Abstract
Multi-parameter flow cytometry was used to monitor cell intrinsic light scatter, viability, and lipid content of Rhodotorula glutinis CCMI 145 cells grown in shake flasks. Changes in the side light scatter and forward light scatter were detected during the yeast batch growth, which were attributed to the different yeast growth phases. A progressive increase in the proportion of cells stained with PI (cells with permeabilized cytoplasmic membrane) was observed during the yeast growth, attaining 79% at the end of the fermentation. A high correlation between the Nile Red fluorescence intensity measured by flow cytometry and total lipid content assayed by the traditional gravimetric lipid analysis was found for this yeast, making this method a suitable and quick technique for the screening of yeast strains for lipid production and optimization of biofuel production bioprocesses. Medium growth optimization for enhancement of the yeast oil production is now in progress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial oils can be used as feedstock for biodiesel production. Compared to other vegetable oils and animal fats, the production of microbial oil has many advantages: short life cycle, less labor required, not affected by season and climate, and easier to scale-up [1]. Therefore, microbial oils might become one of potential oil.
At present, most of the works aiming at microbial biodiesel production uses autotrophic microalgae [2]. However, it is difficult to achieve high biomass concentration and microalgal oil productivities. This is due to unsolved problems, namely light limitation and oxygen accumulation, in photoautotrophic cultures. Although microalgae have faster growth rates than oil seed crops, their rate of growth is much slower than bacteria or fungi. Biodiesel production from microorganisms other than microalgae may have additional benefits, such as higher growth rates and lipid yields, which can reduce the costs of microbial biomass and oil productions. The yeast Rhodoturula glutinis has been reported as a promising oil source for biodiesel production [3, 4].
Despite all of its benefits, microbial lipid production is strongly dependent on environmental factors, such as the carbon source, C/N ratio, oxygen availability, etc. Therefore, it is essential to monitor cell lipid content during the microbial lipid production process. At present, most of the published works studying the production of biodiesel from yeasts used traditional microbiological techniques to monitor the yeast lipid content [1, 3–5], which are time consuming and generate high amounts of waste (organic solvent) which are harmful to the environment if not recycled by distillation. In addition, enough amounts of biomass must be obtained for subsequent lipid extraction and derivatization [6]. Importantly, lipid content data is usually only available a considerable time after the sample is taken, too late for alterations to be made to process control. Therefore, the question that must be answered is how to measure accurately and quickly cellular oil content when optimizing microbial oil production or scaling up biofuel production bioprocesses so that informed decisions on process control can be made.
Multi-parameter flow cytometry can monitor total cell lipid content, in situ, near real time and with a high degree of statistical resolution during the yeast growth. If the cell lipid content information is available during the time course of the bioprocess, decisions on process control strategy can be made (for example, adjusting the carbon/nitrogen ratio of the medium based on the output from the flow cytometry) so that lipid productivity can be increased. In addition, the selection of oleaginous yeast strains, usually a time-consuming step, can be performed in a faster way, if using flow cytometry to quantify intracellular lipids.
Lipid measurement has been previously proposed using the Nile Red fluorescent stain for quantification of microbial lipids [6–10]. Its fluorescence is produced in highly hydrophobic environments and quenched in hydrophilic ones.
So far, most of the works aiming at the biodiesel production from microorganisms, including yeasts, used classical microbiological techniques to monitor cell proliferation [1, 3–5] which beset a number of problems. Optical density and dry cell weight measurements, although indicative of proliferation, provide no information on cell physiological state and rarely take into account changes in cell size [11, 12]. Indeed, throughout the course of any microbial process, it is essential to monitor cell viability, as a high proportion of dead cells present in any part of the bioprocess will be also detrimental, as such cells, although containing lipids in their structure, do not accumulate oil as the metabolically active cells, thus decreasing the process yield. Such information allows decisions on process control in order to reduce the dead cells’ proportion in the culture. This allows products to be harvested at optimum concentrations, so that high oil yields can be achieved. Multi-parameter flow cytometry is capable of providing such information rapidly with a high degree of accuracy using propidium iodide, a fluorescent strain which is usually used to study the cell membrane integrity [10].
Therefore, when producing oil from microorganims, it is crucial to know their physiological response to different environmental growth conditions. Flow cytometry has been widely used for determination of viability and physiological states in yeast using different dyes [13–16]. Regarding the yeast lipid content, Raschkle and Knorr [17] reported the use of flow cytometry to monitor Waltomyces lipofer cell vitality and internal lipid droplets, the latter being a qualitative method of cellular lipid content evaluation using Nile Red. Kimura et al. [8] reported a quantitative spectrophotometric method of lipid content assessment in oleaginous fungi and yeasts using Nile Red. However, no information on microbial physiological states was given in this work.
In the present work, R. glutinis batch growth was monitored by flow cytometry. During the yeast growth, biomass, cell viability and lipid content were monitored. A correlation between the Nile Red fluorescence intensity measured by flow cytometry and the yeast lipid content assayed by the traditional extraction method was established. As far as we are aware, this is the first work reporting the use of the fluorescent stain Nile Red to quantify the yeast lipid content by flow cytometry.
Materials and Methods
Organism
The yeast R. glutinis was deposited in the Industrial Micro-organisms Culture Collection (Instituto Nacional de Engenharia, Tecnologia e Inovação, Lisbon, Portugal), with the reference code CCMI 175. Lyophilized cell cultures of the yeast were also used to provide control data.
Growth Conditions
The yeast R. glutinis was maintained in yeast malt agar. Cultures were grown in 1,000 ml baffled shake flasks containing 100 ml of a medium containing KH2PO4 0.4 g/l, NH4Cl 2 g/l, yeast extract 1 g/l, glucose 28 g/l, maintained at 30°C, 180 rpm, in darkness. After 1 day (exponential growth phase), this culture was used to inoculate the shake flasks. Baffled shake flasks (1,000 ml) contained 100 ml of culture medium with 10% (v/v) of exponential-growing cultures in the same conditions as described above. A control experiment was carried out under the same conditions, but without microorganism. R. glutinis growth was followed for 48 h.
Growth Evaluation
R. glutinis growth was evaluated as average of three independent replicates and was periodically evaluated by optical density at 540 nm, which was converted into dry cell weight per liter of culture by a regression equation.
Residual Glucose Concentration
The residual glucose concentration in filtered (0.45 µm) samples was evaluated as average of three independent replicates using the 3,5-dinitrosalicylic acid method [18].
Lipid Content
Total lipid content was evaluated by the traditional method according to Xiong et al. 2008 [19]. In each sampling, the culture of three independent Erlenmeyers was collected, centrifuged, and freeze dried for further lipid analysis, which was performed in triplicate.
Multi-parameter flow cytometry used Nile Red (Riedel de Haën, Buchs SG, Switzerland) according to the method described by da Silva et al. [10]. A working solution of Nile Red (10 µl) in acetone (0.033 mg/ml) were added to 1 ml of a cell suspension (∼106 cells/ml). This mixture was gently vortex and incubated for 2 min at 37°C in darkness. For this stain concentration, it was observed that R. glutinis cells stained with Nile Red displayed the highest fluorescence after incubation for 2 min at 37°C. After 2 min, the fluorescence faded. Therefore, 2 min staining period was chosen for further experiments.
Nile Red florescence was determined using a FACScan flow cytometer (Becton-Dickinson Instruments, Erembodegem, Belgium) equipped with a 488-nm argon laser. Upon excitation by a 488-nm argon laser, Nile Red exhibits yellow-gold and red fluorescence when dissolved in neutral and polar lipids which are detected by the FL2 and FL3 channels, respectively. Non-stained cells were used as an autofluorescence control measured in FL2 and FL3 (AF2 and AF3, respectively) which were always set to the same pre-fixed fluorescence value in all experiments. The Nile Red fluorescence intensity average was obtained from three independent replicates. The total normalized fluorescence corresponding to total cellular lipids was determined as the sum of the ratios FL2/AF2 and FL3/AF3. A correlation between total yeast cell lipid content assayed by traditional Soxhlet extraction method [19] and Nile Red fluorescence intensity was established by analyzing yeast samples taken at different times of the batch growth.
Cell Viability
Flow cytometry coupled with propidium iodide (PI) (Invitrogen, Carlsbad, USA) was used in this study in order to monitor R. glutinis cytoplasmic membrane integrity. PI binds to DNA but cannot cross an intact cytoplasmic membrane. Samples taken from the culture were immediately diluted (at least 1:2,000 v/v) with phosphate buffer solution (PBS, pH 7.0) and stained with PI. PI stock solution was made up at 2 mg/ml in distilled water. The working concentration of PI was 10 μg/ml. PI was excited at 488 nm and measured at 585 nm (FL2).
All solutions used in flow cytometry were passed through a 0.2-µm filter immediately prior to use to remove particulate contamination. In addition, the control software was set on both the light-scattering properties in the forward angle direction (FS) signal and the right angle direction (SS) signal.
Results and Discussion
Cell Viability Flow Cytometry Controls
In order to demonstrate that it is possible to use multi-parameter flow cytometry to characterize the cell membrane integrity of individual cells of R. glutinis fermentations, it was necessary to establish a positive control. This was then used for comparison with data produced from cultivation experiments.
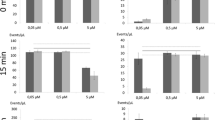
R. glutinis cells collected from a 48-h slant and resuspended in PBS were stained with PI (Fig. 1a). One major population (A) (88%), no staining, could be seen corresponding to cells with an intact cytoplasmic membrane, and a minor population (B) (11%), PI stained, was identified as dead cells. Such proportion of PI negative cells showed that most of the yeast cells removed from the slant were still viable.
a Cells collected from a 48-h slant, resuspended in PBS and stained with PI. One population exist (a), corresponding to cells with intact cytoplasmic membrane, no staining. bR. glutinis cells collected from a 48-h slant, resuspended in PBS and killed in a water bath at 100°C for 10 min and stained with PI. One main subpopulation (b) could be distinguished, corresponding to cells with permeabilized cytoplasmic membrane. cR. glutinis cells grown in shake flasks for 96 h and stained with PI. Two subpopulations could be distinguished. These correspond to cells with intact cytoplasmic membrane (a), no staining (9%), and cells with permeabilized cytoplasmic membrane (b), PI staining, (92%). The subpopulation percentages of three independent replicates were affected by a relative error not exceeding 10% (n = 3)
R. glutinis cells removed from the slant and resuspended in PBS were heat treated in a water bath at 100°C during 10 min and stained with PI. One major population (B) (97%), PI staining, corresponding to cells with permeabilized cytoplasmic membranes, could be identified (Fig. 1b).
In the same way, cells from a R. glutinis starved old culture (96 h) were stained with PI (Fig. 1c). Again, it was observed that a major population (B) (91%), PI staining, was composed of permeabilized cells.
The heat treatment and the starvation induced the permeabilization of the yeast cytoplasmic membrane, although the first treatment resulted in a higher proportion of permeabilized cells (97%) as compared to the starvation (91%).
Shake Flask Fermentations
Figure 2 shows the dry cell weight and glucose uptake during the yeast fermentation. In order obtain a number of samples with different lipid contents, cells were harvested during the time course of the batch growth. In this way, cells would have experienced different nutrient environments (abundance, limitation, and starvation), inducing different cellular lipid contents.
Glucose concentration decreased from 28 (t = 0 h) to 21 g/l (t = 28.25 h) and afterwards slightly decreased until the end of the fermentation, suggesting that a nutrient other than carbon limited the yeast growth after t = 28.25 h. Biomass concentration slowly increased even during the stationary phase, suggesting that oxygen could be limiting the yeast growth. The specific growth rate calculated during the exponential phase was µ = 1.2/h, and the maximum biomass concentration attained was 3 g/l at t = 28.25 h.
Light Scatter Measurements
It has been reported that bacteria, yeast, and microalgae can be detected from the background on the basis of their intrinsic light-scattering properties in forward angle light scatter (FS) and right angle light scatter (SS) [20–22].
In the present work, light scatter measurements of R. glutinis were carried out during the time course of the batch growth. Three dimensional (FS and SS) plots and microscopical observations of samples taken at different times are depicted in Fig. 3a–j.
a, c, e, g, i Intrinsic light-scattering data for R. glutinis cells taken during the batch growth at t = 19, 24.25, 28.25, 42.25, and 47.25 h, respectively. b, d, f, h, j Microscope observations for R. glutinis during the batch growth at t = 19, 24.25, 28.25, 42.25, and 47.25 h, respectively (magnification ×1,000)
In all cases, a broad distribution with respect to FS and SS density plot was observed, suggesting cell size and shape heterogeneity, which may be due to the heterogeneity of the asynchronous exponential-growing yeast population [23]. However, different FS/SS profiles were shown according to the yeast growth phase.
At the exponential phase (t = 24.25 h and t = 28.25 h), two well-defined peaks A and B were observed (Fig. 3c, e) in contrast with the lag phase FS/SS plot (Fig. 3a). These two peaks most probably correspond to single yeast cells (A) and budding cells (B) larger in size and internal complexity due to their higher DNA content which has been previously reported [13]. This is supported by Fig. 3d, f wherein a high proportion of budding cells can be observed, in contrast with the lag phase optical observation showing just a few budding cells (Fig. 3b).
As the culture entered the stationary phase, cells became depleted in a nutrient, and the yeast cell division slowed down. The two peaks, A and B, observed during the exponential phase became progressive in one single peak (Fig. 3g, i). Indeed, the optical observations taken during the stationary phase (t = 40.25 h and t = 47.25 h) also showed a gradual reduction of budding cells (Fig. 3h, j) which may explain the more homogeneous distribution observed at t = 47.25 h.
In this way, it was possible to identify a typical FS/SS profile for each yeast cell growth phase. Such finding may be useful when monitoring industrial or scale-up yeast processes.
The FS and SS profiles detected during the time course of the yeast fermentation are shown in Fig. 4. While the FS signal decreased during the time course of the fermentation, the SS signal attained its maximum at t = 24.25 h and afterwards started decreasing until the end of the fermentation. The SS maximum could be due to the presence of a high proportion of budding yeast cells present at t = 28.25 h as such cells would depict a higher refractive index due to the higher DNA content, as already stated.
R. glutinis Cell Viability
Figure 5 shows the PI fluorescence density plots of R. glutinis samples taken during the shake flask fermentation at different times. A progressive increase in the proportion of PI staining cells with permeabilized membranes was observed. At the stationary phase (t = 47.25 h), the proportion of PI positive cells attained 79%. Such proportion at this stage could be due to the oxygen limitation mentioned above, as R. glutinis, known as an obligate aerobic, is dependent on oxygen for its energy metabolism and synthesis of cellular components [24].
a–e Density plots of cells taken at t = 19, 24.25, 28.25, 40.25, 47.25 h, respectively, during the yeast batch growth and staining with PI. Two main subpopulations could be distinguished. These correspond to cells with intact cytoplasmic membrane(a), no staining, and cells with permeabilized cytoplasmic membrane (b), PI staining. The subpopulation percentages of three independent replicates were affected by a relative error not exceeding 10% (n = 3)
The increase in the proportion of dead cells detected during the yeast growth could also explain the decrease in FS and SS average signals, particularly after t = 24.25 h (Fig. 4), as dead cells have their cytoplasmic membrane permeabilized, leaking out their internal content, therefore becoming shrunk and smaller. In fact, back-gating analysis confirmed that the region (similar to a tail) depicting the lowest FS/SS signals observed in all the FS/SS density plots (Fig. 3a, c, e, g, i) corresponded to dead cells.
Correlation Between Nile Red Fluorescence and Yeast Lipid Content
In order to obtain a correlation between Nile Red fluorescence and the yeast cellular lipid content, cells taken during the yeast batch growth were stained with Nile Red, and the fluorescence was measured by flow cytometry. The yeast lipids from R. glutinis were extracted in a Soxhelet apparatus using n-hexane as solvent [19].
Figure 6a shows the cellular lipid content determined by the gravimetric method and by Nile Red fluorescence. The maximum lipid content was detected at t = 24.25 h (11% w/w). Interestingly, this lipid maximum coincided with the SS maximum signal (Fig. 4) which could also be due to the production of intracellular lipids which may increase the refractive index, thus the SS signal.
a Total cellular lipid content assayed by the gravimetric method [19] and R. glutinis cell fluorescence when stained with Nile Red, assessed by flow cytometry during the yeast batch growth. Lipid content obtained from three independent replicates by the gravimetric extraction method was affected by a relative error not exceeding 10% (n = 3). Nile Red fluorescence measured by flow cytometry and obtained from three independent replicates was affected by a relative error not exceeding 10% (n = 3). b Correlation between the normalized Nile Red fluorescence signal and the yeast oil content obtained by gravimetric extraction (lipid content (percent w/w dry cell) = 0.014 × fluorescence + 1.14; r = 0.913)
The lipid content attained ∼7% (w/w) at t = 28.25 h, remaining almost constant until the end of the batch fermentation. The lipid content reduction might be due to the likely oxygen limitation. In fact, the cellular lipid synthesis in R. glutinis is depending on oxygen availability [24]. On the other hand, oxygen deficiency leads to an essential requirement for lipids itself, principally as sterols and unsaturated fatty acids, as Coccucci et al. [25] have reported for R. glutinis. Therefore, oxygen limitation might have contributed for the lipid decrease observed at t = 28.25 h. Another factor that might have contributed for the cellular lipid content drop after t = 24.25 h was the increasing percentage of dead cells as the culture aged. Due to their permeabilized cytoplasmic membranes, the cells might have lost their internal content including lipid storage material. Therefore, such cells would not contribute for the total lipid content of the culture. Nevertheless, after t = 28.25 h, the lipid content remained almost unchanged (∼7% w/w), suggesting that the yeast cells could still produce lipids at that stage under nutritional limiting conditions.
The normalized Nile Red fluorescence depicted a similar profile as that one observed for cellular lipid content assayed by the gravimetric method (Fig. 6a). For the biomass concentration range studied, a high linear correlation between the Nile Red fluorescence intensity measured by flow cytometry and the total lipid content assayed by the traditional lipid extraction method was found for this yeast [Lipid content (percent w/w dry cell weight) = 0.014 × fluorescence + 1.14] (Fig. 6b). The corresponding relation coefficient (r = 0.921) was higher than that determined by spectrophotometry for Rhodosporidium toruloides IFO-0559 (r = 0.861; [9]), the sexual state of R. glutinis.
Therefore, the flow cytometric method using the Nile Red fluorescent stain can be used for the screening of yeast strains for lipid production, optimization of biofuel production bioprocesses, and scale-up studies.
Conclusions
Despite the low lipid content depicted by R. glutinis CCMI 145 during the batch fermentation, it was possible to monitor the yeast growth by light scatter measurements, cell viability, and cellular lipid content using multi-parameter flow cytometry. Such technique proved to be suitable for screening of oil producer yeasts strains, optimization of the yeast growth conditions, or monitoring industrial or scale-up yeast processes for lipid production.
Studies aiming at the improvement of the yeast’s growth conditions leading to higher yeast’s lipid contents are now in progress.
References
Li, Q., Du, W., & Liu, D. (2008). Applied Microbiology and Biotechnology, 80, 749–758.
Chisti, Y. (2007). Biotechnology Advances, 25, 294–306.
Easterling, E. M., French, W. T., Hernandez, R., & Lich, M. (2009). Bioresource Technology, 199, 356–361.
Dai, C., Tao, J., Xie, F., Dai, Y., & Zhao, M. (2007). Africana Journal of Biotechnology, 6, 2130–2134.
Xue, F., Miao, J., Zhang, X., Luo, H., & Tan, T. (2008). Bioresource Technology, 99, 5923–5927.
Elsey, D., Jameson, D., Raleigh, B., & Cooney, M. J. (2007). Journal of Microbiological Methods, 68, 639–642.
Tornabene, T. G. (1983). Enzyme and Microbial Technology, 5, 435–440.
Kimura, K., Yamaoka, M., & Kamisaka, Y. (2004). Journal of Microbiological Methods, 56, 331–338.
de la Jara, A., Medonza, H., Martel, A., Molina, C., Nordströn, L., de la Rosa, V., et al. (2003). Journal of Applied Phycology, 15, 433–438.
Lopes da Silva, T., Santos, C. A., & Reis, A. (2009). Biotechnology and Bioprocess Engineering, 14, 330–337.
Hewitt, C. J., & Nebe-Von-Caron, G. (2001). Cytometry, 44, 179–187.
Hewitt, C. J., & Nebe-Von-Caron, G. (2004). Advances in Biochemical Engineering/Biotechnology, 89, 197–223.
Abe, F. (1998). Journal of Microbiological Methods, 79, 178–183.
Attfield, P. V., Kletsas, S., Veal, D. A., van Rooijen, R., & Bell, P. J. L. (2000). Journal of Applied Microbiology, 89, 207–214.
Breeuwer, P., Drocourt, J. L., Brunschoten, N., Zwietering, M. H., Rombouts, F. M., & Abee, T. (1995). Applied and Environmental Microbiology, 61, 1614–1619.
Müller, S., & Lösche, A. (2004). Journal of Food Engineering, 63, 375–381.
Raschke, D., & Knorr, D. (2009). Journal of Microbiological Methods, 79, 178–183.
Miller, G. L. (1959). Analytical Chemistry, 31, 426–428.
Xiong, W., Xiufeng, L., Xiang, J., & Wu, Q. (2008). Applied Microbiology and Biotechnology, 78, 29–36.
Yeung, P. K. K., & Wong, J. T. Y. (2003). Protoplasma, 220, 173–178.
Premazzi, G., Buonaccorsi, G., & Zilio, P. (1989). Water Research, 23, 431–442.
Stauber, J. L., Franklin, N. M., & Adams, M. S. (2002). TIBTECH, 20, 141–143.
Ludovico, P., Sansonety, F., & Corte-Real, M. (2001). Microbiology, 147, 3335–3343.
Choi, S. Y., Ryu, D. D., & Rhee, J. S. (1982). Biotechnology and Bioengineering, 24, 1165–1172.
Cocucci, M. C., Belloni, G., & Gianani, L. (1975). Archives of Microbiology, 105, 17–20.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
da Silva, T.L., Feijão, D. & Reis, A. Using Multi-parameter Flow Cytometry to Monitor the Yeast Rhodotorula glutinis CCMI 145 Batch Growth and Oil Production Towards Biodiesel. Appl Biochem Biotechnol 162, 2166–2176 (2010). https://doi.org/10.1007/s12010-010-8991-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-010-8991-3