, which is employed to get the solution of system of equations with initial boundary conditions.
Results and discussion
The numeric amounts and units for a variety of parameters are represented in Table 1.
To validate the proper functioning of the present model, the basic findings of the neuronal spatiotemporal calcium and IP3 distribution at different times and locations in the presence of NCX and VGCC are exhibited in Fig. 1, that were also reported by previous researchers in the absence of NCX and VGCC for interactions between two systems of calcium and IP3 in neuronal cells. It is noted that the [Ca2+] concentration falls spatially from the source to another side of neurons due to the removal of cytosolic [Ca2+] to ER by the SERCA pump, [Ca2+] binding by the buffer and transport of calcium from location of the source to another end of cells as illustrated in Fig. (1A). The [Ca2+] source inflow provides notable amounts of [Ca2+] ions, which causes the increase in the cytosolic [Ca2+] levels with time growth at different positions as depicted in Fig. (1B). The nonlinear behavior of spatial [Ca2+] distribution tends towards linearity with time because the [Ca2+] control mechanism balances different regulatory processes in neurons. Also, the spatial IP3 levels decrease with locations from boundary x = 0 µm upto 5 µm due to the diffusion of IP3 and different IP3 degradation fluxes in neurons. The IP3 control mechanism attempts to maintain a balance between disturbances induced by IP3-elevating mechanisms and IP3-reducing mechanisms with time thereby resulting the nonlinear behavior of IP3 distribution tends towards linearity with increase in time as exhibited in Fig. (1 C & 1D). The temporal IP3 levels rise over time growth and accomplish the steady state at several locations as depicted in Fig. (1D). Thus, the diffusion process, degradation fluxes and other mechanisms regulate the calcium and IP3 distribution in neuronal cells.
Also, Fig. 2 depicts the basic outcomes of the spatial NO distribution in neuronal cells for t = 0.02, 0.05, 0.1 and 0.5 seconds in the presence of NCX and VGCC. With the distant site from the source (0 µm), the nitric oxide concentration decreases for different time instants due to the effects of NO diffusion and NO degradation in neuron cells. With passage of time, the nitric oxide concentration also increases as exhibited in Fig. 2. Thus, several NO-associated mechanisms affect the nitric oxide distribution in neuron cells.
Figure 3 exhibits the novel findings regarding the impacts of higher buffer amounts on the [Ca2+] concentration and IP3 and NO generation fluxes in the presence of NCX and VGCC are illustrated in Fig. 3 at x = 0 µm for the cooperation of three dynamical systems of [Ca2+], IP3 and nitric oxide in neuronal cells. The higher amounts of buffer lead to a higher reduction in the cytosolic [Ca2+] levels by fixing additional [Ca2+] ions in cells, while other [Ca2+]-elevating mechanisms try to increase [Ca2+] levels in the cell. The mismatches among calcium-elevating mechanisms and buffering process cause disturbances in the form of fluctuations in neuron cells. However, the [Ca2+] control mechanism maintains equilibrium among these processes with time and the oscillations in the calcium concentration reduce and achieve a steady state in cells as exhibited in Fig. (3A). The effects of disturbances in calcium concentration also cause disturbances in the [Ca2+]-dependent IP3 and NO formation fluxes in the form of oscillations and these fluxes achieve the steady state with time as depicted in Fig. (3B, C) in neurons. Thus, the dysregulation in the calcium-associated mechanism may be responsible for the disturbances in neuronal [Ca2+], IP3 and NO levels.
The consequences of NO/cGMP pathways on the cytosolic calcium concentration through the plasmalemmal [Ca2+]-ATPase (PMCA) are illustrated as novel results in Fig. 4 at 2.0 sec and 0 µm in neuronal cells. When the PMCA channel is active, the spatiotemporal [Ca2+] levels are notable decreases in neuronal cells. Since, the NO-induced cGMP activation significantly lowers the cytosolic [Ca2+] concentration and elevating the [Ca2+] efflux into the extracellular environment by utilizing the PMCA channels in neuronal cells. Also, the decrease in the neuronal [Ca2+] concentration due to the presence of PMCA channel leads to the reduction in the spatiotemporal [Ca2+]-dependent IP3 and NO productions in neuronal cells as exhibited in Fig. 5 at different times and positions. It is noted that the IP3-induced calcium release via IP3R elevates the cytosolic calcium concentration, resulting the NO production and NO concentration levels also elevates in neuronal cells. In the two-way feedback mechanism between calcium and nitric oxide exhibits that the enhancement in the NO concentration leads to the elevation in cGMP levels, which further lowers the cytosolic calcium concentration via PMCA channel, and these reduced calcium concentration causes decrease in the [Ca2+]-dependent NO production in neuron cells. Thus, one can conclude that the NO signaling may reduce the elevated NO levels by decreasing [Ca2+] concentration in neuronal cells by using cGMP and PMCA mechanisms.
Figure 6 shows the novel and interesting insights into the behavior of different cellular mechanisms like IP3-receptor, which has been significantly differed for the cooperation of three dynamical systems of calcium, IP3 and NO as compared to the cooperation of two dynamical systems of [Ca2+] and NO in neuronal cells. Figure 6A and C represent results for interactions of three systems of calcium, IP3 and NO in neuronal cells. Figure 6B and D represent results for interactions of two systems of calcium and NO in neuronal cells as IP3 is kept constant. The elevated spatiotemporal calcium levels are noted in the consideration of the IP3R as it causes the IP3-induced [Ca2+] release to the cytosol from the ER, resulting in the [Ca2+] levels elevation in neurons. In the absence of the IP3R, the spatiotemporal [Ca2+] level decreases in the cell. The different IP3R states cause higher changes in calcium levels near the center of cells. It is noted in Fig. 6 that when the interactions of three systems namely [Ca2+], IP3 and NO are considered, the IP3-receptor causes significant changes in calcium concentration levels. But, when the interactions of two systems namely [Ca2+] and NO are considered in neurons, the effect of the IP3R decreases on [Ca2+] concentration since IP3 dynamics is required for proper activation of the channel in neurons.
For different IP3R states, the cooperation of three systems of [Ca2+], IP3 and NO bring significant changes in the IP3 and NO formations as compared to the interactions of two systems of calcium and NO in neuronal cells. The novel outcomes about the influences of IP3R on [Ca2+]-associated spatial IP3 and NO formations in the presence of NCX and VGCC in neuron cells are depicted in Fig. 7 at 2.5 sec. Figure 7A and C represent results for interactions of three systems of calcium, IP3 and NO in neurons. Figure 7B and D represent results for interactions of two systems of calcium and NO in neurons as IP3 is kept constant. The [Ca2+] levels increase when IP3R is in an active state as illustrated in Fig. 5, similarly the [Ca2+]-associated spatial IP3 and NO formation fluxes also elevate in the presence of IP3R and decrease in the absence of IP3R in neuron cells as illustrated in Fig. 7. The variations in the spatial IP3 and NO formation fluxes in Fig. 7 are higher near the center of the cell for active and inactive IP3R states. The interaction of dynamic IP3 with systems of calcium and NO activates the IP3-receptor channel for regulating calcium and [Ca2+]-dependent activities like IP3 and NO production at appropriate levels in neuron cells. When the IP3 is not considered dynamic, the IP3R effects on the calcium and [Ca2+]-dependent processes are not significant in the cell as exhibited in Fig. 7. Thus, the IP3R regulates the IP3 and NO formation through [Ca2+] signaling in neurons.
The neuronal IP3 levels are regulated by different IP3R states in neurons as depicted in Fig. 8 at various times and locations. For different IP3R states, the spatial difference curves of IP3 concentration initially increase to the center of the cell and then start decreasing to the other end of neurons concerning various time instants as illustrated in Fig. 8A. The temporal difference curves of IP3 for different IP3R states increase over time growth and accomplish equilibrium at distinct locations in neuron cells. Thus, the different IP3-receptor states notably increase or decrease the IP3 levels in neuron cells.
The neuronal nitric oxide concentration depends on different IP3R receptor states as depicted in Fig. 9 at t = 0.2 sec. It is noted in Fig. 9 that the NO concentration increases when the IP3R is active and decreases when the IP3R is in an inactive state in neurons. This relationship in Fig. 9 is due to the reason that the formation of NO is regulated by the [Ca2+]-associated IP3R mechanism in the cell. Thus, the dysregulation in the IP3-receptor may contribute to the neurotoxic levels of NO in neuronal cells.
Figure 10 displays the novel insights into the effects of different RyR states on the [Ca2+] distribution for the cases of the interactions of two dynamical systems of calcium and NO, and the interactions of the three dynamical systems of [Ca2+], IP3 and nitric oxide in neuronal cells at 2.5 sec and 2.5 µm. When the entirely open RyR state (P0 = 1.0) is considered, the elevated spatiotemporal calcium concentration is noted in neuron cells, since the RyR causes the calcium-induced [Ca2+] release to the cytosol from the ER and the [Ca2+] levels increase in neurons. When the completely closed (P0 = 0) RyR state is considered, the lower neuronal spatiotemporal calcium levels are noted than those for the entirely open RyR state case. The higher impacts of different RyR states on the [Ca2+] concentration are noticed near the center of neuronal cells as depicted in Fig. 10 (A & B). The variations in the [Ca2+] concentration for the active and inactive RyR states are less when the dynamic interactions of the three systems namely calcium, IP3 and NO are considered. But, when the interactions of calcium and NO are considered, there is a higher variation in the [Ca2+] concentration for distinct RyR states in neuron cells. This signifies that the interaction of IP3 dynamics with calcium and NO increase the capabilities of cell control mechanisms to manage the dysregulatory effects of different processes in neuron cells. Thus, the RyR states regulate the [Ca2+] concentration in neurons and the disturbances in the RyR activities may lead to neurotoxicity by elevating neuronal calcium levels.
The novel information about the effects of different ryanodine receptor states on the spatial IP3 and NO production in neurons are shown in Fig. 11 at 2.5 sec with NCX and VGCC channels. In the presence of a completely open RyR state (P0 = 1.0), the spatial IP3 and NO formation fluxes are highly elevated in neurons as compared to the completely closed (P0 = 0.0) RyR state in cells. The high influences of the different RyR states on the spatial IP3 and NO formation fluxes are noticed near the center of neuronal cells as depicted in Fig. 11. When the IP3 is not considered dynamic, the ryanodine receptor highly influences the generation of IP3 and NO in neuron cells, while the interactions of the three systems namely calcium, IP3 and NO reduce the effects of the ryanodine receptor on the production of IP3 and NO in neuron cells. It is noted from Fig. 11B and D that the dominance of concentration-reducing mechanisms and concentration-elevating mechanisms is high in the case of the interactions of two systems of [Ca2+] and NO in neurons. Thus, the RyR mechanism affects the regulation of IP3 and NO formation through neuronal calcium signaling.
The errors were calculated and exhibited in Tables 2–4 for the [Ca2+], IP3 and nitric oxide respectively. E depicts the number of elements. For calcium, the present model’s accuracy is sequentially 99.52%, 99.71%, 99.938708% and 99.989417%, for IP3 concentration, the accuracy is respectively 99.966925%, 99.965469%, 99.969886% and 99.977338% and for NO concentration, the accuracy is respectively 99.99967686%, 99.99963293%, 99.99960978% and 99.99960185% at time 0.1, 0.2, 0.5 and 1.0 sec. Therefore, for [Ca2+], IP3 and NO distributions, the minimum accuracy and maximum error are correspondingly 99.52% and 0.48%. Thus, the solution is grid-independent due to its minimal grid sensitivity.
If determined spectral radius (SR) concerning stability analysis is less or equal to unity then the system is deemed stable [77]. In our case, the SR is 0.9999. Therefore, the FEM used in the current circumstance is stable.
The presented data in Table 5 has been computed for interdependent [Ca2+], IP3 and NO dynamics and compared to past studies [66] at 50 sec, and results are consistent as depicted in Table 5. Also, the calculated root mean square errors between earlier findings and present findings are 0.0000085539 and 0.00000000029159 respectively for [Ca2+] and IP3, which are negligible. However, no experimental outcomes are observed under the circumstances of the current study, but the acquired outcomes are consistent with the biological facts.
Conclusion
The FEM with the Crank-Nicholson method is very effective and flexible in generating information on the interactions of the three systems of [Ca2+], IP3 and NO in neurons. Different regulatory and dysregulatory mechanisms like buffer, IP3R, RyR, NCX, VGCC, NO/cGMP pathways with PMCA channel, etc. have been incorporated to explore the cross talk dynamics of calcium, IP3 and nitric oxide in neuronal cells. Few studies on cooperation of calcium signaling with either nitric oxide or IP3 are reported in the past, but in these studies, the researchers have incorporated one-way feedback between [Ca2+] and NO and two-way feedback between [Ca2+] and IP3 in neuronal cells. Also, in these reported models, the research workers did not take into accounts the sodium-calcium exchanger (NCX), voltage-gated calcium channel (VGCC), and two-way feedback between [Ca2+] and nitric oxide through NO/cGMP pathways incorporating PMCA channel in neuronal cells. In the present work, few fundamental results of the calcium, IP3 and NO dynamics have been included in the current work to illustrate the validation of the current model. The novel findings of the current study are the behavior and functioning of different crucial mechanisms, which significantly differs for interactions of two systems of [Ca2+] and NO and the cooperation of three systems of [Ca2+], IP3 and nitric oxide in neuronal cell.
The following conclusions have been made on the basis of obtained findings.
-
i.
Calcium signaling regulates the concentration levels of IP3 and NO in neurons. Any dysregulation in the calcium concentration may lead to dysregulation in IP3 and nitric oxide production as well as in IP3 and nitric oxide levels in neuron cells, which may further contribute to neurotoxic conditions and cell death.
-
ii.
The higher amounts of buffer concentration cause alterations in the [Ca2+] signaling in the form of fluctuations, which further causes disturbances in the IP3 and NO generations in neuron cells. The [Ca2+] control mechanism regulates the equilibrium among different concentration-elevating and concentration-reducing mechanisms in the cell and provides stable dynamics. Any alterations either in the calcium control mechanism or buffering process may result in neurotoxic conditions in the form of multiple neuronal illnesses including Alzheimer’s.
-
iii.
In the two-way feedback mechanism between calcium and nitric oxide exhibits that the enhancement in the NO concentration leads to the elevation in cGMP levels, which further lowers the cytosolic calcium concentration via PMCA channel, and this reduced calcium concentration causes decrease in the [Ca2+]-dependent NO production in neuron cells. Thus, one can conclude that the NO signaling may reduce the elevated NO levels by decreasing [Ca2+] concentration in neuronal cells by using cGMP and PMCA mechanisms.
-
iv.
The IP3-receptor regulates the [Ca2+], IP3 and NO concentration levels in neurons. The IP3 dynamics are responsible for the proper activation of the IP3R channel in neurons. In the absence of IP3 dynamics, the IP3-receptor exhibits negligible effects on the [Ca2+], IP3 and NO levels in neuron cells. Any disturbances in the IP3R mechanism may be responsible for the alterations in the calcium, IP3 and NO signaling and may be associated with different neuronal illnesses including Ischemia, Alzheimer’s etc.
-
v.
The ryanodine receptor is highly sensitive during disease-associated conditions. The dysregulatory effects of the ryanodine receptor decrease in the case of the cross-talk of three systems namely calcium, IP3 and NO in neuron cells as compared to the case of the interactions among two systems namely calcium and NO in neurons. The interactive IP3 dynamics with system dynamics of calcium and NO provide additional stability to the system to balance the dysregulatory effects of different processes like RyR in neuron cells and reduce the risk of the condition of disease-affected neurons like Ischemic neurons, Alzheimeric neurons etc.
Thus, one may conclude that the cross-talk of three systems calcium, IP3 and nitric oxide regulates the concentration levels of each other in neurons. The loss of the interactions of [Ca2+] with other system dynamics like IP3 or NO may be responsible for the disturbances in the [Ca2+], IP3 and NO concentration levels due to the disturbances in different processes like buffer, IP3-receptor, ryanodine receptor, NCX, NO/cGMP kinetics with PMCC, etc. in neurons and may lead to the conditions of multiple neurological illnesses including Ischemia, Alzheimer’s etc.
The finite element method is effective in generating novel insights into the regulatory as well as dysregulatory processes of interactive systems of [Ca2+], IP3 and NO in neurons. The acquired information regarding the constitutive processes of the interactions of [Ca2+], IP3 and NO in neuron cells can be utilized by biomedical scientists for advancement of the diagnostic and therapeutic procedures.
Appendix: Model Equations Description
The shape functions concerning each element of [Ca2+], IP3 and NO concentration are exhibited as,
Utilizing the nodal conditions in Eq. (35),
Where,
By the Eq. (37), we get
And
Putting \({{\rm{p}}}^{({\rm{e}})},\,{{\rm{q}}}^{({\rm{e}})}\) and \({{\rm{r}}}^{({\rm{e}})}\) from Eq. (39) in (35), we have
Discretized form of Eq. (1, 11 & 17) are represented by the integral \({{\rm{I}}}_{1}^{({\rm{e}})},\,{{\rm{I}}}_{2}^{({\rm{e}})},\,{\rm{and}}\,{{\rm{I}}}_{3}^{({\rm{e}})}\) and represented in this formation as shown below,
Where
Now,
and
Numerous parameters \({{\rm{\alpha }}}_{1},\,{{\rm{\alpha }}}_{2},\,{{\rm{\alpha }}}_{3},\,{{\rm{\kappa }}}_{1},\,{{\rm{\kappa }}}_{2},\,{{\rm{\beta }}}_{1},\,{{\rm{\beta }}}_{2},\,{{\rm{\mu }}}_{1},\,{{\rm{\mu }}}_{2},\,{{\rm{\delta }}}_{1},\,{{\rm{\delta }}}_{2},\,{{\rm{\delta }}}_{3},\,{{\rm{\theta }}}_{1},\,{{\rm{\theta }}}_{2},\,{{\rm{\tau }}}_{1},\,{\rm{and}}\,{{\rm{\tau }}}_{2}\) are obtained by linearization of nonlinear [Ca2+], IP3 and NO terms. After analyzing and incorporating the boundary conditions, the following system of equations are acquired as depicted below,
Where,
Concerning the system matrices A and B along system vectors F, the Crank-Nicholson scheme was utilized to solve the temporal derivatives in the FEM.
Data availability
Data exchange/sharing does not apply to the present manuscript.
References
Garthwaite, J. (1991). Glutamate, nitric oxide and cell-cell signalling in the nervous system. Trends Neurosci, 14, 60–7.
Ahmed, Z., & Connor, J. A. (1988). Calcium regulation by and buffer capacity of molluscan neurons during calcium transients. Cell Calcium, 9, 57–69.
Bootman, M. D., & Berridge, M. J. (1995). The elemental principles of calcium signaling. Cell, 83, 675–8.
Wagner, J., & Keizer, J. (1994). Effects of rapid buffers on Ca2+ diffusion and Ca2+ oscillations. Biophys. J., 67, 447–56.
Smith, G. D., Wagner, J., & Keizer, J. (1996). Validity of the rapid buffering approximation near a point source of calcium ions. Biophys. J., 70, 2527–39.
Sneyd, J., Girard, S., & Clapham, D. (1993). Calcium wave propagation by calcium-induced calcium release: an unusual excitable system. Bull. Math. Biol., 55, 315–44.
Falcke, M. (2003). Buffers and oscillations in intracellular Ca2+ dynamics. Biophys. J., 84, 28–41.
Egelman, D. M., & Montague, P. R. (1999). Calcium dynamics in the extracellular space of mammalian neural tissue. Biophys. J, 76, 1856–67.
Brini, M., Calì, T., Ottolini, D., & Carafoli, E. (2014). Neuronal calcium signaling: function and dysfunction. Cell. Mol. Life Sci., 71, 2787–814.
Pathak, K., & Adlakha, N. (2016). Finite element model to study two dimensional unsteady state calcium distribution in cardiac myocytes. Alexandria J. Med, 52, 261–8.
Singh, N & N Adlakha, (2019). A mathematical model for interdependent calcium and inositol 1, 4,5-trisphosphate in cardiac myocyte. Netw. Model. Anal. Heal. Informatics Bioinforma., 8.
Singh, N., & Adlakha, N. (2019). Nonlinear dynamic modeling of 2-dimensional interdependent Calcium and Inositol 1,4,5-Trisphosphate in cardiac myocyte. Math. Biol. Bioinforma., 14, 290–305.
Kotwani, M., Adlakha, N., & Mehta, M. N. (2014). Finite element model to study the effect of buffers, source amplitude and source geometry on spatio-temporal calcium distribution in fibroblast cell. J. Med. Imaging Heal. Informatics, 4, 840–7.
Kothiya, A. B., & Adlakha, N. (2023). Cellular nitric oxide synthesis is affected by disorders in the interdependent Ca2+ and IP3 dynamics during cystic fibrosis disease. J. Biol. Phys., 49, 133–58.
Kothiya, A. B., & Adlakha, N. (2023). Simulation of biochemical dynamics of Ca2+ and PLC in fibroblast cell. J. Bioenerg. Biomembr., 55, 267–87.
Jha, B. K., Adlakha, N., & Mehta, M. N. (2013). Two-dimensional finite element model to study calcium distribution in astrocytes in presence of VGCC and excess buffer. Int. J. Model. Simulation, Sci. Comput., 4.
Jha, B. K., Adlakha, N., & Mehta, M. N. (2014). Two-dimensional finite element model to study calcium distribution in astrocytes in presence of excess buffer. Int. J. Biomath., 7, 1–11.
Bhardwaj, H., & Adlakha, N. (2022). Radial Basis Function Based Differential Quadrature Approach to Study Reaction Diffusion of Ca 2+ in T Lymphocyte. Int. J. Comput. Methods., 20.
Bhardwaj, H., & Adlakha, N. (2023). Model To Study Interdependent Calcium And IP3 Distribution Regulating NFAT Production in T Lymphocyte. J. Mech. Med. Biol.
Tewari, S. G., & Pardasani, K. R. (2010). Finite element model to study two dimensional unsteady state cytosolic calcium diffusion in presence of excess buffers. IAENG Int. J. Appl. Math., 40, 1–5.
Tewari, S. G., & Pardasani, K. R. (2012). Modeling effect of sodium pump on calcium oscillations in neuron cells. J. Multiscale Model., 04, 1250010.
Tripathi, A., & Adlakha, N. (2012). Two dimensional coaxial circular elements in FEM to study calcium diffusion in neuron cells. Appl. Math. Sci., 6, 455–66.
Tripathi, A., & Adlakha, N. (2011). Closed form solution to problem of calcium diffusion in cylindrical shaped neuron cell. World Acad. Sci. Eng. Technol., 80, 739–42.
Pawar, A., & Pardasani, K. R. (2023). Computational model of calcium dynamics-dependent dopamine regulation and dysregulation in a dopaminergic neuron cell. Eur. Phys. J. Plus, 138, 30.
Pawar, A., & Pardasani, K. R. (2022). Simulation of disturbances in interdependent calcium and β-amyloid dynamics in the nerve cell. Eur. Phys. J. Plus, 137, 1–23.
Pawar, A., & Pardasani, K. R. (2023). Fractional order interdependent nonlinear chaotic spatiotemporal calcium and Aβ dynamics in a neuron cell. Phys. Scr., 98, 085206.
Jagtap, Y., & Adlakha, N. (2018). Finite volume simulation of two dimensional calcium dynamics in a hepatocyte cell involving buffers and fluxes. Commun. Math. Biol. Neurosci., 2018, 1–16.
Mishra, V., & Adlakha, N. (2023). Numerical simulation of calcium dynamics dependent ATP degradation, IP3 and NADH production due to obesity in a hepatocyte cell. J. Biol. Phys., 49, 415–42.
Mishra, V., & Adlakha, N. (2023). Spatio temporal interdependent calcium and buffer dynamics regulating DAG in a hepatocyte cell due to obesity. J. Bioenerg. Biomembr., 55, 249–66.
Manhas, N., & Pardasani, K. R. (2014). Mathematical model to study IP3 Dynamics dependent calcium oscillations in pancreatic acinar cells. J. Med. Imaging Heal. Informatics, 4, 874–80.
Manhas, N., & Pardasani, K. R. (2014). Modelling mechanism of calcium oscillations in pancreatic acinar cells. J. Bioenerg. Biomembr., 46, 403–20.
Vaishali, N., & Adlakha, J. (2023). Disturbances in system dynamics of Ca2+ and IP3 perturbing insulin secretion in a pancreatic β -cell due to type-2 diabetes. Bioenerg. Biomembr., 55, 151–67.
Naik, P. A., & Pardasani, K. R. (2018). 2D finite-element analysis of calcium distribution in oocytes. Netw. Model. Anal. Heal. Informatics Bioinforma., 7, 1–11.
Naik, P. A., & Pardasani, K. R. (2019). Three-Dimensional Finite Element Model to Study Effect of RyR Calcium Channel, ER Leak and SERCA Pump on Calcium Distribution in Oocyte Cell. Int. J. Comput. Methods, 16, 1–19.
Joshi, H., & Jha, B. K. (2021). On a reaction–diffusion model for calcium dynamics in neurons with Mittag–Leffler memory. Eur. Phys. J. Plus, 136, 623.
Dave, D. D., & Jha, B. K. (2021). Mathematical modeling of calcium oscillatory patterns in a neuron. Interdiscip. Sci. Comput. Life Sci., 13, 12–24.
Michell, R. H. (1975). Inositol phospholipids and cell surface receptor function. BBA - Rev. Biomembr., 415, 81–47.
Berridge, M. J., & Irvine, R. F. (1984). Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature, 312, 315–21.
Berridge, M. J., Lipp, P., & Bootman, M. D. (2000). The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol., 1, 11–21.
Bezprozvanny, I., Watras, J., & Ehrlich, B. E. (1991). Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature, 351, 751–4.
Allbritton, N. L., Meyer, T., & Stryer, L. (1992). Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science, 258, 1812–5.
Young, G. W. D. E., & Keizer, J. (1992). A single-pool inositol 1,4,5-trisphosphate-receptor-based model for agonist-stimulated oscillations in Ca2+ concentration. Biophysics, 89, 9895–9.
Li, Y. X., & Rinzel, J. (1994). Equations for InsP3 receptor-mediated [Ca2+]i oscillations derived from a detailed kinetic model: a Hodgkin-Huxley like formalism. J. Theor. Biol., 166, 461–73.
Falcke, M., Huerta, R., Rabinovich, M. I., Abarbanel, H. D. I., Elson, R. C., & Selverston, A. I. (2000). Modeling observed chaotic oscillations in bursting neurons: the role of calcium dynamics and IP3. Biol. Cybern., 82, 517–27.
Emilsson, L., Saetre, P., & Jazin, E. (2006). Alzheimer’s disease: mRNA expression profiles of multiple patients show alterations of genes involved with calcium signaling. Neurobiol. Dis., 21, 618–25.
Jagtap, Y., & Adlakha, N. (2019). Numerical study of one-dimensional buffered advection–diffusion of calcium and IP 3 in a hepatocyte cell. Netw. Model. Anal. Heal. Informatics Bioinforma., 8, 1–9.
Pawar, A., & Pardasani, K. R. (2022). Effects of disorders in interdependent calcium and IP3 dynamics on nitric oxide production in a neuron cell. Eur. Phys. J. Plus, 137, 543.
Pawar, A., & Pardasani, K. R. (2022). Effect of disturbances in neuronal calcium and IP3 dynamics on β-amyloid production and degradation. Cogn. Neurodynamics, 17, 239–56.
Pawar, A., & Pardasani, K. R. (2023). Mechanistic insights of neuronal calcium and IP 3 signaling system regulating ATP release during ischemia in progression of Alzheimer’s disease. Eur. Biophys. J., 52, 153–73.
Salter, M., & Knowles, G. (1991). Widespread tissue distribution, species distribution and changes in activity of Ca(2+)-dependent and Ca(2+)-independent nitric oxide synthases. FEBS Lett., 291, 145–9.
Garthwaite, J., Garthwaite, G., Palmer, R. M. J., & Moncada, S. (1989). NMDA receptor activation induces nitric oxide synthesis from arginine in rat brain slices. Eur. J. Pharmacol. Mol. Pharmacol., 172, 413–6.
Wood, P. L., Emmett, M. R., Rao, T. S., Cler, J., Mick, S., & Iyengar, S. (1990). Inhibition of nitric oxide synthase blocks N-methyl-D-aspartate-, quisqualate-, kainate-, harmaline-, and pentylenetetrazole-dependent increases in cerebellar cyclic GMP in vivo. J. Neurochem., 55, 346–8.
Vincent, S. R., & Neurobiol, Prog (2010). Nitric oxide neurons and neurotransmission. Prog. Neurobiol., 90, 246–55.
Kohlmeier, K. A., & Leonard, C. S. (2006). Transmitter modulation of spike-evoked calcium transients in arousal related neurons: muscarinic inhibition of SNX-482-sensitive calcium influx. Eur. J. Neurosci., 23, 1151–62.
Bolotina, V. M., Najibi, S., Palacino, J. J., Pagano, P. J., & Cohen, R. A. (1994). Nitric oxide directly activates calcium-dependent potassium channels in vascular smooth muscle. Nature, 368, 850–3.
Kourosh-Arami, M., Hosseini, N., Mohsenzadegan, M., Komaki, A., & Joghataei, M. T. (2020). Neurophysiologic implications of neuronal nitric oxide synthase. Rev. Neurosci., 31, 617–36.
Brenman, J. E., Xia, H., Chao, D. S., Black, S. M., & Bredt, D. S. (1997). Regulation of neuronal nitric oxide synthase through alternative transcripts. Dev. Neurosci., 19, 224–31.
Iino, M. (2006). Ca2+-dependent inositol 1,4,5-trisphosphate and nitric oxide signaling in cerebellar neurons. J. Pharmacol. Sci., 100, 538–44.
Chung, J. W., Ryu, W. S., Kim, B. J., & Yoon, B. W. (2015). Elevated calcium after acute ischemic stroke: association with a poor short-term outcome and long-term mortality. J. Stroke, 17, 54–9.
Moro, M. A., Cárdenas, A., Hurtado, O., Leza, J. C., & Lizasoain, I. (2004). Role of nitric oxide after brain ischaemia. Cell Calcium, 36, 265–75.
Gibson W. G, Farnell L., & Bennett M. R. (2007). A computational model relating changes in cerebral blood volume to synaptic activity in neurons, 70, 1674-79.
Mo, E., Amin, H., Bianco, I. H., & Garthwaite, J. (2004). Kinetics of a cellular nitric oxide/cGMP/phosphodiesterase-5 pathway. J. Biol. Chem., 279, 26149–58.
Sriram, K., Laughlin, J. G., Rangamani, P., & Tartakovsky, D. M. (2016). Shear-induced nitric oxide production by endothelial cells. Biophys. J., 111, 208–21.
Pawar, A., & Pardasani, K. R. (2022). Study of disorders in regulatory spatiotemporal neurodynamics of calcium and nitric oxide. Cogn. Neurodyn., 17, 1661–82.
Kothiya A., & Adlakha N. (2023). Impact of Interdependent C a 2+ and I P 3 Dynamics On ATP Regulation in A Fibroblast Model Cell. Biochem. Biophys., 1-17.
Wagner, J., Fall, C. F., Hong, F., Sims, C. E., Allbritton, N. L., Fontanilla, R. A., Moraru, I. I., Loew, L. M., & Nuccitelli, R. (2004). A wave of IP3 production accompanies the fertilization Ca2+ wave in the egg of the frog, Xenopus laevis: theoretical and experimental support. Cell Calcium, 35, 433–47.
J Keener and J Sneyd, Mathematical Physiology, Second Edi (Springer. https://doi.org/10.1007/978-0-387-75847-3, 2009).
Tewari S., & Pardasani K. R. (2008). Finite Difference Model to Study the Effects of Na+ Influx on Cytosolic [Ca2+] Diffusion. World Acad. Sci. Eng. Technol., 670-75.
Panday, S., & Pardasani, K. R. (2013). Finite element model to study effect of advection diffusion and Na +/Ca2+ exchanger on Ca2+ distribution in oocytes. J. Med. Imaging Heal. Informatics, 3, 374–79.
Nelson D. L., & Cox M. M. (2005). Lehninger Principles of Biochemistry. Fourth Ed.
Bugrim, A., Fontanilla, R., Eutenier, B. B., Keizer, J., & Nuccitelli, R. (2003). Sperm initiate a Ca2+ wave in frog eggs that is more similar to Ca2+ waves initiated by IP3 than by Ca2+. Biophys. J., 84, 1580–90.
Condorelli, P., & George, S. C. (2001). In vivo control of soluble guanylate cyclase activation by nitric oxide: a kinetic analysis. Biophys. J., 80, 2110–9.
Smith, G. D. (1996). Analytical steady-state solution to the rapid buffering approximation near an open Ca2+ channel. Biophys. J., 71, 3064–72.
Brown, S. A., Morgan, F., Watras, J., & Loew, L. M. (2008). Analysis of phosphatidylinositol-4,5-bisphosphate signaling in cerebellar Purkinje spines. Biophys. J., 95, 1795–812.
Edwards, A., Cao, C., & Pallone, T. L. (2011). Cellular mechanisms underlying nitric oxide-induced vasodilation of descending vasa recta. Am. J. Physiol. - Ren. Physiol., 300, 441–56.
Kavdia, M., Tsoukias, N. M., & Popel, A. S. (2002). Model of nitric oxide diffusion in an arteriole: impact of hemoglobin-based blood substitutes. Am. J. Physiol. - Hear. Circ. Physiol., 282, 2245–53.
Öziş, T., Aksan, E. N., & Özdeş, A. (2003). A finite element approach for solution of Burgers’ equation. Appl. Math. Comput., 139, 417–28.
Author contributions:
Both authors made equal contributions to the current study regarding the formulation of the problem and its solution, correction of data, review of relevant literature, and interpretations of the outcomes. The MATLAB code is created by Author (1).
Funding
There is no funding to report.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pawar, A., Pardasani, K.R. Modelling Cross Talk in the Spatiotemporal System Dynamics of Calcium, IP3 and Nitric Oxide in Neuron Cells. Cell Biochem Biophys 82, 787–803 (2024). https://doi.org/10.1007/s12013-024-01229-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-024-01229-5