Abstract
Eighty-nine cultured Pseudomonas species isolated from the sediment and water samples collected from five industrial Red Sea regions that have been affected by petroleum and industry. Genotypic (exoT, exoS, exoU, exoY, lasA, lasB, rhlA, rhlB, Pf1, PAGI-1, -2, and -3) and phenotypic (DNase, elastase, lipase, protease, siderophore, antibiotic resistance patterns) characteristics were determined. Out of these isolates, nine Pseudomonas isolates were selected as the hyperactive virulence factors producers along with highly resistant pattern against all antibiotics of different classes included in this study. They were subjected to phenotypic and chemotypic characterization as well as molecular identification through 16S rRNA gene amplification and sequencing. The bioactive metabolites of these nine strains were extracted by ethyl acetate followed by evaluating their cytotoxic activity toward liver tissues, kidney tissues, and other biochemical activities in rat. Both EGY6 and EGY8 caused the highest significant reduction in the levels of packed cell volume (PCV), red blood cell count (RBC), and hemoglobin (Hb), which indicate that these Pseudomonas strain metabolites could cause anemia and toxic effects on hematological values in animals that were infected with them. Rats treated with the most toxic extract, EGY8, showed severe histopathological alterations in liver and kidney.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Red Sea is one of the most spectacular coastal and marine environments in Egypt and has a rich biodiversity. Critical pollution of the Red Sea has been stated multiple times from Egypt, Yemen, Saudi Arabia, Jordan, Sudan, and Eritrea [1]. The causes of pollution in the Red Sea contain land-based (comprising urban development, industrial activities, dredging and filling, tourism and agriculture activities) and oceanic sources (shipping, fishing, marine traffic and petroleum industries) as well as industries or port activities. Such dangerous pollution is likely to affect biological life and disrupt the Red Sea’s natural ecosystems. One of the major topics that have not been well studied is the influence of Red Sea pollution on microbial life especially human pathogens along the Red Sea coast which are leading factors in the emergence of new pathogens involving those associated with the water systems [2].
Pseudomonas species are eminent members of emerging waterborne pathogens that are usually associated with high resistance to antibiotic treatment and high mortality rates [3]. The Pseudomonas’ intrinsic and acquired resistance of many structurally unrelated antibiotics is because of several adaptations containing active efflux systems, decreased cell wall permeability, plasmid acquisition, and expression of different enzymes or by biofilm creation [4]. Pseudomonas pathogenesis includes production of both extracellular and cell-linked virulence factors that are responsible for causing inflammation leading to septic shock or organ failure [5]. During treatment of infection, high amount of virulence factors are released in blood circulation due to immediate lyses of bacteria, and since liver helps in clearing them from the body, it remains the primary target to be stimulated by virulence factors releasing vigorous amount of inflammatory molecules leading to liver injury [6, 7].
Here, we aimed to isolate and identify cultured Pseudomonas species from sediment and water samples collected from industrial Red Sea locations that have been affected by petroleum and industry. Moreover, determining their genotypic (exoT, exoS, exoU, exoY, lasA, lasB, rhlA, rhlB, Pf1, PAGI-1, -2, and -3) and phenotypic characteristics (DNase, elastase, lipase, protease, siderophore, and antibiotic resistance patterns), extracting their bioactive metabolites by ethyl acetate and evaluating their cytotoxic activity toward liver and kidney tissues as well as other biochemical activities in rat. According to our knowledge, neglected investigations have been conducted on pathogenic microbiota in Red Sea (sediments or water) along with determining the ability of their ethyl acetate extracts to induce toxicity in different tissues in rats.
Materials and Methods
Sample Collection, Isolation, and Identification of Cultural Pseudomonas sp. Isolates
Five Egyptian coastal sites on the Red Sea were selected for sampling and isolation of Pseudomonas species based on the industrial impacts affecting those including Safaga, Abu Ghoson, Qusseir, Hamrawein, and Sharm El-Maya ports. The seawater samples and sediments of these sites were transferred to laboratory in ice tank and kept at 4 °C until processing. The cultural Pseudomonas-like organisms were isolated by serial dilution technique using Luria-Bertani (LB) agar medium containing glucose, cetrimide, 50% seawater supplemented with 5% human blood (obtained from VACSERA, Egypt), and ampicillin, which are typically suited for the growth of Pseudomonas isolates [8] and Pseudomonas isolation agar (PIA; BD Diagnostic Systems, USA). The inoculated plates were incubated at 37 °C for 24 to 48 h until the colonies appeared. All presumptive typical Pseudomonas colonies that can grow in LB agar medium and appearing blue-green on PIA agar plates were picked up from agar plates and purified. The selected nine isolates were characterized according to Cowan and Steel’s manual for the identification of medical bacteria [9], the criteria of Bergey’s manual of determinative bacteriology [10], Koneman et al. [11], McFaddin [12], and Kiska and Gilligan [13]. Moreover, fatty acids of Pseudomonas sp. were extracted and analyzed according to Sasser [14].
Characterization of Isolated Cultural Pseudomonas sp. Isolates
Biofilm Formation
Biofilm was generated on sterile Foley’s catheter under static conditions for 7 days. Catheter pieces were transferred to fresh medium after every 24 h. Each day, catheter pieces in duplicate were removed, and rinsed with phosphate-buffered saline (PBS; pH 7.2). Cells were scrapped from catheter surface with sterile scalpel blade, disrupted and dispersed sample was centrifuged, and the pellet was suspended in 1 mL PBS. Biofilm cells were cultured on nutrient agar plates to determine log CFU by standard tube dilution method [15].
Detection of Virulence Factors by Phenotypic Method
Different virulence factors elaborated by Pseudomonas sp. in vitro were estimated in the culture supernatant of biofilm cells. DNase production was determined after spot inoculation of DNase test agar (Difco) plates, incubation at 37 °C for 24 to 48 h, flooding the plate surface with 1.0 N HCl. DNase secreted by the bacteria colonies hydrolyze the DNA in the medium resulting in clearance around the bacterial growth [16]. Elastase activity was determined by the elastin Congo red assay. Overnight growth culture in liquid LB medium was centrifuged at 13,000 rpm for 15 min, and then, 100 μL of the supernatant was added to 900 μL of 100 mM Tris-1 mM CaCl2 (pH 7.5) containing 1 mg of elastin Congo red. The mixture was incubated with shaking at 37 °C for 24 h followed by centrifugation, and then, optical density of the supernatants was measured at 495 nm [17]. Moreover, phospholipase C activity was measured by diameter of hydrolysis of egg yolk around the colony when egg yolk agar inoculated with colonies from 18- to 24-h culture, incubated at 37 °C for 24–48 h [18]. Proteolytic activity was measured according to the method of Prasad et al. [17]. The protease activity was detected in the supernatants at 595 nm.
On the other hand, siderophore production was determined according to the method of Sokol et al. [19]. Cell-free supernatants (CFSs) were extracted with ethyl acetate. For pyochelin, 1 mL each of 0.5 N HCl, nitrite molybdate reagent, and 1 N NaOH were added. Final volume was made to 5 mL with sterile distilled water and absorbance was read at 510 nm. For pyoverdin estimation, fluorescence of 3 mL of culture supernatant in 50 mM Tris-HCl (pH 7.4) was measured at 460 nm while the samples were excited at 400 nm [20]. Quantitative determination of both cell-free and cell-bound hemolysin using 2% suspension of washed human erythrocytes was done following the method of Linkish and Vogt [21], and then, absorbance of supernatant was read at 545 nm and hemolysin in mg mL−1 was calculated by using lyophilized hemoglobin as standard. Alginate was precipitated by mixing culture supernatant with 2% (w/v) cetylpyridinium. Amount of alginate was determined using a borate/carbazole method [22]. To determined cell surface hydrophobicity, cell culture supernatant was mixed with equal volume of p-xylene, and after phase separation, initial optical density and optical density of the aqueous phase were taken and surface hydrophobicity was expressed as percent [23].
PCR Analysis of Virulence-Associated Genes
The presence of several virulence-associated genes encoding exotoxins (T, exoT; S, exoS; U, exoU; and Y, exoY), elastase (lasA and lasB), quorum-sensing mediators (rhlA and rhlB), the presence of prophage (Pf1), and genomic islands (PAGI-1, -2, and -3) were assessed by PCR amplification as previously described [24], except the primers for rhlA (rhlAF, 5′-GCGCGAAAGTCTGTTGGTAT and rhlAR, 5′-CAGGTGATTGACCTCGAAGC) and rhlB, (rhlBF, 5′-GAGCGACGAACTGACCTACC and rhlBR, 5′-GTTGAACTTGGGGTGTACCG). Amplification of virulence-associated genes was performed using puReTaq™ Ready-To-Go™ PCR Beads (GE Healthcare) and carried out using the following protocol: 94 °C for 1 min, 35 cycles consisting of 94 °C for 1 min, 54 °C for 2 min, and 72 °C for 2 min and a final extension step at of 72 °C for 8 min [24].
Each gene was amplified in a separate reaction to obviate possible competition among primers. The presence of genomic islands was investigated using the following parameters: initial denaturation at 96 °C for 5 min, 35 cycles at 94 °C for 30 s, 47 to 64 °C at 30 s, 1 to 4 min at 72 °C, and final extension for 8 min. PCR products were separated in a 1% agarose gel for 1 h at 100 V, stained with ethidium bromide, and detected by UV trans-illumination. Amplified genes were identified on the basis of fragment size, and reference strain PAO1 was used as the positive control [25].
Antibiotic Sensitivity Patterns and Multi-Antibiotic Resistant Value Index
Values interpreted into susceptible (S), intermediate (I), or resistant (R) to 30 antimicrobial agents were determined on Mueller-Hinton agar using standard disk diffusion technique according to the criteria recommended by the Clinical and Laboratory Standards Institute (CLSI, 2010) [26]. The antibiotics used in this study are cefixime (CEF), cedipime (CED), cefradoxil (CEFR), ceftazidime (CEFT), cefuroxime (CEFU), imipenem (IMI), piperacillin (PIP), carbenicillin (CAR), ciprofloxacin (CIP), ofloxacin (OFL), gentamycin (GEN), amikacin (AMI), penicillin G (PEN-G), ampicillin (AMP), oxacillin (OXA), cefotaxime (CEFO), cefepime (CEFE), cephalothin (CEP), sulfamethoxazole (SULP), rifampin (RIF), nalidixic acid (NAL), ampicillin/sulbactam (AM/SUL), chloramphenicol (CHL), tetracycline (TET), minocycline (MIN), erythromycin (ERY), vancomycin (VAN), nitrofurantoin (NIT), and clindamycin (CLI) (Oxoid, UK). Multi-antibiotic resistant (MAR) value index for each isolate was calculated by following the procedure described by Olayinka et al. [27] as follows: MAR = a/b, where a is the number of antibiotics to which the isolate was resistant and b is the total number of antibiotics against which individual isolate was tested.
Molecular Identification of Selected Pseudomonas sp. Isolates Through 16S Ribosomal RNA Gene Sequencing
Genomic DNA extraction, PCR amplification of the 16S ribosomal RNA (rRNA) gene, purification of PCR products, gel electrophoreses, and the 16S rRNA gene sequencing were done as mentioned previously [28,29,30]. The 16S rRNA gene sequences of the selected nine Pseudomonas sp. isolates were aligned with published sequences in the NCBI GenBank database of 16S rRNA gene sequences. The tree topologies were evaluated by maximum-likelihood and bootstrap analyses based on 1000 replications with MEGA6 [31, 32].
Extraction of Secondary Metabolites
The isolated strains were cultivated in 500-mL Erlenmeyer flasks each containing 100 mL of LB medium supplemented with 5% blood, and fermentation was carried out for 2 days at 37 °C with shaking at 180 rpm. After fermentation, the whole broth of each isolate was extracted twice with ethyl acetate at pH 4 and then evaporated to dryness. The crude extracts were took a number similar to the number of its own Pseudomonas strain.
Animals and Experimental Design
Two-hundred and seventy adult male rats weighting between 100 and 120 g used in this experiment were maintained and had free access to clean freshwater at the Animal Division, Biological Sciences Department, Faculty of Sciences, King Abdulaziz University. These animals were fed on standard diet and divided into nine groups of 30 rats per group for each Pseudomonas strain extract used in this study. In each group, the 30 animals were divided into six subgroups, each consists of five rats. While the first three subgroups corresponded to the treatment with such extract for 10, 20, and 40 days of daily treated with the extract supplemented diet of oral dose 0.08 g kg−1 diet individually, the remaining subgroups served as control for these treatments after 10, 20, and 40 days.
Technique for Obtaining Blood and Serum Samples
Blood was collected by cardiac puncture from chloroform-anesthetized rats into heparinized bottles for hematological studies. A further blood sample was collected into a clean bottle (non-heparinized) and allowed to clot. The serum was separated and centrifuged according to groups into clean bottles for biochemical analysis.
Determination of Hematological Parameters
Hemoglobin concentration, erythrocyte count, total leukocyte, and leukocyte differential counts were estimated using the method of Jain [33]; packed cell volume (PCV) was determined according to Schalm et al. [34]. Erythrocyte indices were determined from values obtained from RBC, hemoglobin, and PCV values. The results obtained were statistically analyzed by the method of Kurtz [35].
Determination of Serum Biochemical Parameters
Following decapitation of each animal, blood was collected in clean centrifuge tube placed at 4 °C for 2 to 3 h and then centrifuged at 3000 rpm for 15 min. Serum was separated and kept at −20 °C till biochemical analysis begin. Total protein was calculated by Biuret reaction; aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were determined according to Reitman and Frankel [36], but alkaline phosphatase was determined by Burtis et al. [37]. Serum creatinine and urea concentration were determined by the method of Patton and Crouch [38]. Tissue and serum malondialdehyde (MDA) levels were measured according to the method of Ohkawa et al. [39]. Absorbance of the product of reaction was measured at 532 nm and expressed as nmol mL−1 in serum and nmol mg−1 of protein in liver tissues. Paraoxonase (PON) and arylesterase (AE) activities were measured by using paraoxon and phenylacetate as the substrates, respectively, as previously described [40]. Paraoxonase activity was taken at 412 nm and calculated as U L−1 in serum and mU mg−1 protein in liver tissues, while arylesterase activity was taken at 270 nm and calculated as kU L−1 in serum and U mg−1 protein in liver tissues. Whereas lactate dehydrogenase (LDH) was determined as U L−1 in serum, caspase-3 activity was measured as μM pNA/h/mg protein in liver tissues with commercial micro plate kit according to the manufacturer’s instructions (Chemicon International Temecula Co., USA) as described by Beltowski et al. [40]. The results obtained were statistically analyzed by the method of Kurtz [35].
Histopathological Examination
After collection of blood for biochemical estimation, the rats were sacrificed and the tissue samples viz. kidneys and liver were carefully dissected, cleaned of extraneous tissue, and fixed in 10% neutral buffered formalin (NBF) for at least 24 h. Then, the paraffin sections were prepared (automatic tissue processor, auto-technique) and cut into 5-μm-thick sections using a rotary microtome. The tissue sections of 5 μm size were stained by hematoxylin and eosin dye and then examined for histopathological changes as suggested by Chouhan et al. [41].
Results and Discussions
Isolation and Distribution of Pseudomonas Isolates in Some Industrial Zones of the Red Sea
Sediments and water samples collected from five coastal locations that have been affected by petroleum and industry along the Egyptian Red Sea coast were analyzed for the distribution of Pseudomonas species. A total of 89 Pseudomonas colonies which are Gram-negative, oxidase-positive, catalase-positive, aerobic, rod-shaped, and had the ability to grow on LB agar medium having glucose and cetrimide appended with ampicillin were chosen (Table 1). The highest number of isolates was obtained from the sediments of Quesser port (19 strains, 21.35%) followed by sediments of Safaga aluminum (13 strains, 14.61%), sediments of Hamrawein (11 strains, 12.36%), and the water of Hamrawein ports (10 strains, 11.24%) but the lowest number obtained from the water samples of Sharm El-Maya (5 strains, 5.62%) and Abu Ghoson ports (4 strains, 4.49%). Seventy five (84.27%) of all isolates produced pigment. The pigment formation ability was ranging from 100% in isolates derived from the sediments of Sharm El-Maya and Abu Ghoson ports as well as isolates obtained from the water of Quesser and Abu Ghoson ports to 50% in isolates obtained from water of Hamrawein port (Table 1). Finlayson and Brown [42] reported that pigment productivity seemed to be more considerably connected with multi-drug resistance, existence of virulence-related genes, and expression of proven virulence factors. Overall, except for isolates from sediment and water samples of Quesser and Abu Ghoson ports, respectively, producers for the virulence factor pyoverdin (69 isolates, 77.53%) were more frequently and significantly higher than pyocyanin producers (43 isolates, 48.32%; p < 0.05) (Table 1). Cheluvappa et al. [43] suggested that Pseudomonas aeruginosa virulence factor, pyocyanin, has toxic effect on the liver sinusoidal endothelial cell, while Wagner et al. [4] reported that pyoverdin is a virulence factor encoded by the pvd genes in P. aeruginosa to act as a siderophore, which implicated in a complex iron acquisition system and to be a critical component in biofilm creation.
Distribution of Virulence Factors in Pseudomonas Isolates
Data in Table 2 indicated that the isolates that yielded either pyocyanin or pyoverdin were capable to yield additional virulence factors, containing hemolysin, biofilm production, DNase, phospholipase, elastase, protease, and siderophore. Whereas all isolates from the Quesser port expressed biofilm production, only 94.74, 57.14, 53.85, and 20% of isolates derived from Safaga aluminum, Hamrawein, Sharm El-Maya, and Abu Ghoson ports, respectively, were biofilm producers. Moreover, all 89 isolates obtained in this study exhibited phospholipase C activity, but among them, only 73 (82.02%), 61 (68.55%), 69 (77.53%), 77 (86.52%), and 68 (76.41%) isolates showed hemolysin, DNase, elastase, protease, and siderophore activities, respectively. The pathogenesis of P. aeruginosa is mainly due to production of several cell-associated and secreted extracellular virulence factors which include pyocyanin, pyoverdin, hemolysin, phospholipase, elastase (metallo-protease), and alkaline protease that can destroy a large variation of tissue components such as proteinaceous elements of connective tissue and cell surface receptors on neutrophils [44]. Our data are in line with Khalil et al. [45], in which 95.2, 81, and 78% of Pseudomonas species were positive for hemolysin, phospholipase, and gelatinase, respectively, and Pramodhini et al. [44] who reported that 80.3, 70, and 71.4% of P. aeruginosa isolates were positive for hemolysin, phospholipase, and gelatinase.
Detection of Virulence Genes in Pseudomonas Isolates
Isolates were evaluated for the existence or nonexistence of 12 genetic regions linked with virulence in Pseudomonas species, and we found that 11 (12.36%), 21 (23.6%), 36 (40.45%), 53 (59.55%), 65 (73.07%), 12 (13.84%), 7 (7.87), 5 (5.62%), and 4 (4.49%) of total isolates were PCR-positive for lasA, lasB, rhlA, rhlB, exoS, exoT, PAGI-1, PAGI-2, and exoU genes, respectively (Table 2). Furthermore, only three isolates (3.37%) obtained from Safaga aluminum and Hamrawein ports were positive for both pf1and PAGI-3 genes, but exoY was found in one isolate obtained from Hamrawein port (Table 2). The detection of these virulence genes in Pseudomonas isolates highly important due to the lasA protein has been revealed to be implicated in the final processing of elastase. lasB locus is answerable for the regulation of elastase and protease. rhl system regulates productivity of rhamnolipid, which has hemolytic properties, and it is essential for full elastase activity. exoS gene has been associated with the capability of Pseudomonas isolates to prevalence from epithelial colonization spots to the blood stream, and it reason effects on multiple cellular processes with inhibition of DNA synthesis and interference of cell matrix adherence. exoT has been involved in increased cellular and toxic effects in trial models and human infections. PAGI-1, PAGI-2, and PAGI-3 are virulence genes located in the chromosome, analogous to plasmid-encoded regulators of chromosome genes [4, 46]. Finlayson and Brown [42] reported that the analysis and comparison of virulence genes provide an overall understanding of the disease causing factors and their genetic regulation as well as providing a means of identifying the subsets of parameters in characterizing a pathogen.
Antimicrobial Receptivity Types of Pseudomonas Isolates
Table 3 displays the resistance types of Pseudomonas isolates to each antibiotic used. All Pseudomonas isolates showed complete (100%) resistance to penicillin G, ampicillin, oxacillin, cefotaxime, cephalothin, sulfamethoxazole, rifampin, and nalidixic acid. Moreover, the isolates of Safaga aluminum and Hamrawein ports exhibited complete resistant (100%) against ampicillin/sulbactam while isolates of Quesser and Abu Ghoson ports showed complete resistant (100%) against both ampicillin/sulbactam and chloramphenicol antibiotics. On the other hand, Pseudomonas isolates under study showed the highest sensitivity profile among Sharm El-Maya isolates (76.92, 92.31, 76.92, 84.61, 100, 76.92, and 84.62%) against seven antibiotics in common use including cefuroxime, imipenem, ciprofloxacin, gentamycin, erythromycin, nitrofurantoin, and clindamycin, respectively, followed by isolates of Safaga aluminum port (89.47, 78.95, and 73.58% of isolates were sensitive to imipenem, gentamycin, and erythromycin, respectively), isolates of Quesser port (80.77 and 73.08% of isolates were sensitive to gentamycin and nitrofurantoin, respectively), and Abu Ghoson port isolates (70 and 80% of isolates showed sensitivity to ciprofloxacin and nitrofurantoin, respectively) (Table 3). The high level of resistant toward all antibiotics that detected in Hamrawein port might be attributed to the highest content of virulence factors detected in these isolates. Proportionate with the opinion of this investigation, Olayinka et al. [27] and Odjadjare et al. [47] reported that Pseudomonas species were reported to be frequently resistant to antibiotics with high resistance to ampicillin, penicillin G, oxacillin, rifampin, sulfamethoxazole, cefepime, and cephems, but they exhibited high sensitivity to gentamicin, clindamycin, erythromycin, and erythromycin.
Out of 89 Pseudomonas isolates tested, nine marine Pseudomonas isolates designated as Pseudomonas sp. EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9 were selected for further studies depending on they were the hyperactive virulence factor producers (hemolysin, biofilm production, DNase, phospholipase, elastase, protease, and siderophore; p < 0.05) and their high degree of resistant against different antibiotics. Then, their MAR indices, quantitative production of some severe virulence factors involved in pathogenesis such as UEC adhesion, alginate, pyochelin, cell surface hydrophobicity, cell-bound hemolysin, cell-free hemolysin, phospholipase C, and malondialdehyde formation, were evaluated and they were subjected to phenotypic and chemotypic characterization and molecular identification. Moreover, the assessment of the biochemical and cytotoxic effects of their ethyl acetate extracts in rats was performed.
The MAR Index of Selected Pseudomonas Isolates
The MAR index was varied from 0.47 to 7.0 (Table 4). The ratio of isolates with MAR index greater than 0.5 was 77.78%, while those with MAR index equal to or less than 0.5 was 11.11% (Table 4). Compared to genotypic characterization, the MAR indexing method is simple, cost-effective, rapidly easy to perform, and does not need specialized training and expensive equipment, and tracking MAR analysis can been applied to differentiate bacteria from different sources using antibiotics [47]. Furthermore, Navon-Venezia et al. [48] reported that MAR index values larger than 0.2 indicate high-risk source of contamination. Consequently, the higher MAR index in our study more than 0.2 limits indicates that the isolates in this investigation originated from high-risk sources of contamination, where antibiotics are often used. The observation was not surprising in these polluted areas of the Red Sea.
Characterization and Identification of Selected Pseudomonas Isolates
All isolates were individual, motile by means of a polar flagellum, Gram-negative, non-endospore forming, aerobic, rod-shaped, and had the capability to raise on MacConkey, Mueller Hinton, blood agar, and LB agar medium containing glucose and cetrimide added with ampicillin (Table 5). Moreover, they were positive for oxidase, catalase, and cytochrome oxidase as well as hydrolyze casein. On the other hand, they gave negative results for H2S, and indole production, Voges-Proskauer reaction, and starch hydrolysis. Whereas all strains were unable to grow on lactose, rhamnose, trehalose, sucrose, raffinose, and inositol, they assimilate glucose, fructose, N-acetylglucosamine, gluconate, and malonate (Table 5). The diameter of the cell of these isolates was ranged between 0.7 × 2.5- and 1.0 × 4-μm diameters, and the G-C content of genomic DNA (mol%) of these isolates was within the range of 59.2 to 64.5 that was previously measured in species of Pseudomonas [49]. Moreover, they showed that colony color ranged between light creamy, light yellow, pale yellow, to greenish yellow. Some other physiochemical properties of strains are presented in Table 5.
From data in Table 6, a total of 14 altered patterns of fatty acids were observed in EGY4 and EGY5, while 10, 9, 12, 11, 12, 11, and 13 fatty acid types were identified from EGY1, EGY2, EGY3, EGY6, EGY7, EGY8, and EGY9, respectively. The fatty acid compositions of all isolates were much different, which was observed in the concentrations of the saturated fatty acids C10:0 3OH (11.5, 9.5, 3.3, 6.0, 10.6, 3.5, 2.5, 5, and 4.72%), C12:0 3OH (5.1, 4.7, 3.9, 3.4, 4.0, 4.7, 2.93, 1.22, and 5.00%), C12:1 3OH (2.1, 1.6, 0.1, 1.6, 1.2, 0.0, 2.22, 0.76, and 0.0%), C12:0 2OH (5.5, 5.7, 3.4, 4.3, 3.8, 6.0, 7.00, 4.38, and 7.10%), C12:0 (0.0, 0.0, 3.0, 2.0, 1.9, 4.1, 0.0, 0.0, and 6.1%), C14:0 (0.4, 0.5, 0.2, 13.3, 0.3, 3.42, 17.40, 0.90, and 15.36%), C16:0 (19.6, 14.25, 10.1, 10.5, 23.5, 20.0, 27.81, 32.11, and 23.20%), and C18:0 (0.3, 0.0, 0.2, 19.9, 0.6, 0.0, 2.0, 1.18, and 6.88%) and unsaturated fatty acids 18:1ω7c (0.0, 0.0, 22.4, 5.2, 23.5, 9.13, 6.19, 11.25, and 4.0%), 17:0 cyclo (2.8, 12.4, 1.0, 1.15, 2.8, 0.0, 0.0, 0.0, and 2.30%), 19:0 cyclo ω8c (0.0, 0.0, 0.0, 5.2, 0.2, 2.0, 1.2, 0.0, and 5.23%), 16:1 ω7c (20.31, 19.65, 36.4, 20.2, 25.0, 14.0, 18.16, 13.0, and 10.11%), Is0–15:0 2OH (0.0, 0.0, 16.0, 4.79, 1.2, 9.0, 3.15, 12.0, and 4.00%), and C18:1(cis∆7/ cis∆9/ cis∆12) (11.22, 31.7, 0.0, 2.64, 1.4, 24.1, 10.0, 18.2, and 1.2%) detected in the total fatty acid contents of EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7 EGY8, and EGY9, respectively (Table 6).
According to their morphology, pigmentation, Gram reaction, specific confirmatory biochemical tests, and fatty acid analysis, these isolates were characterized as members of Pseudomonas species (Tables 5 and 6). Kiska and Gilligan [13] reported that the principles for identifying an isolate as Pseudomonas are pigment production, growth at 42 °C, oxidase, and catalase positivity.
Phylogenetic Analyses of the Selected Nine Pseudomonas Isolates
The 16S rRNA gene sequences of the selected Pseudomonas isolates EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9 (accession numbers MF093523, MF093524, MF093525, MF093526, MF093527, MF093528, MF093529, MF093530, and MF093531, respectively) were compared to the sequences of members of the order Pseudomonadales and family Pseudomonadaceae. It was detected that the EGY1, EGY4, and EGY9 strains show 99.78, 99.79, and 100% homology to P. migulae, P. putida 1106, and P. aeruginosa (CEMC_P1 and RS1), respectively (Fig. 1). On the other hand, the isolates EGY2 and EGY8 showed 99.79 and 99.73% homology to P. fluorescens LMG 14677, respectively. The sequence results indicate that the four isolates EGY3, EGY5, EGY6, and EGY7 were only distantly related to their closest described relatives, which include 99.19, 99.66, 99.43, and 99.65%, respectively, identity to P. protegens (strain-type Darke, Clinton, Wayne, and 38G2). While the isolates EGY5 and EGY7 showed 100% homology to Pseudomonas sp. LD 53 and Pseudomonas sp. LM 63, respectively. Moreover, the isolate EGY7 exhibited 99.93% similarly to P. syringae KB11 (Fig. 1). Based on the combination of morphological, physiological, and chemotaxonomic as well as16S rRNA gene analyses and phylogenetic data, it was concluded that the isolates EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9 are species of the genus Pseudomonas and hence were given the names as Pseudomonas sp. EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9, respectively.
Quantitative Expression of Some Virulence Factors Implicated in Pathogenesis of Selected Marine Pseudomonas Isolates
In this study, phenotypic expression of virulence factors involved into pathogenesis of these nine strains was determined quantitatively. Biofilm was generated on the catheter surface in all strains, and its formation was maximum on the third day of time course of biofilm formation in EGY8 followed by EGY6 and EGY5 (log CFU equal to 17.1, 15.8, and 15.6, respectively) but the lowest biofilm formation was detected in EGY 4 (log CFU 11.9) (Fig. 2). Whereas there was slight decline on the fourth day of biofilm formation in EGY1, EGY3, EGY5, EGY6, EGY7, and EGY8 strains, in strains under the isolation code EGY2, EGY4, and EGY9, it was reduced on day 5. Biofilm formation on the catheter surface increases the susceptibility of biofilm cells to the action of antibiotics and hostile factors [24]. However, production of QS signals was ranged between 129 and 178 (Miller units) in all strains under study and it was declined significantly after 5 days (Fig. 3). Formation of biofilm and production of virulence factors are largely regulated by cell-to-cell communication, which is known as quorum sensing (QS). Without QS, the expression of virulence factor reduces significantly and results in no virulence of the organism [50].
Data in Table 7 showed that all selected Pseudomonas strains had high ability to adhere to the UECs, and the average number of cells adhered was detected to be 57 ± 2.0, 45 ± 1.3, 60 ± 1.8, 58 ± 1.6, 61 ± 2.0, 66 ± 1.8, 51 ± 1.5, 71 ± 2.2, and 69 ± 2.0 in EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9, respectively, but alginate production as an important component of biofilm exopolysaccharide that contributes to external morphology of biofilm and tissue colonization during infection was ranged between 488 ± 13.1 μg mL−1 in EGY1 strain and 612 ± 9.3 μg mL−1 in EGY8 strain. Moreover, siderophore (pyochelin) that trap iron from the host tissues was produced in high amounts in all strains but the highest absorbance was detected in EGY8 (0.34 ± 0.03) followed by EGY6 (0.31 ± 0.01), while for cell surface hydrophobicity (another mechanism of bacterial adherence), all isolates showed potent adherence abilities (Table 7), which might result in the success of establishment of infection in the urinary tract and liver. Significantly, amounts of both cell-bound hemolysin (3.9 ± 0.16, 4.1 ± 0.20, 4.3 ± 0.17, 5.0 ± 0.21, 4.6 ± 0.19, 5.1 ± 0.21, 4.8 ± 0.23, 5.5 ± 0.25, and 4.7 ± 0.24 mg mL−1) as well as cell-free hemolysin (3.2 ± 0.10, 4.2 ± 0.14, 4.0 ± 0.14, 3.6 ± 0.09, 4.0 ± 0.13, 4.2 ± 0.11, 3.9 ± 0.10, 4.5 ± 0.14, and 4.1 ± 0.11 mg mL−1) were detected in EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9, respectively. Whereas the highest phospholipase C activity was detected in EGY8 (32.1 ± 0.35 nm) followed by EGY6 (30.3 ± 0.30 nm), the lowest amount was observed in EGY7 (22.7 ± 0.27 nm) and EGY2 (19.8 ± 0.20 nm) (Table 7). By comparing levels of these virulence factor formation detected in these isolates, the virulence potential of EGY8 followed by EGY6 were the highest among all strains which refer to powerful virulence processes in these strains that lead to the development of antibiotic resistance and toxicity. Our data are in line with Harjai et al. [50], who reported that this organism produces several cell-associated and extracellular virulence factors including alginate, protease, elastase, rhamnolipids, exotoxin A, exoenzyme S, siderophores (pyochelin, pyoverdin), and hemolysins, in addition to biofilm formation on the catheter surfaces, which are usually resistant to conventional antibiotics and host immune defense mechanism [3].
Effect of Ethyl Acetate Extracts on Hematological Parameters in Rat
Data in Table 8 indicated that ethyl acetate extracts of all Pseudomonas strains cause a gradual decrease in hematological values (packed cell volume, red blood cell counts, and hemoglobin concentration). Both EGY6 and EGY8 caused the highest statistically significant reduction (p < 0.05) in the levels of PCV (reduced from 37.1% in control group to 31.1 and 31.9%, respectively), RBC (reduced from 7.1 U L−1 in control group to 5.2 and 5.0 U L−1, respectively), and Hb (reduced from 11.9 g dL−1 in control group to 7.9 and 7.6 g dL−1, respectively) (Table 8), which indicate that these Pseudomonas strain metabolites could cause anemia and toxic effects on hematological values in animals that were infected with them. Moreover, EGY5, EGY8, and EGY9 extracts caused a significant decrease (p < 0.05) in TWBC (from 5.1 × 103/mL in control group to 3.9, 3.5, and 2.6 × 103/mL, respectively) as well as lymphocyte level (from 3.0 × 103/mL in control group to 2.3, 2.1, and 1.5 × 103/mL, respectively). Therefore, the continual contact to these bacteria may then drive to lymphopenia, which may have an immunosuppressive effect and affect phagocyte function, which is to defend against invading microorganisms. Thus, participation to cellular inflammatory processes may be compromised [3]. It should be noted that some extracts have significant toxic effects on neutrophils, which play an essential role in the pathogenesis. Extracts of EGY6 and EGY8 induced a significant elevation in the levels of total protein (p > 0.05), which may be attributed to the refusal of animals to drink water as a result of inclusion of these extracts, which can itself lead to dehydration. Hall et al. [3] previously reported that Pseudomonas virulence factors such as PCN caused neutrophilia, inhibit the release of interleukin (IL)-2, and reduce the expression of IL-2 receptors on T cells, which lead to inhibition of lymphocyte proliferation that resulting in a reduced of immune response and hence potentially protecting Pseudomonas species from the immune system.
Effect of Ethyl Acetate Extracts on Serum and Some Tissue Parameters
The extracts of the selected nine strains induced a significant increase in the serum levels of AST, ALT, and alkaline phosphatase (ALP) after the administration of each Pseudomonas extract supplemented diet for 10, 20, and 40 days of treatments, respectively (Table 9). It seems that the changes in serum ALT and AST activities are due to cellular degradation by Pseudomonas extracts on the liver or heart muscle. The elevation in the level of AST activity produced by these extracts is an indication of damaged organs and cell necrosis of many tissues [4]. Since ALT is existent in liver and it is one of the specific assayable liver enzymes, its elevated level in this investigation may indicate hepatic damage induced by these Pseudomonas strain extracts. ALP has many physiological functions in bone cells; it splits inorganic phosphates from organic phosphate which is a powerful inhibitor of mineralization [4, 51]. The results of this experiment exhibited significant increase in ALP activities for Pseudomonas extracts that can be attributed to the defect in liver function [3]. This study stating that these strains have toxic potentials and then the continuous exposure to infection by them may lead to morbidity and mortality.
However, Table 9 shows that urea and creatinine levels were significantly elevated following repeated administration of each extract dose of each Pseudomonas sp. that attributed to impairment of renal function. Furthermore, there was markedly continuous increase in serum levels of urea, creatinase, ALT, AST, and ALP activities with increasing the period of treatment.
As shown in Table 10, whereas MDA and AE activities in serum significantly decreased, they were significantly increased in liver tissue of all test groups treated with EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9 extracts individually. They were significantly increased (p < 0.05) compared to the control group, but PON was significantly decreased in both serum and liver tissue of all treated groups with the extracts under study (p < 0.05). Moreover, LDH in serum was significantly decreased but caspase-3 activity in liver tissue was significantly increased in all test groups after administration of extracts (Table 10). Cheluvappa et al. [52] mentioned that P. aeruginosa produces a number of virulence factors containing pyocyanin that induce liver sinusoidal endothelial cells and acute non-oxidative hepatic injury beside another activities including redox activity, immunomodulation, pro-inflammatory effects, generation of reactive oxidative species, succinic dehydrogenase enzyme inactivation, cytotoxicity, pro-apoptotic effects, and induction of senescence. Interestingly, Hall et al. [3] reported that whereas the highest concentration of the toxic material extracted from Pseudomonas occur in the kidney where it is degraded, the greatest toxic effect takes place in the liver.
Histopathological Effects of Pseudomonas sp. EGY8 Extract on Liver and Kidney of Treated Rats
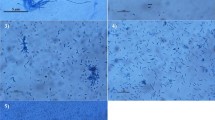
Histopathological effects of the most toxic extract, EGY8, on the liver and kidney of treated rats are presented in Figs. 4a, b and 5a, b, respectively. Rats treated with EGY8 extract showed severe histopathological alterations. The microscopic examination of liver showed derangement of hepatic cords with granular changes in cytoplasm, multifocal swelling of hepatocytes with congestion of central and portal blood vessels, and focal degenerative and necrotic changes along with mononuclear cell infiltration (Fig. 4b). On the other hand, histological evaluation of kidney of treated rats with EGY8 extract (Fig. 5b) showed moderate to severe nephropathic changes and distinct cellular alterations with marked degenerative, necrobiotic changes with inflammatory reaction, tubular alterations like diffuse tubular swelling, and loss of tubular epithelium. Focal areas showed degenerative changes in glomeruli with atrophy were observed. Similar cytotoxic observations were reported [3, 4, 43, 52, 53].
Photomicrographs for liver sections stained with hematoxylin and eosin. a Control rat showing the normal histological architecture of rat liver. b Rats liver treated with Pseudomonas sp. EGY8 extract, which showed severe hepatic lesions, necrotic hepatocytes, cell infiltration lymphocyte, and fatty changes with hydropic degeneration
Photomicrographs of kidney tissue stained with hematoxylin and eosin. a Control rat showing the normal histological structure of rat kidney. b Rats kidney treated with Pseudomonas sp. EGY8 extract showed severe nephropathic changes with distinct cellular alterations, degenerative and necrobiotic changes, and loss of tubular epithelium in both proximal and distal tubules
Conclusion
Sediments and water samples collected from five coastal locations that have been affected by petroleum and industry along the Egyptian Red Sea coast were analyzed for the distribution of Pseudomonas species. Out of 89 Pseudomonas isolates tested, nine marine Pseudomonas isolates designated as Pseudomonas sp. EGY1, EGY2, EGY3, EGY4, EGY5, EGY6, EGY7, EGY8, and EGY9 were selected. They were the hyperactive virulence factor producers (hemolysin, biofilm production, DNase, phospholipase, elastase, protease, and siderophore) and their high degree of resistant against different antibiotics. They were subjected to phenotypic and chemotypic characterization as well as molecular identification.
Moreover, the assessment of the biochemical and cytotoxic effects of their ethyl acetate extracts in rats were determined. Ethyl acetate extracts of all Pseudomonas strains caused a gradual decrease in hematological values. Both EGY6 and EGY8 caused the highest significant reduction in the levels of PCV, RBC, and Hb, which indicate that these Pseudomonas strain metabolites could cause anemia and toxic effects on hematological values in animals that were infected with them. Rats treated with the most toxic extract, EGY8, showed severe histopathological alterations in liver and kidney.
References
Mustafa, G. A., Abd-Elgawad, A., Abdel-Haleem, A. M., & Siam, R. (2014). Egypt’s Red Sea coast: phylogenetic analysis of cultured microbial consortia in industrialized sites. Frontiers in Microbiology, 5, 363. doi:10.3389/fmicb.2014.00363.
Thompson, L. R., Field, C., Romanuk, T., Ngugi, D. K., Siam, R., Dorry, H. E., et al. (2013). Patterns of ecological specialization among microbial populations in the Red Sea and diverse oligotrophic marine environments. Ecology and Evolution, 3, 1780–1797. doi:10.1002/ece3.593.
Hall, S., McDermott, C., Anoopkumar-Dukie, S., McFarland, A. J., Forbes, A., Perkins, A. V., Davey, K. A., Chess-Williams, R., Kiefel, M. J., Arora, D., & Grant, G. D. (2016). Cellular effects of pyocyanin, a secreted virulence factor of Pseudomonas aeruginosa. Toxins, 8, 236. doi:10.3390/toxins8080236.
Wagner, V. E., Filiatrault, M. J., Picardo, K. F., & Iglewski, B. H. (2008). Pseudomonas aeruginosa: virulence and pathogenesis issues. In P. Cornelis (Ed.), Pseudomonas: Genomics and molecular biology (pp. 129–158). Norfolk: Caister Academic Press.
Luczkiewicz, A., Kotlarska, E., Artichowicz, W., Tarasewicz, K., & Fudala-Ksiazek, S. (2015). Antimicrobial resistance of Pseudomonas spp. isolated from wastewater and wastewater-impacted marine coastal zone. Environmental Science and Pollution Research, 22, 19823–19834.
El-Gendy, M. M. A. A., Abdel-Wahhab, K. G., Mannaa, F. A., Farghaly, A. A., & El-Bondkly, A. M. A. (2017). Carcinogenic activities and sperm abnormalities of methicillin resistance Staphylococcus aureus and inhibition of their virulence potentials by ayamycin. Applied Biochemistry and Biotechnology. doi:10.1007/s12010-017-2467-7.
Kumar, L., Chhibber, S., & Harjai, K. (2014). Hepatoprotective effect of zingerone [4-(4-hydroxy-3-methoxyphenyl) butan-2-one] in lipopolysaccharide induced liver injury mouse model through down regulation of inflammatory mediators. International Journal of Pharmacognosy and Phytochemical Research, 6(2), 308–316.
Brown, V. I., & Lowbury, E. J. L. (1965). Use of an improved cetrimide agar medium and of culture methods for Pseudomonas aeruginosa. Journal of Clinical Pathology, 18, 752–756.
Barrow, G. I., & Feltham, R. K. A. (1993). Cowan and Steel’s manual for the identification of medical bacteria (3rd ed.). Cambridge: Cambridge University Press. doi:10.1017/CBO9780511527104.
Holt, J. G., Krieg, N. R., Sneath, P. H. A., Staley, J. T. & Williams, S. T. (1994). Bergey’s manual of determinative bacteriology, 9th edition. Baltimore: Williams & Wilkins.
Koneman, E. W., Allen, S. D., Janda, W. M., Schrenkenberger, P. C., & Winn, W. C. (1997). Colour atlas and textbook of diagnostic microbiology (5th ed.). PA: Lippincott, Williams & Wilkins.
McFaddin, J. F. (2000). Biochemical tests for the identification of medical bacteria (3rd ed.pp. 170–182). Philadelphia, Pa: The Lippincott, Williams and Wilkins Co..
Kiska, D. L., & Gilligan, P. H. (2003). Pseudomonas. In P. R. Murray (Ed.), Manual of clinical microbiology (Vol. 47, 8th ed., pp. 719–728). Washington DC: ASM Press.
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. Newark, DE: MIDI Inc..
Mittal, R., Chhibber, S., Sharma, S., & Harjai, K. (2006). Effect of macrophage secretary products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comparative Immunology, Microbiology and Infectious Diseases, 29, 12–26.
Colle, J. G., Miles, R. S., & Wan, B. (1996). Tests for the identification of bacteria. In J. G. Collee, A. G. Fraser, B. P. Marmion, & A. Simmons (Eds.), Mackie and Mc Cartney practical medical microbiology (14th ed., pp. 131–150). Churchill Livingstone: Edinburg.
Prasad, S. V., Ballal, M., & Shivananda, P. G. (2009). Slime production a virulence marker in Pseudomonas aeruginosa strains isolated from clinical and environmental specimens: a comparative study of two methods. Indian Journal of Pathology & Microbiology, 52, 191–193.
Pavlov, D., De Wet, C. M. E., Grabow, W. O. K., & Ehlers, M. M. (2004). Potentially pathogenic features of heterotrophic plate count bacteria isolated from treated and untreated drinking water. International Journal of Food Microbiology, 92, 275–287.
Sokol, R. J., Ager, J., Martier, S., Debanne, S., Ernhart, C., Kuzma, J., & Miller, S. I. (1986). Significant determinants of susceptibility to alcohol teratogenicity. Annals of the New York Academy of Sciences, 477, 87–102.
Ankenbauer, R., Sriyosachati, S., & Cox, C. D. (1985). Effects of siderophores on the growth of Pseudomonas aeruginosa in human serum and transferrin. Infection and Immunity, 49, 132–140.
Linkish, P. G., & Vogt, W. (1972). Direct haemolytic activity of phospholipase A. Biochimica et Biophysica Acta, 270, 241–247.
Mathee, K., Ciofu, O., Sternberg, C., Lindum, P. W., Campbell, J. I., Jensen, P., et al. (1999). Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology, 145, 1349–1357.
Rosenberg, M., Gutinick, D., & Rosenberg, E. (1980). Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiology Letters, 9, 29–33.
Head, N. E., & Yu, H. (2004). Cross sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infection and Immunity, 72, 133–144.
Finnan, S., Morrissey, J. P., O’Gara, F., & Boyd, E. F. (2004). Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. Journal of Clinical Microbiology, 42, 5783–5792.
Clinical and Laboratory Standard Institute (CLSI). (2010). Performance standards for antimicrobial susceptibility testing; 20 th informational supplement. CLSI document M100-S20. CLSI, Wayne, PA: Clinical and Laboratory Standard Institute.
Olayinka, B. O., Olonitola, O. S., Olayinka, A. T., & Agada, E. A. (2004). Antibiotic susceptibility pattern and multiple antibiotic resistance index of Pseudomonas aeruginosa urine isolates from a University Teaching Hospital. African Journal of Clinical and Experimental Microbiology, 5(2), 198–202.
El-Bondkly, A. M., El-Gendy, M. M. A. A., Wiese, J., & Imhoff, J. F. (2012). Phylogenetic diversity and antimicrobial activities of culturable endophytic actinobacteria isolated from different Egyptian marine sponges and soft corals. Australian Journal of Basic and Applied Sciences, 6(4), 25–33.
El-Bondkly, A. M. A., El-Gendy, M. M. A. A., & Bassyouni, R. H. (2012). Overproduction and biological activity of prodigiosin-like pigments from recombinant fusant of endophytic marine Streptomyces species. Antonie Van Leeuwenhoek, 102, 719–734.
El-Gendy, M. M. A. A., & El-Bondkly, A. M. A. (2016). Evaluation and enhancement of heavy metals bioremediation in aqueous solutions by Nocardiopsis sp. MORSY1948, and Nocardia sp. MORSY2014. Brazilian Journal of Microbiology, 47, 571–586.
Roth, A., Andrees, S., Kroppenstedt, R. M., Harmsen, D., & Mauch, H. (2003). Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals under speciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. Journal of Clinical Microbiology, 41, 851–856.
Saitou, N., & Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425.
Jain, N. C. (1986). Schalm’s veterinary haematology (4th ed.pp. 564–572). Philadelphia: Lea and Fabiger.
Schalm, O. W., Jain, N. C., & Caroll, E. J. (1975). Veterinary haematology (3rd ed.). Philadelphia: Lea and Fabiger.
Kurtz, N. R. (1983). Introduction of social statistics (p. 163). N.Y: McGraw, Hill Book Co..
Reitman, S., & Frankel, S. (1957). A colorimetric method for determination of serum glutamic oxaloacetate and glutamic pyrumic transminase. American Journal of Clinical Pathology, 28, 26–34.
Burtis, C. A., Ashwood, E. R., & Bruns, D. E. (2008). Tietz fundamentals of clinical chemistry. Sawyer, B. G. (Ed.), Saunders. St. Louis: Elsevier.
Patton, C. J., & Crouch, S. R. (1977). Spectrophotometric and kinetics investigation of the Berthelot reaction for the determination of ammonia. Analytical Chemistry, 49(3), 464–469. doi:10.1021/ac50011a034.
Ohkawa, H., Ohishi, N., & Yagi, K. (1979). Assay for lipid peroxides animal tissues by thiobarbituric acid reaction. Analytical Biochemistry, 95, 351–358.
Beltowski, J., Jamroz-Wiśniewska, A., Borkowska, E., & Wójcicka, G. (2005). Differential effect of antioxidant treatment on plasma and tissue paraoxonase activity in hyperleptinemic rats. Pharmacological Research, 51, 523–532.
Chouhan, R. S., Ahmad, B., Chauhan, R., & Shrivastava, V. K. (2014). Ameliorative role of ascorbic acid against hepatotoxicity induced by chemotherapeutic drug cyclophosphamide in male albino rat. International Journal of Biological & Pharmaceutical Research, 5(11), 830–842.
Finlayson, E. A., & Brown, P. D. (2011). Comparison of antibiotic resistance and virulence factors in pigmented and non-pigmented Pseudomonas aeruginosa. The West Indian Medical Journal, 60(1), 24.
Cheluvappa, R., Jamieson, H. A., Hilmer, S. N., Muller, M., & Le Couteur, D. G. (2007). The effect of Pseudomonas aeruginosa virulence factor, pyocyanin, on the liver sinusoidal endothelial cell. Gastroenterología y Hepatología, 22(8), 1350–1351.
Pramodhini, S., Umadevi, S., & Seetha, K. S. (2016). Detection of virulence determinants and its association with drug resistance in clinical isolates of Pseudomonas aeruginosa. International Journal of Research in Medical Sciences, 4(9), 3917–3923.
Khalil, M. A. E. F., Sonbol, F. I., Mohamed, A. F. B., & Sameh, S. A. (2015). Comparative study of virulence factors among ESβL-producing and nonproducing Pseudomonas aeruginosa clinical isolates. Turkish Journal of Medical Sciences, 45, 60–69.
Feltman, H., Schulert, G., Khan, S., Jain, M., Peterson, L., & Hauser, A. R. (2001). Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Journal of Microbiology, 147, 2659–2669.
Odjadjare, E. E., Igbinosa, E. O., Mordi, R., Igere, B., Igeleke, C. L., & Okoh, A. I. (2012). Prevalence of multiple antibiotics resistant (MAR) Pseudomonas species in the final effluents of three municipal wastewater treatment facilities in South Africa. International Journal of Environmental Research and Public Health, 9, 2092–2107.
Navon-Venezia, S., Ben-Ami, R., & Carmeli, Y. (2005). Update on Pseudomonas aeruginosa and Acinetobacter baumannii infections in the healthcare setting. Current Opinion in Infectious Diseases, 18, 306–313.
Zzaman, M. H., Umadhay-Briones, K. M., Zsiros, S. M., Nodasaka, Y., Yumoto, I., & Okuyama, H. (2004). Isolation, identification, and characterization of a novel, oil-degrading bacterium, Pseudomonas aeruginosa T1. Current Microbiology, 49, 108–114.
Harjai, K., Gupta, R. K., & Sehgal, H. (2014). Attenuation of quorum sensing controlled virulence of Pseudomonas aeruginosa by cranberry. The Indian Journal of Medical Research, 139, 446–453.
Charles, P., Hasting, C., Ristelli, J., Mosekilde, L., & Eriksen, E. F. (1992). Assessment of bone formation by biochemical markers in metabolic bone disease: separation between osteopathic activity at the cell and tissue level. Calcified Tissue International, 51, 406–411.
Cheluvappa, R., Cogger, V. C., Kwun, S. Y., Reilly, J. N., Couteur, D. G. L., & Hilmer, S. N. (2008). Liver sinusoidal endothelial cells and acute non-oxidative hepatic injury induced by Pseudomonas aeruginosa pyocyanin. International Journal of Experimental Pathology, 89, 410–418.
Schümann, J., Angermüller, S., Bang, R., Lohoff, M., & Tiegs, G. (1998). Acute hepatotoxicity of Pseudomonas aeruginosa exotoxin A in mice depends on T cells and TNF. Journal of Immunology, 161, 5745–5754.
Contributors
El-Gendy M. M. A. A., Al-Zahrani H. A. A., Abozinadah N. Y., and El-Bondkly A. M. designed the study, preformed the experiments, managed the literature searches and data analysis, and wrote the manuscript. All the authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
El-Gendy, M.M.A.A., Al-Zahrani, H.A.A., Abozinadah, N.Y. et al. In Vivo Evaluation of the Toxic Effect of Ethyl Acetate Extracts of Marine Antibiotic Resistance Pseudomonas Species Derived from the Red Sea. Appl Biochem Biotechnol 184, 323–349 (2018). https://doi.org/10.1007/s12010-017-2553-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2553-x