Abstract
Purpose of Review
The Runx family genes (Runx1, Runx2, Runx3, and Cbfb) are important transcriptional regulators in the development of various tissues. We herein highlight the roles of the Runx family genes in morphogenesis in the craniofacial regions and in the pathogenesis of congenital morphological problems in these regions.
Recent Findings
A recent analysis using conditional Runx mutant animals and a human genetic study identified the novel roles of Runx genes in the development of the tooth, salivary glands, and the palate. In an animal study, Runx1/Cbfb signaling was found to regulate the Lgr5 expression and maintain the stem cells in the dental epithelium in the growing incisors. Aberrant Runx1/Cbfb signaling induced male-specific involution of the convoluted granular cell differentiation of the submandibular gland. In palatogenesis, Runx1/Cbfb signaling regulated the Tgfb3 expression in the fusing palatal epithelium through Stat3 activation.
Summary
The combination of a human genetic study and a phenotype analysis of mutant animals revealed the various roles of Runx genes in the development of the tooth, palate, and salivary glands. Runx genes have functional redundancy in various tissues, which still hinder the roles of Runx genes in morphogenesis. Future studies may reveal the novel roles of Runx signaling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Runx2 is a master gene for bone development [1] and haploinsufficiency of Runx2 causes cleidocranial dysplasia [2]. As suggested by the function of Runx2, this disease is characterized by problems in skeletogenesis, such as missing clavicles and opened anterior fontanelle. A recent study further showed that osteoporosis and osteopenia are detected in this syndrome [3•]. Supernumerary teeth are also among the characteristic features of this syndrome [4]. Interestingly, Runx2 deletion disturbed tooth development in mice [5], whereas haploinsufficiency induces extra tooth generation in humans [4]. The tooth is a mineralized tissue, like bone; however, unlike bone, teeth are classified as skin appendages and their morphogenesis is regulated by epithelial-mesenchymal interaction. Indeed, tooth development arrest in Runx2 deletion is due to disturbed Runx2-regulated Fgf signaling in the process of epithelial-mesenchymal interaction [6]. A recent study further demonstrated that other Runx family genes are also involved in the morphogenesis, differentiation, and maintenance of stem cells in various organs. In the present review, we introduce novel roles of Runx signaling in the craniofacial region.
Runx genes encode transcription factors sharing a conserved region of 128 amino acids termed the runt domain (RD) [7]. There are three members of RUNX gene family in mammals: RUNX1, RUNX2, and RUNX3. These Runx family transcription factors form a heterodimeric transcription complex with Core binding factor β (Cbfb), a cofactor of the Runx family, which enhances the binding affinity of the complex for DNA.
Runx1, Runx3, and Cbfb, a cofactor for the Runx genes, are also involved in skeletal development. Runx3 is expressed in osteoblasts and Runx3 deletion results in congenital osteopenia in mice [8]. Runx1 is coexpressed in early stages of intramembranous ossification in craniofacial bones [9]. The conditional deletion of Runx1 demonstrates a delay in sternal development and further double null mutation of Runx1 and Runx2 results in complete lack of a sternum [10]. Furthermore, as is observed in Runx1 mutants, Cbfb null mutation has been demonstrated to be associated with skeletal defects [11]. Taken together, Runx2 is dominant in bone development, but other Runx genes also have redundant and regulatory roles.
The null mutant phenotypes in each Runx gene display distinct tissue-specific roles in mice; RUNX1 is required for hematopoiesis; RUNX2 is essential for osteogenesis; and RUNX3 is crucial for the development of dorsal root ganglia neurons and T-lymphocytes (reviewed in [12]). Both Runx1 and Runx2 null mutants are lethal in early embryogenesis [1, 13], and Runx3 mutants cannot survive long after birth. A recent phenotype analysis of their conditional mutants or conditionally rescued mutants further revealed the additional and various roles of the Runx family genes in various biological events (e.g., cancer development, organogenesis, and morphogenesis).
The aim of this review is to summarize the findings from current studies that showed the importance of the Runx genes in the development of tissues and organs in the craniofacial regions and Runx-related human disease.
CCD and Supernumerary Teeth
The etiology of the supernumerary teeth in Runx2 mutant mice remains to be elucidated at the molecular and cellular levels. In rodents, Runx2 null mutant demonstrated extra buds at the lingual side of the upper tooth buds [14]. This evidence directly suggests that Runx2 inhibits the initiation of extra tooth formation in early tooth development. However, the tooth development does not proceed beyond the cap stage in Runx2 null mutants and supernumerary teeth are not evident in these mutants [5]. Furthermore, the Runx2 mouse CCD model is not ideal for the study of supernumerary teeth since mice are monophydont. Humans are diphydont and the supernumerary teeth in man have been shown to be supernumerary to the second dentition [15]. Unlike humans, rodents do not show supernumerary teeth with Runx2 haploinsufficiency.
The budding of the tooth germs is regulated by various molecules. Activation of canonical Wnt signaling in the dental and oral epithelium induces extra tooth buds as well as supernumerary teeth in rodents [16, 17]. Interestingly, treatment with BIO, a small-molecule GSK3 inhibitor and, therefore, activator of canonical Wnt signaling, prevents the induction of supernumerary teeth in epithelial Wnt-activated mutants ex vivo [18•]. Furthermore, BIO also inhibits the successive formation of M2 molars in WT mice molar in vitro [18•]. Taken together, mesenchymal Wnt signaling prevents the formation of successive or extra tooth, unlike the stimulatory roles of epithelial Wnt signaling in the induction of tooth budding. In this study, it was demonstrated that Wnt inhibitors, such as Axin2 or Drapc1, were activated in Runx2 null mutants. Hence, it is likely that antagonistic interactions between Runx signaling and Wnt inhibitor at the dental mesenchyme surrounding the tooth buds may—at least in part—explain the molecular etiology of the supernumerary teeth in CCD patients [18•].
The Involvement of Runx Genes in Amelogenesis and the Maintenance of the Dental Epithelium
Some CCD patients demonstrate enamel hypoplasia [19]. Runx2 is expressed in ameloblasts, which is derived from the dental epithelium, during the late secretory and maturation stages [20], and the epithelial-specific ablation of Runx2 results in hypomaturation of the enamel matrix with Klk4 downregulation in mice. In these mutants, the tooth morphology was not affected; however, the enamel matrix was remarkably abraded in aging with significant downregulation of the AMELX, AMBN, and ENAM expression [21•].
Runx1 is also expressed in ameloblasts and the dental epithelium [22]. Humans with Braddock-Carey syndrome, in which A 21q22 contiguous gene syndrome encompasses RUNX1, show enamel hypoplasia [23]. In mice, epithelial deletion of Runx1 results in the absence of enamel formation in the incisors and enamel hypoplasia with tipping in the molars [24••]. Similar defective phenotypes in the enamel matrix were also demonstrated in Cbfb mutants [25].
On the other hand, Runx3 null mutants did not show phenotypes associated with tooth development [22]. Collectively, Runx1 and Runx2 with Cbfb, cofactor, are involved in enamel formation in amelogenesis. Since the expression of Runx1 and Runx2 partly overlap in ameloblast differentiation, their function could be redundant. Hence, the deletion of both Runx1 and Runx2 may possibly cause more severely defective amelogenesis, which would further elucidate the distinct roles of Runx signaling in enamel formation.
The Involvement of Runx Genes in the Maintenance of Stem Cells
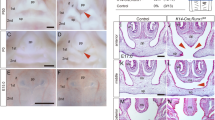
The incisors of rodents continuously elongate throughout their lifetime and specifically contain an epithelial stem cell compartment at the cervical loop. In conditional null mutants of each Runx1 and Cbfb, the incisors show remarkable shortening with shrinkage of the cervical loop and with a significant reduction in the proliferative activity of the ameloblast precursors [24••, 25]. In the cervical loop of mutant incisors, the expression of Lgr5, a stem cell marker, is remarkably disturbed, indicating that Runx1 is involved in the maintenance of the epithelial stem cells in the growing incisors [24••] (Fig. 1).
K14-Cre; Runx1fl/fl mice showed extremely short incisors and enamel defects (A) in comparison to control mice (C). On the other hand, there was no significant difference in the molar root length or morphology. Double immunostaining for Ki67and K14 revealed that the epithelium in the cervical loop of the Runx1 mutant (B) was significantly less proliferative than that in the control (D). Nuclei were counterstained with DAPI
This regulatory role of Runx1 in stem cell maintenance is also evident in hair follicles [26]. In the growing incisors, Runx2 is expressed in the cervical loop and Runx3 was observed in ameloblast precursors [24••]; however, neither Runx2 nor Runx3 null mutants showed shortening of the incisors [21•]. Taken together, among the Runx genes, Runx1 is dominantly involved in the maintenance of stem cells.
The molecular mechanisms underlying the regulation of the Lgr5 expression by Runx remain to be elucidated. Lgr5 is known to be a Wnt target gene; however, an analysis using the canonical Wnt signaling reporter mouse demonstrated that Lgr5 is not directly regulated by Wnt signaling in the mouse incisors [27]. In our study, Runx1 abrasion disturbed Stat3 phosphorylation specifically at the cervical loop regions. In vitro, Stat3 inhibitors (e.g., AG490 or S3I-201) disturbed the Lgr5 expression at the WT cervical loop and Stat3 activation using ZnCl2 rescued the disturbed Lgr5 expression in the Runx1 mutant incisors [24••]. Interestingly, a ChIP analysis demonstrated that both Runx and Stat transcriptional factors bind to the promoter region of Lgr5, suggesting that Runx1-Stat3 regulatory network could directly—at least in part—regulate the expression of Lgr5.
As stated above, Runx1 affects ameloblast differentiation; however, it was curious that the enamel phenotypes were remarkably more severe in incisors than in molars. Such a difference could be due to the disturbance of the supply of the ameloblast precursors in the Runx1 mutant incisors.
The Involvement of Runx in Exocrine Gland Development
Salivary glands are regulated by epithelial-mesenchymal interaction, as well as teeth. In development of the salivary gland, Runx1 is specifically expressed in the ductal epithelium of the submandibular glands. Interestingly, in male mice, Runx1 conditional null mutants exhibited involution of the granular convoluted tubules (GCT). GCT is a secretory canal lying between the intercalated and striated ducts and is specifically induced in the postnatal stage under the control of androgens [28]. Such androgen-specific characteristics are also supported by its involution by castration in male mice [29]. In these Runx1 mutants, serum androgen levels remained unchanged and testis development was not affected, however, the expression of Crisp3, a biomarker gene of the AR pathways, is significantly downregulated, and it was assumed that Runx1 directly regulates the sensitivity of GCTs cells to androgen and contributes to the induction of the GCTs in male mice [30•]. Epithelial Cbfb null mutant mice demonstrate similar ductal phenotypes with the involution of GCT, indicating that Cbfb is also an indispensable cofactor in Runx signaling in postnatal development of the salivary glands [31]. Taken together, Runx1/Cbfb signaling is involved in sexual dimorphism in the induction of GCTs in the presence of androgen. To date, no reports have demonstrated the possible association between genetic Runx1 or Cbfb mutations and salivary gland disease.
Runx3 is also expressed in both ductal and serous or mucous acinous cells of the parotid, submandibular, and sublingual salivary glands and in adenoid cystic carcinoma cells [32]. Runx3 is a tumor suppressor gene in gastric cancer [33] and the expression of Runx3 was downregulated in carcinoma cells, which could be associated with tumor progression [32]. A CpG island analysis demonstrated that Runx3 is methylated in these carcinoma cells.
Methylation of the runt-related transcription factor-3 CpG island spreads the most from the 5′ region to the transcription start site in adenoid cystic carcinoma tissues, and the transcription start site may be a critical region for runt-related transcription factor-3 methylation. The spreading pattern of the runt-related transcription factor-3 methylation may play a role in the progression of adenoid cystic carcinoma [34].
The Involvement of Runx in Cleft Palate
The expression of Runx2 is evident in the palatal mesenchyme and Runx2 deletion results in complete cleft palate in mice [35]. In humans, association analyses of four populations identified single nucleotide polymorphisms (SNPs) that showed significant excess maternal transmission and suggested that RUNX2 may influence the risk of CL/P [36]. On the other hand, there is no evident association between CCD and cleft palate; however, a clinical report described that CCD patients with mutation of p.R225Q, which is located closely to the Runt binding region, had a median pseudo-cleft palate [37]. The deletion of Hoxa2, a homeobox gene, results in cleft palate with the upregulation of Runx2 in mice, suggesting that palatal Runx2 is regulated by Hoxa2, presumably by regulating the osteogenic differentiation of the palatal mesenchyme [38].
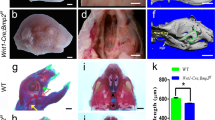
Runx1 is specifically expressed at the fusing palatal epithelium [22, 39••]. In humans, microdeletion encompassing RUNX1 causes Braddock-Carey syndrome and, as well as thrombocytopenia, cleft palate is among the characteristic features of this disease [40]. In mice, epithelial abrasion of Runx1 results in anterior-specific cleft palate due to disintegration of the fusing epithelium; however, cleft palate is not induced in the secondary palate of the mutants [39••]. Previous studies have also demonstrated that the molecular and the cellular mechanism in the palatal fusion do not differ along the anterior-posterior axis and some molecules are specific to the either anterior or posterior palate [41]. Anterior cleft is a rare cleft phenotype in mutant mice and some signaling molecules and transcription factors, such as Bmp4, Msx1, Shh, and Shox2 are specific to the anterior regions. Shox2 null mutants and Bmp4-rescuing Msx1 null mutants also showed anterior cleft [41]. In Runx1 mutants, the expression of previously known anterior-specific molecules, such as Bmp4, Msx1, or Shox2, was not deviated; however, Tgfb3 was significantly downregulated at the primary palate. Tgfb3 is strongly expressed in the fusing palatal epithelium and null mutation of Tgfb3 results in complete cleft palate in mice (Fig. 2) [42]. On the other hand, the epithelial-specific ablation of Tgfb3 results in anterior cleft [43], which is similar to the phenotypes in Runx1 mutants. TGFB3 protein rescues Runx1 mutants from anterior cleft, which suggests that Tgfb3 could be a critical target molecule for Runx signaling in anterior palatal development [39••] and the Runx-Tgfb3 signaling axis is independent from molecular networks that have been previously reported to regulate palatogenesis.
Occlusal views of K14-Cre; Runx1fl/fl and control mouse palate demonstrate anterior cleft palate at the boundary between the primary and secondary palate at P50 (A, D). Frontal histological sections at 16.0 reveal failed palatal fusion in the Runx1 mutant shelves (B, E). Whole-mount in situ hybridization analyses show that the expression of Tgfb3 is markedly disturbed at the primary palate regions in Runx1 mutant mice (C, F)
Noteworthy, Runx1 regulates the phosphorylation of Stat3, which further regulates the expression of Tgfb3 in the fusing epithelium. The application of Stat3 inhibitor remarkably downregulates the expression of Tgfb3 and also disturbs the fusion between the primary and secondary palate. Interestingly, the inhibition of Stat3 also downregulates the expression of Runx1, indicating that Runx1 and Stat3 phosphorylation reciprocally regulate their activity. On the other hand, Tgfb3 and Runx1 are both expressed in all regions of the palatal fusion and the reason why Runx1 deficiency causes anterior-specific cleft palate remains to be elucidated. One possible explanation is the involvement of the Socs3 expression in the anterior palate [39••]. Socs3 is a direct target molecule for Runx transcription factors and a negative regulator of Stat3 phosphorylation. Interestingly, Socs3 is localized at the anterior palate regions and Runx1 deficiency results in the remarkable upregulation of Socs3, which may subsequently attenuate the phosphorylation of Stat3 [44••]. Another explanation is the functional redundancy between Runx1 and Runx2. Indeed, Runx2 is expressed at the fusing palatal epithelium as well as the palatal mesenchyme; however, its expression at the palatal epithelium of the primary palate is not obvious [45]. To evaluate this hypothesis, a further analysis using double mutants is needed to confirm the roles of Runx signaling in palatal fusion.
Like Runx1, Cbfb is essential to mouse anterior palatogenesis. Epithelial Cbfb deletion results in anterior cleft palate, specifically due to the failed disintegration of the fusing epithelium, as is observed in Runx1 mutants. CBFb haploinsufficiency due to interstitial deletion was shown to cause cleft palate and congenital heart anomalies in humans [46, 47]. In these mutants, the expression of Tgfb3 is also disrupted in the area of failed palatal fusion, in which Stat3 phosphorylation is also affected. Stat3 signaling is activated in a number of cancers [48], in the immune response, and also in response to various environmental signals [49]. Hyper-IgE syndrome is caused by heterozygous mutations of STAT3 and demonstrates primary immune deficiency and cleft palate is often observed in this syndrome [50]. However, the possible mechanisms underlying the involvement of Stat3 in the pathogenesis of the cleft palate remain be elucidated. In in vitro cultures of isolated primary palate tissue, a Stat3 inhibitor disturbed epithelial fusion with the remarkable downregulation of the expression of Tgfb3. Taken together, Stat3 signaling could mediate the functional roles of Runx1/Cbfb signaling in the induction of Tgfb3 at the fusing palatal epithelium in the anterior palate [39••, 44••].
Prevention is the ultimate objective with regard to cleft palate. Non-syndromic cleft palate is a multifactorial genetic disease and the combination of genetic susceptibility factors and environmental risk factors can trigger pathologic palatogenesis. In our study, we identified Stat3 signaling as a downstream pathway of Runx1/Cbfb signaling. It is also known that Stat3 is activated by various molecular signals, such as EGF signaling and IL-6 signaling, and further activated by various environmental modifications and the application of various pharmaceutical agents. Indeed, maternal folic acid application attenuates the risk of cleft palate and also activates Stat3 phosphorylation [44••]. Actually, we demonstrated that folic acid rescues the cleft palate phenotype of the Runx1 mutants with the activation of Tgfb3, suggesting that pharmaceutical modulation of Stat3 signaling could be used for preventing CP in parents with genetic mutations that affect Stat3 signaling [44••].
Summary and Future Directions
Regarding craniofacial development, the analysis of mutant phenotypes clarified various roles of Runx family genes in tooth, craniofacial, and salivary gland development. Furthermore, recent findings on the involvement of Runx signaling in cancer cells have provided a key understanding of the roles of Runx signaling in cellular differentiation, stem cell maintenance, and signal transduction in morphogenesis. Each gene of the Runx family often has the functional redundancy, which potentially complicates the evaluation of the roles of redundant genes and which also hinders the elucidation of the unknown roles of Runx genes. Future studies using tissue-specific multiple null mutant animals might reveal novel roles of Runx signaling.
Genetic studies have demonstrated that Runx1 SNPs are associated with methacholine responsiveness, which is further modified by a history of IUS exposure [51]. Runx2 SNPs also influence susceptibility to CL/P through interaction with environmental tobacco smoke [52]. As discussed above, Runx1/Cbfb signaling affects Stat3 activity, and Runx1 might have some regulatory roles in the pathogenesis of disease. In the future, an understanding of the etiology and the mechanisms of Runx gene family–related diseases in combination with genetic and environmental factors might be useful for molecular diagnostics and pharmaceutical modulation for the prevention of such diseases.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64.
Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–9.
Dinçsoy Bir F, Dinçkan N, Güven Y, Baş F, Altunoğlu U, Kuvvetli SS, et al. Cleidocranial dysplasia: clinical, endocrinologic and molecular findings in 15 patients from 11 families. Eur J Med Genet. 2017;60:163–8. Osteoporosis is common in CCD patients.
Mundlos S. Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet. 1999;36:177–82.
D’Souza RN, Aberg T, Gaikwad J, Cavender A, Owen M, Karsenty G, et al. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–20.
Aberg T, Wang XP, Kim JH, Yamashiro T, Bei M, Rice R, et al. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev Biol. 2004;270:76–93.
Wheeler JC, Shigesada K, Gergen JP, Ito Y. Mechanisms of transcriptional regulation by Runt domain proteins. Semin Cell Dev Biol. 2000;11:369–75.
Bauer O, Sharir A, Kimura A, Hantisteanu S, Takeda S, Groner Y. Loss of osteoblast Runx3 produces severe congenital osteopenia. Mol Cell Biol. 2015;35:1097–109.
Yamashiro T, Wang XP, Li Z, Oya S, Aberg T, Fukunaga T, et al. Possible roles of Runx1 and Sox9 in incipient intramembranous ossification. J Bone Miner Res. 2004;19:1671–7.
Kimura A, Inose H, Yano F, Fujita K, Ikeda T, Sato S, et al. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137:1159–67.
Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–8.
Ito Y. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells. 1999;4:685–96.
Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–30.
Wang XP, Aberg T, James MJ, Levanon D, Groner Y, Thesleff I. Runx2 (Cbfa1) inhibits Shh signaling in the lower but not upper molars of mouse embryos and prevents the budding of putative successional teeth. J Dent Res. 2005;84:138–43.
Jensen BL, Kreiborg S. Development of the dentition in cleidocranial dysplasia. J Oral Pathol Med. 1990;19:89–93.
Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2006;103:18627–32.
Wang XP, O’Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, et al. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–49.
Järvinen E, Shimomura-Kuroki J, Balic A, Jussila M, Thesleff I. Mesenchymal Wnt/β-catenin signaling limits tooth number. Development. 2018;145 Activated canonical Wnt signaling disturbs the sequential tooth formation.
Golan I, Baumert U, Hrala BP, Müssig D. Dentomaxillofacial variability of cleidocranial dysplasia: clinicoradiological presentation and systematic review. Dentomaxillofac Radiol. 2003;32:347–54.
Bronckers AL, Engelse MA, Cavender A, Gaikwad J, D’Souza RN. Cell-specific patterns of Cbfa1 mRNA and protein expression in postnatal murine dental tissues. Mech Dev. 2001;101:255–8.
Chu Q, Gao Y, Gao X, Dong Z, Song W, Xu Z, et al. Ablation of Runx2 in Ameloblasts Suppresses Enamel Maturation in Tooth Development. Sci Rep. 2018;8:9594 Runx2 is essential in enamel maturation.
Yamashiro T, Aberg T, Levanon D, Groner Y, Thesleff I. Expression of Runx1, -2 and -3 during tooth, palate and craniofacial bone development. Mech Dev. 2002;119(Suppl 1):S107–10.
Braddock SR, South ST, Schiffman JD, Longhurst M, Rowe LR, Carey JC. Braddock-Carey syndrome: a 21q22 contiguous gene syndrome encompassing RUNX1. Am J Med Genet A. 2016;170:2580–6.
Sarper SE, Inubushi T, Kurosaka H, Ono Minagi H, Kuremoto KI, Sakai T, et al. Runx1-Stat3 signaling regulates the epithelial stem cells in continuously growing incisors. Sci Rep. 2018;8:10906 Runx1 is involved in the maintenance of the stem cells through regulation of Lgr5 in the cervical loop of the growing incisors.
Kurosaka H, Islam MN, Kuremoto K, Hayano S, Nakamura M, Kawanabe N, et al. Core binding factor beta functions in the maintenance of stem cells and orchestrates continuous proliferation and differentiation in mouse incisors. Stem Cells. 2011;29:1792–803.
Osorio KM, Lee SE, McDermitt DJ, Waghmare SK, Zhang YV, Woo HN, et al. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development. 2008;135:1059–68.
Suomalainen M, Thesleff I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev Dyn. 2010;239:364–72.
Minetti CA, Valle LB, Oliveira-Filho RM, Fava-De-Moraes F. Differential actions of testosterone and its metabolites on mice submandibular gland. J Biol Buccale. 1985;13:205–13.
Caramia F. Ultrastructure of mouse submaxillary gland. II. Effect of castration in the male. J Ultrastruct Res. 1966;16:524–36.
Ono Minagi H, Sarper SE, Kurosaka H, Kuremoto KI, Taniuchi I, Sakai T, et al. Runx1 mediates the development of the granular convoluted tubules in the submandibular glands. PLoS One. 2017;12:e0184395 Runx1 regulates the development of the granular convoluted tubules in the submandibular glands.
Islam MN, Itoh S, Yanagita T, Sumiyoshi K, Hayano S, Kuremoto K, et al. Runx/Cbfb signaling regulates postnatal development of granular convoluted tubule in the mouse submandibular gland. Dev Dyn. 2015;244:488–96.
He JF, Ge MH, Zhu X, Chen C, Tan Z, Li YN, et al. Expression of RUNX3 in salivary adenoid cystic carcinoma: implications for tumor progression and prognosis. Cancer Sci. 2008;99:1334–40.
Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–24.
Ge MH, Chen C, Xu JJ, Ling ZQ. Critical regions and spreading of runt-related transcription factor-3 C-phosphate-G (CpG) island methylation in human salivary gland adenoid cystic carcinoma. Hum Pathol. 2011;42:1862–72.
Aberg T, Cavender A, Gaikwad JS, Bronckers AL, Wang X, Waltimo-Sirén J, et al. Phenotypic changes in dentition of Runx2 homozygote-null mutant mice. J Histochem Cytochem. 2004;52:131–9.
Sull JW, Liang KY, Hetmanski JB, Fallin MD, Ingersoll RG, Park J, et al. Differential parental transmission of markers in RUNX2 among cleft case-parent trios from four populations. Genet Epidemiol. 2008;32:505–12.
Wu LZ, Su WQ, Liu YF, Ge X, Zhang Y, Wang XJ. Role of the RUNX2 p.R225Q mutation in cleidocranial dysplasia: a rare presentation and an analysis of the RUNX2 protein structure. Genet Mol Res. 2014;13:1187–94.
Iyyanar PPR, Nazarali AJ. Inhibits bone morphogenetic protein signaling during osteogenic differentiation of the palatal mesenchyme. Front Physiol. 2017;8:929
Sarper SE, Kurosaka H, Inubushi T, Ono Minagi H, Kuremoto KI, Sakai T, et al. Runx1-Stat3-Tgfb3 signaling network regulating the anterior palatal development. Sci Rep. 2018;8:11208. Runx1 regulates the epithelial fusion at the boundary between the primary and the secondary palate.
Braddock SR, Carey JC. A new syndrome: congenital thrombocytopenia, Robin sequence, agenesis of the corpus callosum, distinctive facies and developmental delay. Clin Dysmorphol. 1994;3:75–81.
Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat. 2005;207:655–67.
Taya Y, O’Kane S, Ferguson MW. Pathogenesis of cleft palate in TGF-beta3 knockout mice. Development. 1999;126:3869–79.
Lane J, Yumoto K, Azhar M, Ninomiya-Tsuji J, Inagaki M, Hu Y, et al. Tak1, Smad4 and Trim33 redundantly mediate TGF-β3 signaling during palate development. Dev Biol. 2015;398:231–41.
Sarper SE, Inubushi T, Kurosaka H, Ono Minagi H, Murata Y, Kuremoto KI, et al. Anterior cleft palate due to Cbfb deficiency and its rescue by folic acid. Dis Model Mech. 2019;12. Cbfb deficiency results in anterior cleft palate and folic acid rescues the mutant phenotype by activation of Stat3 phosphorylation.
Charoenchaikorn K, Yokomizo T, Rice DP, Honjo T, Matsuzaki K, Shintaku Y, et al. Runx1 is involved in the fusion of the primary and the secondary palatal shelves. Dev Biol. 2009;326:392–402.
Khan A, Hyde RK, Dutra A, Mohide P, Liu P. Core binding factor beta (CBFB) haploinsufficiency due to an interstitial deletion at 16q21q22 resulting in delayed cranial ossification, cleft palate, congenital heart anomalies, and feeding difficulties but favorable outcome. Am J Med Genet A. 2006;140:2349–54.
Tsoutsou E, Tzetis M, Giannikou K, Syrmou A, Oikonomakis V, Kosma K, et al. Array-CGH revealed one of the smallest 16q21q22.1 microdeletions in a female patient with psychomotor retardation. Eur J Paediatr Neurol. 2013;17:316–20.
Reich NC. STATs get their move on. JAKSTAT. 2013;2:e27080.
Stark GR, Darnell JE. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–14.
Grimbacher B, Holland SM, Gallin JI, Greenberg F, Hill SC, Malech HL, et al. Hyper-IgE syndrome with recurrent infections--an autosomal dominant multisystem disorder. N Engl J Med. 1999;340:692–702.
Haley KJ, Lasky-Su J, Manoli SE, Smith LA, Shahsafaei A, Weiss ST, et al. RUNX transcription factors: association with pediatric asthma and modulated by maternal smoking. Am J Phys Lung Cell Mol Phys. 2011;301:L693–701.
Wu T, Fallin MD, Shi M, Ruczinski I, Liang KY, Hetmanski JB, et al. Evidence of gene-environment interaction for the RUNX2 gene and environmental tobacco smoke in controlling the risk of cleft lip with/without cleft palate. Birth Defects Res A Clin Mol Teratol. 2012;94:76–83.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Takashi Yamashiro, Hiroshi Kurosaka, and Toshihiro Inubush declare no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Craniofacial Skeleton
Rights and permissions
About this article
Cite this article
Yamashiro, T., Kurosaka, H. & Inubush, T. The Association Between Runx Signaling and Craniofacial Development and Disease. Curr Osteoporos Rep 20, 120–126 (2022). https://doi.org/10.1007/s11914-021-00692-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11914-021-00692-w