Abstract
To induce tumor cell apoptosis, a modified 15 kDa actin linked with a peptide NGR “homing” into tumor or tumor vessels was expressed in Escherichia coli. After refolding and purification, this fusion protein NGR-15actin was labeled with FITC to testify whether NGR-15actin could integrate into the cytoskeleton. It was found that this targeted peptide could induce HepG2 and HeLa cells apoptosis through its effect on the cytoskeleton function by binding to cytoskeleton protein. Thus, targeted NGR-15actin could be a candidate molecule for the therapy of cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A strategy for targeting cancer is inhibiting angiogenesis, which makes tumor cell survival, growth, and metastasis possible, hence, posing a problem. To address this concern, research has seen the importance of applying several peptides to target angiogenic endothelial cells; these peptides were selected by in vivo phage display and could bind to receptor expressed within angiogenic vessels [1–3]. In like manner, some anticancer drugs with these peptides, such as RGD-4C and CNGRC, have been used to induce tumor repression more efficiently, not to mention their lower toxicity as compared with drugs alone [4, 5].
Tumor results from the imbalance between apoptosis and anti-apoptosis. One of the characteristic changes in cancer cell is the abnormality in the cytoskeleton, which suggests some roles of cytoskeletal proteins in tumorigenesis or the maintenances of tumor cells [6, 7]. Research results say that several cytoskeleton proteins, such as Gas2, gelsolin, beta-catenin, fodrin, actin, and PAK-2, are prototypically cleaved by caspase proteases during apoptosis [8, 9]. Moreover, after being cleaved, actin produces several fragments. One of these fragments, about 15 kDa, can induce the morphological changes of apoptosis, especially in some solid tumor cells [10].
Such findings gave this study some push to investigate whether targeted delivery of 15 kDa actin to tumor cell would allow receptor-mediated internalization, and induce tumor cells apoptosis. In this article, we report the salient finding that coupling actin to CNGRC, a CD13 ligand capable of “homing” into tumor vessels, could enter cancer cells, bind to cytoskeleton, hence, induce the morphological changes of apoptosis in cancer cells.
Material and Methods
Construction of Plasmid pET28-NGR-15actin
First strand cDNA was synthesized from the total RNA of 107 fresh HepG2 cells using avian myeloblastosis virus reverse transcriptase (Promega, USA) and random primer (9 mer; Takara, Japan). The 15 kDa actin gene was amplified from the cDNA mixture with two primers: P1: 5′-GGT GTG GCG GCC AGG TCA TCA C-3′, and P2: 5′-GTG AAT TCC TAG AAC ATT TGC GGT G-3′ (which contained an EcoR I site (underlined)). Polymerase chain reaction (PCR) was performed using pfu polymerase (Promega, USA) under the following conditions: first denaturation, 1 min at 95 °C; denaturation, 30 s at 55 °C; annealing, 1 min at 72 °C; extension, 1.5 min at 95 °C for 35 cycles; and lastly, extension for 10 min at 72 °C.
To add the CNGRC and linker GG, 15 kDa actin DNA fragment was amplified by primers P2 and P3 (5′-GCTCCACCATGGCGTGCAACGGGCGGTGTGGCGGCCA-3′). The PCR product was inserted into plasmid pET28a with two restriction enzyme sites NcoI and EcoR I. DH5α competent cells were first transformed and screened in LB medium containing kanamycin (50 µg/ml). The plasmids were then confirmed by endonuclease restriction digestion assay and further confirmed by DNA sequencing. The resulting plasmid was named pET28a/NGR-15actin.
Expression, Refolding, and Purification of Targeted NGR-15actin
Plasmid pET28a/NGR-15actin was transformed into Escherichia coli BL21(DE3); the bacteria were grown at 37 °C in 50 mL LB medium (1% Bacto-tryptone, 0.5% yeast extract, 85 mM NaCl) with 50 μg/ml kanamycin. When the OD600 reached 0.6, 1 mM isopropyl β-d-thiogalactogalactopyranoside (IPTG) was added and the culture was incubated at 37 °C for 5 h. To purify the recombinant NGR-15actin, bacteria were harvested by centrifugation at 8,000×g for 10 min at 4 °C and then suspended in 10 ml washing buffer (50 mM Tris–HCl, 5 mM EDTA, and 1% Triton X-100, pH 8.0).
After the repeated rounds of sonication, the solution was centrifuged at 12,000×g. The supernatant was then decanted and the precipitate pellet was solubilized in 2.5 ml of denaturation buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM imidazole, and 8 M urea; pH 8.0). The cellular debris was removed by centrifugation at 4 °C for 15 min at 12,500×g. The supernatants were applied to Ni2+-IDA His-bind resin and bound antibodies were recovered according to the manufacturer’s instructions. The elution was dialyzed against 1 l refolding buffer (50 mM Tris–HCl, 150 mM NaCl, 2 mM reduced glutathione, and 0.4 mM oxidized glutathione; pH 8.0) at 4 °C for 24 h. It was continued to be dialyzed against 1 l refolding buffer (50 mM Tris–HCl and 150 mM NaCl; pH 8.0) for 24 h. The solutions were concentrated fivefold with a Centricon 30 (Millipore, USA) and then passed through a 0.22 μm pore size membrane.
Targeted NGR-15actin Induce Apoptosis
To evaluate the apoptosis efficacy of targeted NGR-15actin in cancer cells, HepG2 and HeLa cells (5 × 104) were cultured with RPIM1640 containing 10% fetal calf serum (FCS) in 37 °C and 5% CO2. Overnight, the medium was removed. Before RPIM1640 (10% FCS) was added, 10 µg purified NGR-15actin was added to cells in 100 µL PBS and incubated for 20 min. The cell culture medium was aspirated from adherent cells, and the cells were gently washed once with PBS at 37 °C. Then, a 20-fold dilution of the dye mixture (100 µg/ml acridine orange and 100 µg/ml ethidium bromide) in PBS was gently pipetted on the cells, which were viewed on an inverted microscope (Nikon TE 300).
Targeted NGR-15actin Labeled with FITC and Extracting of Cytoskeleton
Whether the targeted NGR-15actin could enter into tumor cells and be integrated into the cytoskeleton was investigated. The researchers labeled the targeted NGR-15actin with fluorescein isothiocyanate (FITC); 30 µg targeted NGR-15actin and 1 µg FITC were resolved in CBS buffer (50 mM Na2CO3, pH 9.0), respectively. Then, two solutions were mixed at room temperature, and after 2 h, 100 mM lysine solution was added. The labeled NGR-15actin solution was dialyzed with PBS overnight. Secondly, the cytoskeleton was extracted to check whether the labeled NGR-15actin interacted with cytoskeleton. The labeled NGR-15actin was incubated with HepG2 and HeLa cells. After 7 h, the medium was removed, and the cells were gently washed with PBS. Subsequently, the tumor cells were incubated with M buffer (50 mM imidazole, 50 mM KCl, 0.5 Mm, 1 mM EGTA, 0.1 mM EDTA, 1 mM mercaptoethanol, and 4 mM glycerol; pH 7.2) containing 1% Triton X-100 at room temperature. After 15 min, the cells were gently washed with M buffer three times and fixed with 3% glutaraldehyde. The extracted cytoskeleton was checked by fluorescence microscopy. Finally, the cytoskeleton was further stained with the 100 ml solution containing 0.2% Coomassie brilliant blue R250, 46.5 ml methanol, 7 ml HAC, and 46.5 ml distilled water. After 30 min, the cytoskeleton was rinsed with water and observed through a microscope.
Results
Construction of Plasmid pET28-NGR-15actin
pET28a plasmid has been widely used in obtaining high expression levels of proteins in E. coli. To express a recombinant protein NGR-15actin, which can be used further, the restricted PCR-generated DNA fragment was subcloned using Ncol and EcoRl cloning sites. The analysis by PCR and restriction endonuclease digestion proved that the insertion of the fusion gene in the recombinant plasmid was correct. The ORF of NGR-15actin cDNA in the recombinant plasmid was sequenced and was confirmed as the correct sequence in the recombinant plasmid.
Expression, Refolding, and Purification of Targeted NGR-15actin
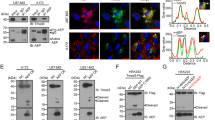
E. coli BL21(DE3) cells harboring pET28-NGR-15actin were induced by IPTG. The expression of the NGR-15actin protein was analyzed by SDS-PAGE with Coomassie brilliant blue R-250 staining, as well as by immune-blotting using mouse anti-His6 IgG. A predominant protein at 15 kDa, corresponding to the expected molecular size of NGR-15actin, was observed (Fig. 1a, lane 1). Most of the NGR-15actin protein remained in the insoluble fraction. Attempts to obtain soluble NGR-15actin by reducing the temperature of cultivation or the concentration of IPTG were unsuccessful. After the renaturation, the NGR-15actin protein was purified in a single purification step on a Ni2+-IDA His-bind resin. The purity of NGR-15actin and its authenticity were confirmed by SDS-PAGE (Fig. 1a, lane 4), and also by immune-blotting with mouse anti-His6 IgG (Fig. 1b). The NGR-15actin appeared as a single 15 kDa band at high purity (more than 95%, as estimated by absorbance scanning). In this expression system, at least 20 mg of the purified NGR-15actin was obtained from 1 l of E. coli cell culture.
SDS-PAGE and Western blotting analysis of NGR-15actin expressed in BL21(DE3) and purified by Ni2+-IDA His-bind resin. a Proteins were resolved on 13% SDS-polyacrylamide gel under reducing conditions and stained with Coomassie blue R-250. M LMW marker of protein, lane 1 induced total bacteria proteins, lane 2 supernatant of induced total bacterial proteins, lane 3 soluble inclusion bodies in 8 M urea, lanes 4 and 5 purified NGR-15actin. b Lane 1 Prestained marker, lane 2 Western blotting analysis of the purified NGR-15actin stained with mouse anti-His6 IgG and peroxidase-conjugated goat anti-mouse IgG
Morphological Quantification of Cellular Apoptosis
The cells with nuclei showing margination and condensation of the chromatin and/or nuclear fragmentation (early-/mid-apoptosis; acridine orange-positive) or with compromised plasma membranes (late apoptosis; ethidium bromide-positive) were considered inviable. At least 500 cells were assessed by morphological change after strain of EB/AO. Percent viability was determined by apoptotic morphology. After 12 h, 50% of HepG2 cells showed apoptosis, whereas, after 20 h, apoptosis appeared in 50% of HeLa cells (Fig. 2).
Targeted NGR-15actin Labeled with FITC and Extracting of Cytoskeleton
To determine whether NGR-15actin could combine to cytoskeleton, the actin was labeled with FITC and was incubated with HepG2 and Hela cells. After 6 h, the cells were washed with Triton/M buffer to rinse off non-cytoskeleton protein, sugar, DNA, and other ingredients. FITC-NGR-15actin still remained in the cytoskeleton (Fig. 3a–d). Furthermore, the cell debris with Coomassie brilliant blue R250 was retained, and an obvious cytoskeleton structure was observed (Fig. 3e–h). The result showed that the targeted NGR-15actin not only entered into the cell but also integrated into the cytoskeleton.
Cytoskeleton of tumor cells was integrated by FITC-labeled NGR-15actin. a The cytoskeleton of HepG2 was extracted after incubating with FITC labeled NGR-15actin for 6 h; b the cytoskeleton of HeLa was extracted after incubating with FITC-labeled NGR-15actin for 6 h; c the cytoskeleton of HepG2 was extracted after incubating without FITC-labeled NGR-15actin treatment; d the cytoskeleton of HeLa was extracted after incubating without FITC-labeled NGR-15actin treatment; e the cytoskeleton of HepG2 was stained with R-250 after incubating with targeted NGR-15actin for 6 h; f the cytoskeleton of HeLa was stained with R-250 after incubating with targeted NGR-15actin for 6 h; g the cytoskeleton of HepG2 was stained with R-250 without incubating with targeted NGR-15actin; h the cytoskeleton of HeLa was stained with R-250 without incubating with targeted NGR-15actin
Discussion
In apoptosis, proteolysis is the central biochemical reaction regardless of cell type. The cleavage process by apoptotic protease is highly selective at the earlier stage of apoptosis [11]. Recent studies show that major proteolysis is caspases and that granzymes and calpain are also involved in the apoptotic signaling process [12–15]. The linkage between the characteristic morphological changes in apoptotic cells and specific degradation of actin was first established by Kayalar [16], who testified that actin in PC12 cells was degraded at 24 h after serum withdrawal. The cleavage of actin results in several fragments, 29 kD, 31 kD, 41 ku, and 15 kD [17]. Thus, the cytoskeleton may be a target in cancer therapy [18–22].
That the anticancer drug should kill or oppress cancer alone but does not hurt normal cell intrigues many researchers [23]. Nevertheless, many scientists have shown promising progress. Among their findings, targeting peptide, such as NGR and RGD, can introduce the drug to home into tumor and enhance the therapy efficiency. In this study, NGR-15actin (consisting of 15 kDa actin fused with the N terminus of CNGRCGG) was prepared by recombinant DNA technology. Two glycine residues were interposed between CNGRC and 15 kDa actin as a linker. In the constructed expression system, the recombinant NGR-15actin without leader sequence was expressed at a high level in the form of inclusion bodies under the T7 promoter. Reducing the temperature of cultivation or the concentration of IPTG had no effect on the formation of these inclusion bodies. Why the expressed recombinant protein NGR-15actin accumulated in inclusion bodies could be due to the overexpression of recombinant protein in prokaryotes and the direct expression without a leader sequence. However, collecting inclusion bodies proved to be a very efficient way of obtaining a large amount of expressed protein. While the recombinant protein in inclusion bodies may be aggregated, it was not difficult to render the inclusion bodies soluble with 8 M urea although it then had to be refolded. To get efficient purification of the NGR-15actin protein, Ni2+-IDA His-bind resin was used to absorb a 6-histidine tag that had been infused to the protein. In the constructed plasmid pET28a/NGR-15actin, there was a 6-histidine sequence between the promoter and the NGR-15actin inserted site. Thus, a highly purified unfolded protein could be isolated from the solubilized inclusion bodies by eluting the protein bound to the Ni2+-IDA His-bind resin with 1 M imidazole. In the preliminary tests, purifying the NGR-15actin before refolding was found to give a higher refolding yield than was the reverse procedure. The reason could be that purification before refolding removed contaminating proteins of bacteria that would form aggregates with the NGR-15actin and decrease the final yield of the active product.
Furthermore, those targeting actin incubated with HepG2 and HeLa cell, and detected cell apoptosis with EB/AO. The time of inducing cell apoptosis is different between HepG2 and HeLa cells, which could depend on the abundance of NGR receptor CD13. The researchers labeled the actin with FITC, which did not affect its function. The researchers found that if they did not block TITC with lysine, the FITC alone could bind cells; in addition, they found out that after Trition/M buffer was washed, the FITC could still bind cells. Subsequently, the researchers observed that the labeled NGR-15actin could enter the tumor cell to integrate into cytoskeleton by extracting cytoskeleton. The researchers concluded that targeting NGR-15actin inserted into cytoskeleton as actin homology–analogy and affected the normal function of actin, thus inducing selective apoptosis of such cells after entry.
Targeted peptides represent a potentially new class of anticancer agents [24–26]. Their activity may be optimized for maximum therapeutic effect by fusing targeted peptide. From a clinical point of view, targeted NGR-15actin has a potential valuation of cancer therapy/intervention. Beyond this, targeted NGR-15actin can also provide a feasible reagent for the research of cytoskeleton.
References
Arap, W., Pasqualini, R., & Ruoslahti, E. (1998). Science, 16, 377–380.
Pasqualini, R., Koivunen, E., & Ruoslahti, E. (1997). Nature Biotechnology, 15, 510–522.
Koivunen, E., Arap, W., Valtanen, H., Rainisalo, A., Medina, O. P., Heikkila, P., et al. (1999). Nature Biotechnology, 17, 768–774.
Brancolini, C., Marzinotto, S., & Schneider, C. (1997). Cell Death and Differentiation, 4, 247–253.
Kothakota, S., Azuma, T., Reinhard, C., Klippel, A., Tang, J., Chu, K., et al. (1997). Science, 278, 294–298.
van Laarhoven, H. W., Gambarota, G., Heerschap, A., Lok, J., Verhagen, I., Corti, A., et al. (2006). Investigational New Drugs, 24, 27–36.
Curnis, F., Gasparri, A., Sacchi, A., Cattaneo, A., Magni, F., & Corti, A. (2005). Cancer Research, 65, 2906–2913.
Curnis, F., Arrigoni, G., Sacchi, A., Fischetti, L., Arap, W., Pasqualini, R., et al. (2002). Cancer Research, 62, 867–874.
Sandoval, C. M., Geierstanger, B. H., Fujimura, S., Balatbat, C., Williams, T., de Unamuno, J., et al. (2006). Chem Biol Drug Des, 67, 417–424.
Ellerby, H. M., Arap, W., Ellerby, L. M., Kain, R., Andrusiak, R., Rio, G. D., et al. (1999). Natural Medicines, 5, 1032–1038.
Wang, L. F., Yu, M., Hansson, E., Protchard, L. I., Shiell, B., Michalsk, W. P., et al. (2000). Journal of Virology, 74, 9972–9979.
Kidd, V. J., Lahti, J. M., & Teitz, T. (2000). Cell and Developmental Biology, 11, 191–201.
Martin, S. J. (1995). Cell, 11, 349–352.
Thornberry, N. A., Rosen, A., & Nicholson, D. W. (1997). Advances in Pharmacology, 41, 155–177.
Jacob, T., Hingorani, A., & Ascher, E. (2005). Vascular, 13, 34–42.
Kayalar, C., Ord, T., Testa, M. P., Zhong, L. T., & Bredesen, D. E. (1996). Proceedings of the National Academy of Sciences of the United States of America, 93, 2234–2238.
Brown, S. B., Bailey, K., & Savill, J. (1997). Biochemical Journal, 323, 233–237.
Neradil, J., Veselska, R., & Svoboda, A. (2005). International Journal of Oncology, 27, 1013–1021.
Mashima, T., Naito, M., & Tsuruo, T. (1999). Oncogene, 18, 2423–2430.
Yamazaki, Y., Tsuruga, M., Zhou, D., Fujita, Y., Shang, X., Dang, Y., et al. (2000). Experimental Cell Research, 259, 64–78.
Senderowicz, A. M., Kaur, G., Sainz, E., Laing, C., Inman, W. D., Rodriguez, J., et al. (1995). Journal of the National Cancer Institute, 87, 46–51.
Statsuk, A. V., Bai, R., Baryza, J. L., Verma, V. A., Hamel, E., Wender, P. A., et al. (2005). Nat Chem Biol, 1, 383–388.
Broxterman, H. J., & Georgopapadakou, N. H. (2002). Drug Resist, 7, 79–87.
Brissette, R., Prendergast, J. K., & Goldstein, N. I. (2006). Current Opinion in Drug Discovery & Development, 3, 363–369.
Ma, X., Zheng, W., Wei, D., Ma, Y., Wang, T., Wang, J., et al. (2006). Journal of Biotechnology, 123, 367–378.
Guelen, L., Paterson, H., Gaken, J., Meyers, M., Farzaneh, F., & Tavassoli, M. (2004). Oncogene, 23, 1153–1165.
Acknowledgments
This work was supported by the grants from the Scientific and Technological Program of Zhejiang Province (No.2008C33062) and supported by Jiangsu Province’s Outstanding Leader Program of Traditional Chinese Medicine.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lei, H., Cao, P., Miao, G. et al. Expression and Functional Characterization of Tumor-Targeted Fusion Protein Composed of NGR Peptide and 15-kDa Actin Fragment. Appl Biochem Biotechnol 162, 988–995 (2010). https://doi.org/10.1007/s12010-009-8901-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-009-8901-8