Abstract
Atherosclerosis (AS) is an inflammatory disease with multiple causes. Multiple circular RNAs (circRNAs) are known to be involved in the pathogenesis of AS. To explore the function and mechanism of circ_0005699 in oxidative low-density lipoprotein (ox-LDL)-induced human umbilical vein endothelial cells (HUVECs) injury. Ox-LDL treatment restrained HUVECs viability, cell proliferation, and angiogenesis ability, and accelerated HUVECs apoptosis, inflammatory response, and oxidative stress. Circ_0005699 was up-regulated in the serum samples of AS patients and ox-LDL-induced HUVECs. Interference of circ_0005699 effectively rescued ox-LDL-induced injury in HUVECs. Additionally, miR-384 could bind to circ_0005699, and miR-384 depletion inverted the effects of circ_0005699 deficiency on ox-LDL-mediated HUVEC injury. Moreover, ASPH was a direct target of miR-384, and the enforced expression of ASPH overturned miR-384-induced effects on ox-LDL-induced HUVECs. Importantly, circ_0005699 regulated ASPH expression via sponging miR-384. Interference of circ_0005699 protected against ox-LDL-induced injury in HUVECs at least partly by regulating ASPH expression via acting as a miR-384 sponge.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atherosclerosis (AS) is a risk factor and basic cause of cardiovascular and cerebrovascular diseases [1], as well as one of the causes of high mortality in the elderly [2]. However, the mechanisms involved in the development of AS are complex and remain unclear at present. The imbalance between endothelial cell injury and repair is considered to be the main factor in the occurrence of AS [3]. In addition, endothelial cell (EC) injury is also the major pathological feature of AS development [4]. Previously published studies showed oxidized low density lipoprotein (ox-LDL) enhanced the progress of AS by inducing endothelial cell dysfunction, including endothelial cell injury, inflammatory response and oxidative stress [5, 6]. Therefore, uncovering the development mechanism of AS by using ox-LDL treated endothelial cells can provide an effective biological target for the clinical treatment of AS.
Circular RNAs (circRNAs) are a class of endogenous RNAs characterized by the lack of 5’ and 3’ free ends [7]. An increasing body of studies have identified that circRNAs participated in the development of AS [8]. For example, Ji et al. demonstrated that circ_0004104 knockdown reversed ox-LDL-induced pro-proliferative, pro-inflammatory and anti-apopHtotic effect in human umbilical vein endothelial cells (HUVECs) [9]. Huang et al. identified inhibition of circUSP36 accelerated the proliferation and migration of HUVECs [10]. As for circ_0005699, it has been confirmed to be overexpressed in ox-LDL-induced macrophages [11]. Nevertheless, its biological action and specific mechanism in ox-LDL-treated HUVECs have not been reported.
Multiple studies have revealed that circRNAs can affect cellular biological processes by acting as an endogenous sponge of microRNAs (miRNAs) [12]. miRNAs are small noncoding RNAs, which are involved in the process of endothelial cell dysfunction during atherosclerotic formation [13]. A previous study verified that the abundance of miR-384 was reduced in Ang II-induced HUVECs, and miR-384 upregulation reversed Ang II-triggered apoptosis in HUVECs by targeting Herpud1 expression [14]. However, the mechanism of miR-384 in AS needs further investigation.
Aspartyl (asparaginyl) β-hydroxylase (ASPH) was demonstrated as a target of miR-384 in our work. ASPH has been verified to be closely related to AS development. Xiao et al. suggested that miR-206 hindered the oxidative stress injury and inflammation progress in ox-LDL-treated macrophages via repressing ASPH [11]. Nevertheless, the functional mechanism of ASPH in ox-LDL-stimulated HUVECs is indistinct and needs to be further explored.
Here, we identified that circ_0005699 expression was evidently upregulated in AS patients and ox-LDL-treated HUVECs, while interference of circ_0005699 ameliorated cell injuries caused by ox-LDL by suppressing the expression of ASPH via sponging miR-384, suggesting that the circ_0005699/miR-384/ASPH axis may provide a new strategy for the clinical treatment of AS.
Materials and Methods
Clinical Blood Samples
This study was approved by the Ethics Committee of Chinese People Liberation Army (PLA) 93864 Military Hospital and was performed in accordance with the Declaration of Helsinki. Serum samples from 21 healthy volunteers and 27 AS patients were collected from Chinese People Liberation Army (PLA) 93864 Military Hospital. Madison ultrasound system was utilized to evaluate plaque site and range. Meanwhile, two professional doctors identified the diseases. AS patients with other clinical diseases were excluded. The clinicopathologic features of these subjects were provided in Table 1. All of them had signed written informed consents.
Cell Culture and Treatment
HUVECs were obtained from the Procell (Wuhan, China), and cultivated in the prescribed Dulbecco's modified Eagle medium (DMEM, Solarbio, Beijing, China) plus 10% fetal bovine serum (FBS, Solarbio) at 37 °C with 5% CO2. Meanwhile, 1% endothelial cell growth supplement was added into DMEM medium. Then, HUVECs were stimulated by ox-LDL for 24 h at various concentrations (0, 25, 50 and 100 μg/mL). Besides that, 50 μg/mL ox-LDL-treated HUVECs were collected for further functional experiments.
Cell Transfection
When HUVECs reached 50% confluences, cell transfection was executed. Small interfering RNA (siRNA) against circ_0005699 (si-circ_0005699), circ_0005699 overexpression plasmids (circ_0005699), miR-384 mimics and inhibitor (miR-384, anti-miR-384), ASPH overexpression vector (ASPH) and their corresponding controls were purchased from GenePharma (Shanghai, China). Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was utilized for cell transfection. 24 h post transfection, HUVECs were treated with 50 μg/mL ox-LDL for 24 h.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol reagent (Invitrogen) were applied to extract the total RNA and RNA concentration and purity were measured by NanoDrop-1000 apparatus. Next, the reversed transcription was conducted by using RT-PCR kit (Invitrogen). QRT-PCR was performed by SYBR Premix Ex Taq II (TaKaRa, Dalian, China). All primer sequences were shown in Table 2. RNA expression was normalized to the expression of GAPDH or U6 and assessed using the 2-ΔΔCt method.
Cell Viability and 5-Ethynyl-2ʹ-deoxyuridine (EdU) Assay
Approximately 5 × 103 HUVECs were cultured in 96-well plates for 24 h, followed by stimulation with different dose of ox-LDL for 24 h. Then, the CCK-8 solution (10 μL) (Solarbio) was added to cells. 4 h later, the cell viability was detected via using a microplate reader.
EdU positive cells were evaluated to detect cell proliferation according to the instructions of BeyoClick™ EdU-488 Kit (Beyotime, Shanghai, China). In short, HUVECs were incubated with EdU solution. After fixation and permeabilization, nuclei was stained by utilizing DAPI. Lastly, EdU positive cells were investigated by the application of fluorescence microscope (Leica, Wetzlar, Germany).
Flow Cytometry
After transfection or treatment, HUVECs were digested with trypsin and collected into a centrifuge tube. After 3 washes with PBS, the cell suspensions was collected and incubated with 5 μL Annexin V-FITC and 5 μL PI (Solarbio) for 15 min in the dark. Subsequently, cell apoptosis rate was gauged by flow cytometer (BD Biosciences, San Jose, CA, USA).
Tube Formation Assay
The angiogenesis ability was evaluated by tube formation assay. In brief, approximately 1 × 104 HUVECs were plated into 96-well plates coated with 70 μL matrigel (Corning, Rochester, NY, USA) and grown for 48 h. The number of capillary-like branches was estimated with a microscope (Leica).
Enzyme-Linked Immunosorbent Assay (ELISA) and Measurement of Malondialdehyde (MDA) and Superoxide Dismutase (SOD)
The supernatant of the HUVECs was harvested by centrifugation and analyzed by ELISA. The levels of Interleukin-1β (IL-1β) and Tumor necrosis factor-α (TNF-α) were detected using the ELISA Kits (Invitrogen) following the instruction of manufacturers. The OD value at 450 nm was measured by the Microplate Reader.
After ox-LDL stimulation and relevant transfection, the cell supernatants were acquired and the MDA and SOD levels were measured using MDA and SOD Assay Kit (Solarbio) according to the instructions, respectively.
Western Blot Analysis
Cell lysis buffer (Beyotime, Shanghai, China) was used to isolate total protein. Subsequently, protein sample was segregated by 10% SDS-PAGE and then transferred to PVDF membranes (Beyotime). The membrane was blocked in 5% non-fat milk for 1 h, and then incubated with antibodies against Bax (ab32503, 1:1000), Cleaved-caspase-3 (ab2302, 1:500), GAPDH (ab9485, 1:2500) and ASPH (SAB1402121, 1:5000) at 4 °C overnight. Next, secondary antibody was used to incubate protein band for 2 h at indoor temperature. Antibodies against ASPH was purchased from (Sigma, St. Louis, MO, USA). Other antibodies were obtained from Abcam (Cambridge, MA, USA). Finally, Enhanced Chemiluminescence Kit (Beyotime) was used to visualize the protein signals.
Dual-Luciferase Reporter Assay and RIP Assays
The targeted relationship was predicted via using circInteractome or starBase v2.0. The wild type circ_0005699 sequences (WT-circ_0005699), mutant type circ_0005699 sequences (MUT-circ_0005699), and ASPH (ASPH-3ʹUTR-WT, ASPH-3ʹUTR-MUT) were constructed and cloned into pmirGLO vector (Promega, Madison, WI, USA). Next, the reporter plasmid and miR-NC or miR-384 was co-transfected into HUVECs. At 48 h post-transfection, the Dual-Luciferase Reporter Assay Kit (Solarbio) was used to quantify luciferase activity.
RIP buffer was used to treat HUVECs. Then, magnetic beads conjugated with Ago2 or IgG antibody were incubated with cell lysate (Millipore, Billerica, MA, USA). The enrichment of circ_0005699 or ASPH and miR-384 in the immunoprecipitated RNAs was analyzed by qRT-PCR.
Statistical Analysis
The experiment results were displayed as mean ± SD and evaluated by GraphPad Prism 7.0. Statistical analysis was conducted by using Student’s t-test or one-way analysis of variance. P-value < 0.05 meant significant difference.
Results
Circ_0005699 was Increased in ox-LDL-Induced HUVECs
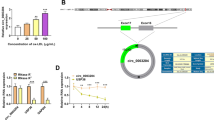
The level of circ_0005699 in the serum of AS patients (n = 27) and healthy volunteers (n = 21) was determined by qRT-PCR, and we demonstrated that circ_0005699 expression was increased in AS patients than that in healthy volunteers (Fig. 1A). Similarly, we confirmed that circ_0005699 was increased in a concentration-dependent manner under ox-LDL-stimulated HUVECs (Fig. 1B). Moreover, we revealed that circ_0005699 was resistant to RNase R (Fig. 1C), and mainly localized in the cytoplasm of HUVECs through the analysis of subcellular localization (Fig. 1D). Taken together, the expression of circ_0005699 was increased in AS.
Circ_0005699 was up-regulated in AS patients and ox-LDL-induced HUVECs. A The level of circ_0005699 in serum of AS patients and healthy volunteers was gauged by qRT-PCR. B The abundance of circ_0005699 was evaluated in HUVECs treated with different concentrations of ox-LDL (0, 25, 50, 100 μg/mL) by qRT-PCR. C Circular form of circ_0005699 was identified by RNase R treatment. D The subcellular distribution of circ_0005699 was verified by qRT-PCR. *P < 0.05, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. Student’s t-test was utilized to analyze the diferences in A, whereas one-way ANOVA was utilized to assess the diferences in B–D
Circ_0005699 Silencing Decreased ox-LDL-Induced Cell Injury in HUVECs
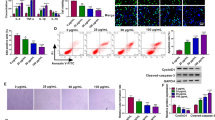
Firstly, our results showed that different concentrations of ox-LDL stimulation contributed to the process of cell injury by regulating growth, apoptosis, angiogenesis, inflammatory responses and oxidative stress in HUVECs, and the specific results were shown in supplementary Fig. 1. In order to explore the biological function of circ_0005699 in the development of AS, we carried out related functional experimental study in vitro. As shown in Fig. 2A, the elevation of circ_0005699 in ox-LDL-induced HUVECs was partly alleviated after interference of circ_0005699. Subsequently, we explored the effects of circ_0005699 silencing on the growth, apoptosis, angiogenesis, inflammatory responses and oxidative stress in HUVECs. The results showed that ox-LDL treatment hindered the viability (Fig. 2B), proliferation (Fig. 2C) and triggered apoptosis (Fig. 2D, E) of HUVECs, but these effects were rescued after circ_0005699 deficiency. Tube formation assay verified that the interference of circ_0005699 elevated the angiogenesis capacity of HUVECs under ox-LDL treatment (Fig. 2F). Meanwhile, ox-LDL treatment of HUVECs resulted in an elevation in the abundance of Bax and Cleaved-caspase-3, while their expression was reduced after the transfection of si-circ_0005699 (Fig. 2G, H). Moreover, circ_0005699 inhibition evidently overturned the increase on IL-1β, TNF-α production in ox-LDL-induced HUVECs (Fig. 2I). In the oxidative stress related experimental tests, we confirmed that the levels of MDA were enlarged while SOD had an opposite result under ox-LDL treatment, while these trends were counteracted by si-circ_0005699 transfection (Fig. 2J, K). To sum up, circ_0005699 inhibition ameliorated ox-LDL-induced HUVEC damage.
Knockdown of circ_0005699 restored ox-LDL-induced cell injury in HUVECs. Cell were transfected with si-circ_0005699 or si-NC before ox-LDL treatment. A The expression of circ_0005699 was determined by qRT-PCR. B Cell viability was analyzed by CCK8 assay. C Cell proliferation was estimated by EdU assay. D and E Cell apoptosis was investigated by flow cytometry. F Angiogenesis ability was determined by tube formation assay. G–H Western blot assay was utilized to assess the protein levels of Bax and Cleaved-caspase-3. I The concentrations of inflammatory cytokines (IL-1β and TNF-α) were counted by ELISA. J–K The level of oxidative stress was estimated by detecting the production of MDA and SOD. **P < 0.01, ***P < 0.001, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. One-way ANOVA was utilized to assess the diferences in A–K
MiR-384 is a Target of circ_0005699 in HUVECs
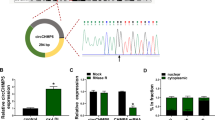
To uncover the molecular mechanism of circ_0005699 in AS progress. The binding sites of circ_0005699 on miR-384 were predicted by circInteractome online database and displayed in Fig. 3A. The expression of miR-384 was strikingly increased in HUVECs transfected with miR-384 (Fig. 3B). To confirm the targeting relationship between circ_0005699 and miR-384, we performed Dual-luciferase reporter assay and results exhibited that miR-384 mimic obviously reduced the luciferase activity in WT-circ_0005699 group, while the luciferase activity in MUT-circ_0005699 group was no apparent difference (Fig. 3C). RIP assay exhibited that miR-384 and circ_0005699 were markedly enriched in anti-Ago2 complexes, which further confirmed their targeting relationship (Fig. 3D). Additionally, the expression of miR-384 was downregulated in AS patients than that in healthy volunteers (Fig. 3E), and the expression of miR-384 was negatively associated with circ_0005699 in AS patients (Fig. 3F). Similarly, we also demonstrated that the expression of miR-384 was reduced in ox-LDL-stimulated HUVECs (Fig. 3G). Together, these data suggested that circ_0005699 acted as a sponge of miR-384 in ox-LDL-induced HUVECs.
Circ_0005699 was verified as a miR-384 sponge. A Circinteractome software online showed the binding sites between miR-384 and circ_0005699. B QRT-PCR was conducted to analyze the transfection efficiency of miR-384. C The relationship between circ_0005699 and miR-384 was identified by Dual-luciferase reporter assay. D RIP assay was employed to confirm the relationship between circ_0005699 and miR-384. E The expression of miR-384 in AS patients and healthy volunteers was gauged by qRT-PCR. F Pearson correlation analysis was executed to assess the correlation between the expression of miR-384 and circ_0005699. G The expression of miR-384 was evaluated in HUVECs treated with different doses of ox-LDL by qRT-PCR. **P < 0.01, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. Student’s t-test was applied to analyze the diferences in (B and E), whereas one-way ANOVA was utilized to assess the diferences in (C, D and G)
Circ_0005699 Mediated ox-LDL-Stimulated HUVEC Damage via miR-384
Next, we performed experiments to measure whether circ_0005699 regulates ox-LDL-induced HUVEC injury via targeting miR-384. The results showed that the elevation of miR-384 in circ_0005699-silenced HUVECs was partially ameliorated by anti-miR-384 introduction in ox-LDL-induced HUVECs (Fig. 4A). In addition, anti-miR-384 introduction effectively rescued the promoting effect of circ_0005699 silencing on the viability and proliferation in ox-LDL-induced HUVECs (Fig. 4B, C). Also, circ_0005699 deficiency mediated the inhibition of apoptosis and the promoting of angiogenesis in ox-LDL-induced HUVECs were overturned after interference of miR-384 (Fig. 4D–F). MiR-384 inhibition elevated the expression of Bax and Cleaved-caspase-3 in circ_0005699-silenced HUVECs upon ox-LDL treatment (Fig. 4G). In addition, interference of miR-384 alleviated the inhibitory effect of circ_0005699 deficiency on the inflammation and oxidative stress (Fig. 4H–J). Collectively, circ_0005699 mediated ox-LDL-stimulated HUVECs damage via sponging miR-384.
Circ_0005699 depletion-mediated impacts in ox-LDL-induced HUVECs were effectively rescued by anti-miR-384 introduction. A–J HUVECs were transfected with si-NC, si-circ_0005699, si-circ_0005699 + anti-miR-NC, or si-circ_0005699 + anti-miR-384, and then treated with ox-LDL (50 μg/mL). A The expression of miR-384 was analyzed by qRT-PCR. B and C Cell viability and proliferation were assessed by CCK8 assay and EdU assay, individually. D and E Cell apoptosis was assessed by flow cytometry. F The angiogenesis capacity was estimated using tube formation assay. G The protein levels of Bax and Cleaved-caspase-3 was assessed by using western blot assay. H The levels of IL-1β and TNF-α was evaluated by ELISA. I and J The levels of MDA and SOD were analyzed by using related detection kit. **P < 0.01, ***P < 0.001, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. One-way ANOVA was utilized to assess the diferences in A–J
MiR-384 Directly Targeted ASPH
StarBase v2.0 was used to predict the targeting relationship between miR-384 and ASPH, and the binding site was displayed in Fig. 5A. Furthermore, the dual-luciferase reporter assay identified the interaction between the ASPH and miR-384 (Fig. 5B). Further, we confirmed that miR-384 and ASPH were enriched in the anti-Ago2 complexes through RIP assays (Fig. 5C), uncovering that ASPH was a target of miR-384 in HUVECs. The expression of ASPH was upregulated in AS patients than that in healthy volunteers (Fig. 5D). Moreover, ASPH expression was negatively correlated with miR-384 expression in AS patients (Fig. 5E). We also evaluated the expression of ASPH in ox-LDL-stimulated HUVECs. As shown in Fig. 5F, with the increased of ox-LDL doses, the expression of ASPH was gradually upregulated in HUVECs. Importantly, we also found that miR-384 deficiency effectively rescued the down-regulation of ASPH caused by circ_0005699 silencing (Fig. 5G).
ASPH was a direct target of miR-384. A The binding site between miR-384 and ASPH was exhibited. B–C The interaction between ASPH and miR-384 was identified by dual-luciferase reporter assay and RIP assay. D QRT-PCR was applied to measure the expression of ASPH in AS patients. E The correlation between the ASPH and miR-384 was assessed by Pearson correlation analysis. F The protein level of ASPH in HUVECs treated with different doses of ox-LDL was gauged by western blot. G Western blot assay was employed to evaluate the protein level of ASPH in ox-LDL-induced HUVECs transfected with si-NC, si-circ_0005699, si-circ_0005699 + anti-miR-NC, or si-circ_0005699 + anti-miR-384. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. Student’s t-test was applied to analyze the diferences in B, whereas one-way ANOVA was utilized to assess the diferences in B, C, F and G
MiR-384 Overexpression Ameliorated ox-LDL-Triggered HUVEC Injury by Down-Regulating ASPH
Given that ASPH was a target of miR-384, we further investigated whether miR-384 regulated ox-LDL-triggered HUVECs injury through ASPH. MiR-384 and ASPH were co-transfected into HUVECs to detect the injury process induced by ox-LDL. Western blot analysis manifested that miR-384 inhibited ASPH expression, and ASPH overexpression abolished miR-384-induced reduce of ASPH expression in ox-LDL-treated HUVECs (Fig. 6A). MiR-384 overexpression led to a notable promotion in cell viability and proliferation (Fig. 6B, C), a significant repression on cell apoptosis (Fig. 6D, E), and a striking enhancement in tube formation (Fig. 6F), as well as a notable reduction in pro-apoptotic protein expression (Fig. 6G), IL-1β and TNF-α production (Fig. 6H). Nevertheless, these effects were apparently attenuated by the overexpression of ASPH level in ox-LDL-treated HUVECs. Finally, the production of MDA was reduced and the production of SOD was elevated in ox-LDL-induced HUVECs transfected with miR-384, whereas overexpression of ASPH neutralized these effects (Fig. 6I, J). Overall, our work demonstrated that miR-384 protected HUVECs from ox-LDL-evoked injury partly via decreasing the expression of ASPH.
Overexpression of ASPH abated the effects of miR-384 in ox-LDL-induced HUVECs. A–J HUVECs were transfected with miR-NC, miR-384, miR-384 + pcDNA, or miR-384 + ASPH, and then stimulated with ox-LDL. A Western blot assay was conducted to evaluate the protein expression of ASPH in HUVECs. B CCK8 assay was executed for cell viability detection. C Cell proliferation were tested by EdU assay. D–E Cell apoptosis was investigated by flow cytometry. F Tube formation assay was executed to explore the angiogenesis ability of HUVECs. G Protein level among Bax and Cleaved-caspase-3 was investigated by western blot assay. H–J The levels of IL-1β, TNF-α, MDA and SOD were tested in HUVECs. **P < 0.01, ***P < 0.001, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. One-way ANOVA was utilized to assess the diferences in A–J
Discussion
AS is a chronic inflammatory disease which leads to the development of cardiovascular and cerebrovascular diseases [15]. As the morbidity and mortality of AS is increasing, it becomes one of the reasons for the high mortality among the elderly [16]. The development of AS is often accompanied by impairment of endothelial cell function [4]. There is increasing evidence that circRNAs are involved in the development of AS [17]. The role of circ_0005699 in AS was also revealed in our work. ox-LDL-stimulated HUVECs were used to establish the injury model, so as to simulate the pathogenesis of AS. Our work first identified the function and mechanism of circ_0005699/miR-384/ASPH regulatory network in ox-LDL-mediated injury of HUVECs.
Previous study uncovered that circ-USP9X interference effectively rescued the effects of ox-LDL on HUVECs viability, angiogenesis, apoptosis, inflammation and oxidative stress [18]. Moreover, interference of circ_0068087 effectively recovered the cell damage in ox-LDL-induced HUVECs [19]. Recently, circ_0005699 was found to be elevated in ox-LDL-induced macrophages [11]. In this study, the result exhibited that circ_0005699 was evidently elevated in AS patient serum and ox-LDL-stimulated HUVECs. In subsequent experiments, we also demonstrated that ox-LDL apparently impeded cell growth and reinforced cell apoptosis, inflammatory reaction, and oxidative stress progress. However, circ_0005699 inhibition in HUVECs distinctly counteracted these effects.
Accumulating evidence suggested that circRNAs influence gene expression via sponging miRNAs. Here, we demonstrated that circ_0005699 influenced AS progression by targeting miR-384. MiR-384 has been verified to be a key player in a variety of cancers, including lung cancer [20], breast cancer [21], prostate cancer [22] and gastric cancer [23]. Furthermore, Zhang et al. claimed that upregulation of miR-384 increased high glucose-evoked HUVECs viability, inhibited apoptosis, release of inflammatory factor and oxidative stress process via down-regulating LIN28B [24]. Fan et al. highlighted overexpression of miR-384 elevated the proliferation and angiogenesis of endothelial progenitor cells by hindering the expression of DLL4 in the pathogenesis of cerebral ischemic stroke [25]. The above studies showed that miR-384 was involved in endothelial cell injury. In AS, we demonstrated that miR-384 level was down-regulated in response to ox-LDL-stimulated HUVECs in a dose-dependent manner. Additionally, rescue experiments identified that circ_0005699 accelerated ox-LDL-stimulated damage in HUVECs partly via abating the level of miR-384. Interestingly, it was first confirmed that circ_0005699 deficiency retarded ox-LDL-evoked cell injury via upregulation of miR-384.
ASPH was verified to interact with miR-384 in HUVECs. ASPH is a type II transmembrane protein belonging to the family of α-ketoglutarate-dependent dioxygenase [26]. Xiao et al. stated that miR-206 hampered inflammation injury and oxidative stress injury via reducing the level of ASPH in the development of AS disease [11]. In our work, we verified that up-regulation of ASPH partially reversed miR-384 mimic-mediated effect on ox-LDL-evoked injury in HUVECs. This further elucidates that ASPH can inhibit the proliferation and tube formation of HUVECs, accelerate the process of apoptosis, inflammation and oxidative stress, and clarify the effect of ASPH on endothelial cells. Collectively, we concluded that circ_0005699 silencing influenced cell viability, proliferation, apoptosis and angiogenesis ability of ox-LDL-induced HUVECs by reducing ASPH expression via elevating the expression of miR-384.
In summary, circ_0005699 was overexpressed in AS patients and ox-LDL-induced HUVECs. Interference of circ_0005699 hindered ox-LDL-treated HUVECs dysfunction via miR-384/ASPH axis, which implied that circ_0005699 might be an effective therapeutic strategy for AS.
Data Availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Wolf, D., & Ley, K. (2019). Immunity and inflammation in atherosclerosis. Circulation Research, 124(2), 315–327.
Kattoor, A. J., Pothineni, N. V. K., Palagiri, D., & Mehta, J. L. (2017). Oxidative stress in atherosclerosis. Current Atherosclerosis Reports, 19(11), 42.
Kosta, T., & Boon, R. A. (2018). Endothelial cell metabolism in atherosclerosis. Frontiers in Cell and Developmental Biology, 6, 82.
Paone, S., Baxter, A. A., Hulett, M. D., & Poon, I. K. (2019). Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis. Cellular and Molecular Life Sciences, 76(6), 1093–1106.
Gao, S., Zhao, D., Wang, M., Zhao, F., Han, X., Qi, Y., & Liu, J. (2017). Association between circulating oxidized LDL and atherosclerotic cardiovascular disease: A meta-analysis of observational studies. Canadian Journal of Cardiology, 33(12), 1624–1632.
Bian, W., Jing, X., Yang, Z., Shi, Z., Chen, R., Xu, A., Wang, N., Jiang, J., Yang, C., Zhang, D., & Li, L. (2020). Downregulation of LncRNA NORAD promotes Ox-LDL-induced vascular endothelial cell injury and atherosclerosis. Aging (Albany NY), 12(7), 6385–6400.
Shang, Q., Yang, Z., Jia, R., & Ge, S. (2019). The novel roles of circRNAs in human cancer. Molecular Cancer, 18(1), 6.
Cao, Q., Guo, Z., Du, S., Ling, H., & Song, C. (2020). Circular RNAs in the pathogenesis of atherosclerosis. Life Sciences, 255, 117837.
Ji, P., Song, X., & Lv, Z. (2021). Knockdown of circ_0004104 alleviates oxidized low-density lipoprotein-induced vascular endothelial cell injury by regulating miR-100/TNFAIP8 Axis. Journal of Cardiovascular Pharmacology, 78(2), 269–279.
Huang, J. G., Tang, X., Wang, J. J., Liu, J., Chen, P., & Sun, Y. (2021). A circular RNA, circUSP36, accelerates endothelial cell dysfunction in atherosclerosis by adsorbing miR-637 to enhance WNT4 expression. Bioengineered, 12(1), 6759–6770.
Wang, X., & Bai, M. (2021). CircTM7SF3 contributes to oxidized low-density lipoprotein-induced apoptosis, inflammation and oxidative stress through targeting miR-206/ASPH axis in atherosclerosis cell model in vitro. BMC Cardiovascular Disorders, 21(1), 51.
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Advances in Experimental Medicine and Biology, 1087, 67–79.
Churov, A., Summerhill, V., Grechko, A., Orekhova, V., & Orekhov, A. (2019). MicroRNAs as potential biomarkers in atherosclerosis. International Journal of Molecular Sciences, 20(22), 5547.
Lin, H., Pan, S., Meng, L., Zhou, C., Jiang, C., Ji, Z., Chi, J., & Guo, H. (2017). MicroRNA-384-mediated Herpud1 upregulation promotes angiotensin II-induced endothelial cell apoptosis. Biochemical and Biophysical Research Communications, 488(3), 453–460.
Cristiano, F., & Martina, M. (2018). Atherosclerosis is an inflammatory disease which lacks a common anti-inflammatory therapy: How human genetics can help to this issue. A narrative review. Frontiers in Pharmacology, 9, 55.
Zhu, Y., Xian, X., Wang, Z., Bi, Y., Chen, Q., Han, X., Tang, D., & Chen, R. (2018). Research progress on the relationship between atherosclerosis and inflammation. Biomolecules, 8(3), 80.
Zhang, F., Zhang, R., Zhang, X., Wu, Y., Li, X., Zhang, S., Hou, W., Ding, Y., Tian, J., Sun, L., & Kong, X. (2018). Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging, 10, 2266–2273.
Peng, H., Sun, J., Li, Y., Zhang, Y., & Zhong, Y. (2021). Circ-USP9X inhibition reduces ox-LDL-induced endothelial cell injury via the miR-599/CLIC4 axis. Journal of Cardiovascular Pharmacology, 78(4), 560–571.
Li, S., Huang, T., Qin, L., & Yin, L. (2021). Circ_0068087 silencing ameliorates oxidized low-density lipoprotein-induced dysfunction in vascular endothelial cells depending on mir-186-5p-mediated regulation of roundabout guidance receptor 1. Frontiers in Cardiovascular Medicine, 8, 650374.
Ma, Q., Huai, B., Liu, Y., Jia, Z., & Zhao, Q. (2021). Circular RNA circ_0020123 promotes non-small cell lung cancer progression through miR-384/TRIM44 axis. Cancer Management and Research, 13, 75–87.
Wang, Q., Liang, D., Shen, P., Yu, Y., Yan, Y., & You, W. (2021). Hsa_circ_0092276 promotes doxorubicin resistance in breast cancer cells by regulating autophagy via miR-348/ATG7 axis. Translational Oncology, 14(8), 101045.
Shi, Z., Zhang, H., Jie, S., Yang, X., Huang, Q., Mao, Y., & Zhang, Y. (2021). Long non-coding RNA SNHG8 promotes prostate cancer progression through repressing miR-384 and up-regulating HOXB7. The Journal of Gene Medicine, 23(3), e3309.
Liu, P., Cai, S., & Li, N. (2020). Circular RNA-hsa-circ-0000670 promotes gastric cancer progression through the microRNA-384/SIX4 axis. Experimental Cell Research, 394(2), 112141.
Zhang, W., & Sui, Y. (2020). CircBPTF knockdown ameliorates high glucose-induced inflammatory injuries and oxidative stress by targeting the miR-384/LIN28B axis in human umbilical vein endothelial cells. Molecular and Cellular Biochemistry, 471, 101–111.
Fan, J., Xu, W., Nan, S., Chang, M., & Zhang, Y. (2020). MicroRNA-384-5p promotes endothelial progenitor cell proliferation and angiogenesis in cerebral ischemic stroke through the delta-likeligand 4-mediated notch signaling pathway. Cerebrovascular Diseases (Basel, Switzerland)., 49(1), 39–54.
Chen, X., Jin, P., Tang, H., & Zhang, L. (2019). miR-135a acts as a tumor suppressor by targeting ASPH in endometrial cancer. International Journal of Clinical and Experimental Pathology, 12(9), 3384–3389.
Funding
None.
Author information
Authors and Affiliations
Contributions
Cangcang Liu designed and supervised the study. Xiaobiao Cao conducted the experiments and drafted the manuscript. Jun Yang collected and analyzed the data. Lujun He contributed the methodology, operated the software and edited the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical Approval
This study was approved by the Ethics Committee of Chinese People Liberation Army (PLA) 93864 Military Hospital.
Consent to Participate
All of them had signed written informed consents.
Consent to Participate
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Daniel Conklin.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12012_2024_9889_MOESM1_ESM.tif
Supplementary Figure 1. Treatment of ox-LDL induced cytotoxicity in HUVECs. (A) CCK8 assay, (B) EdU assay, (C) flow cytometry, (D-E) Tube formation assay were conducted to evaluate the viability, proliferation, apoptosis and angiogenesis ability with different concentrations of ox-LDL (0, 25, 50, 100 μg/mL), respectively. (F) The protein levels of Bax and Cleaved-caspase-3 were tested via western blot. (G) ELISA was used to investigate the production of IL-1β and TNF-α in ox-LDL-induced HUVECs. (H-I) The MDA Assay Kit and SOD Assay Kit were used to analyze the production of MDA and SOD, respectively. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. This experiment was performed for three times with three technical repetitions. one-way ANOVA was utilized to assess the diferences in (A–I). Supplementary file1 (TIF 1416 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, X., Yang, J., He, L. et al. Circ_0005699 Expedites ox-LDL-Triggered Endothelial Cell Injury via Targeting miR-384/ASPH Axis. Cardiovasc Toxicol 24, 1067–1076 (2024). https://doi.org/10.1007/s12012-024-09889-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12012-024-09889-8