Abstract
Purpose of Review
Epigenetic modifications via DNA methylation have previously been linked to blood lipid levels, dyslipidemias, and atherosclerosis. The purpose of this review is to discuss current literature on the role of DNA methylation on lipid traits and their associated pathologies.
Recent Findings
Candidate gene and epigenome-wide approaches have identified differential methylation of genes associated with lipid traits (particularly CPT1A, ABCG1, SREBF1), and novel approaches are being implemented to further characterize these relationships. Moreover, studies on environmental factors have shown that methylation variations at lipid-related genes are associated with diet and pollution exposure.
Summary
Further investigation is needed to elucidate the directionality of the associations between the environment, lipid traits, and epigenome. Future studies should also seek to increase the diversity of cohorts, as European and Asian ancestry populations are the predominant study populations in the current literature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipids are a class of biomolecules that include fats, sterols, phospholipids, and more. Their primary function in humans is to store energy and provide structural support to the cell membrane [1]. However, dysfunction of lipid metabolism, storage, or clearance may result in abnormal blood lipid levels, i.e., dyslipidemia [2]. An independent risk factor for cardiovascular disease, dyslipidemias, arises through both monogenic mutations (e.g., low density lipoprotein receptor (LDLR) in familial hypercholesterolemia) and polygenic influences, such as single nucleotide polymorphisms (SNPs) [3]. This is supported by the high heritability of plasma lipids: estimates for high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, triglycerides (TG), and total cholesterol (TC) range from 40 to 60% [4]. Yet, even with the incorporation of rare variants, known genetic variants account for 10 to 25% of the total variation in lipid levels [5, 6]. This suggests that complex interactions between the genome and environment remain unaccounted for in the pathogenesis of dyslipidemia. Other genomic modifications (e.g., epigenomics) have been theorized to contribute to the “missing heritability” of lipids.
DNA methylation is the most common form of epigenetic modification and involves the addition of methyl (-CH3) groups to cytosine-phosphate-guanine (CpG) sites of genes. Generally, increased methylation in gene promoters—regions of DNA that regulate gene expression via activation of transcription—is associated with decreased gene expression, and decreased methylation is associated with increased gene expression. The relationship between DNA methylation and lipids, as well as pathologies associated with abnormal lipid levels (dyslipidemia, atherosclerosis, etc.), has been extensively studied. Specifically, candidate gene and epigenome-wide association studies (EWAS) have identified differential methylation of genes in connection with lipid phenotypes. However, some questions remain, such as the directionality of the relationship between methylation and lipid levels, as well as whether the observed methylation variations in blood (the predominant tissue sample in these analyses) reflect gene regulation in target tissues. Furthermore, findings are mixed on the effect of environmental factors on lipids via DNA methylation. In this review, we will explore novel methodological approaches to explicate the relationship between lipids and methylation; evaluate the current literature on methylation variations at genes that have been widely replicated in association with plasma lipids; and highlight environmental exposures that have been linked to both methylation changes and lipid traits.
Methodological Approaches
EWAS
Early studies of DNA methylation and lipids predominantly applied candidate gene and EWAS approaches. Candidate gene studies test for associations between methylation and preselected genes of interest. In recent years, they have been used primarily in case–control studies and smaller cohorts (n < 100), and, for the purpose of this review, we will not discuss those smaller studies. Moreover, many EWAS—which use a hypothesis-free approach to identify associations between methylation of individual CpG sites and the trait of interest—since 2016 have included some form of validation (split sample approach or external validation). Meta-analyses are also becoming an increasingly useful tool to increase sample size and statistical power to detect significant CpG sites across cohorts. To date, most EWAS of lipids have been cross-sectional in design and conducted predominantly in populations of European or Asian ancestry. These studies have not only evaluated methylation variations in association with lipid levels directly [7,8,9,10], but also lipidomic profiles [11], plasma lipoprotein A [12], and pathologies such as atherosclerotic stroke [13]. However, a limitation of this approach is that it considers the effects of individual CpG sites, whereas multiple CpGs may be present in a single promoter region. Studies that evaluate the combined effect of methylation variations of CpG sites, e.g., differentially methylated regions (DMRs), are lacking.
Multi- “omics” Approaches
Methylation analyses have begun to integrate other “-omics” data to evaluate the joint effects of both the genome and epigenome on gene expression. While these studies are not as abundant as EWAS, they have been increasing in frequency. For example, some investigators have begun to incorporate genome-wide association study (GWAS) data into their methylation analyses. In a cohort of ~ 700 older African American adults, Wright et al. evaluated the association between serum lipids and DNA methylation sites that were proximal to single nucleotide polymorphisms (SNPs) previously linked to lipid traits [14]. Using a methylation-quantitative trait loci (meQTL) approach, Bandesh et al. identified functional variants in a cohort of ~ 230 Indian adults that were associated with methylation changes in the same genes with those significant variants [15]. In a GWAS meta-analysis, Ghanbari et al. explored associations between long non-coding RNAs (lncRNAs) and cardiometabolic disorders, in which they found that the methylation level of cg17371580 (located in the promoter of LOC157273) was associated with HDL [16]. The lncRNA gene, LOC157273, is an effector transcript located near PPP1R3B, which has also been linked to LDL and coronary artery disease [16,17,18].
In an expression-quantitative trait loci (eQTL) approach, Love-Gregory et al. identified lipid-associated SNPs located near the cluster of differentiation 36 (CD36) gene in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. The association between those SNPs, CpG sites in the CD36 promoter region, and CD36 expression in adipose tissue from the Multiple Tissue Human Expression Resource (MuTHER) was then evaluated [19]. The investigators determined that both SNPs and methylation changes independently influence CD36 expression. CD36 facilitates fatty acid uptake by the cell, and in previous studies, CD36 variants have been shown to influence fasting lipid levels and risk for metabolic syndrome [20, 21]. Furthermore, Tremblay et al. applied a weighted gene correlation network analysis in a cohort of 16 families (n = 48) and identified several genes that were associated with HDL, LDL, and TC, as well as ApoB100 [22].
Tissue-Specific Methylation
While most human methylation studies have performed analyses from whole blood, a few have begun to explore tissue-specific methylation to further evaluate the relationship between DNA methylation and lipid-associated pathologies. In larger cohort studies, these tissues tend to be used for more targeted, functional analyses after identification of significant CpG sites from genome-wide analyses. For example, Pfeiffer et al. used skin (n ~ 400) and adipose (n ~ 650) tissue samples from the MuTHER cohort to evaluate the relationship between CpG sites that were significant in an EWAS of whole blood and gene expression [23]. Similar to the results from blood, increased methylation at sterol regulatory element binding transcription factor 1 (SREBF1) was associated with elevated TG in both adipose and skin. Increased methylation at adenosine triphosphate (ATP) binding cassette subfamily G member 1 (ABCG1) was only associated with elevated TG in adipose. In another analysis, in subcutaneous fat from the TwinsUK registry (which has some overlap with MuTHER), cg20544516 methylation was negatively associated with SREBF1 expression [24••]. These findings are comparable to EWAS in whole blood, which have shown increased methylation of this CpG site in association with elevated lipid levels [7, 24••]. Overall, the concordance between methylation analyses of lipid traits in whole blood and those in other tissues, particularly adipose, suggest that blood-based methylation studies are reflecting functionally important variations in DNA methylation at the tissue level.
While in larger cohort studies, tissue-specific methylation analyses tend to be applied for validation and/or secondary analyses, some smaller studies are using this approach for primary investigation. Adipose tissue samples collected from ~ 200 Canadian adults were analyzed by methylC-capture sequencing (MCC-Seq) to fine-map EWAS signals [25]. Investigators identified and externally validated adipose tissue regulatory regions that were associated with HDL and TG. Moreover, Lacey et al. identified tissue-specific differentially methylated regions (DMRs) in atherosclerosis using smooth muscle cells (SMCs) collected from patient aortas (n = 3); they found that methylation changes in aorta SMCs may downregulate enhancers to facilitate a pro-atherosclerotic phenotype [26]. The same investigators also identified tissue-specific regulation of ANGTP in atherosclerosis, in addition to methylation changes in enhancers regions, thus contributing to ANGPT expression [27]. Furthermore, Wang et al. observed that decreased methylation at genes in atherosclerotic right coronary artery tissue (compared to the great saphenous vein) were associated with pro-inflammatory pathways (e.g., cytokine receptor interactions); genes with increased methylation were associated with fat digestion and absorption pathways [28]. Tristán-Flores et al. characterized a differential methylation motif in human atheromas that was associated with Alu methylation, which is a hallmark of atherosclerosis [29]. Most studies of methylation and lipids are still based on whole blood samples, but some are beginning to evaluate methylation variations in vascular and adipose tissues with respect to lipids.
Finally, other approaches to evaluating DNA methylation and lipids are broadening from discovery and validation of individual genes or CpG sites of interest. More recent studies have included pathway [30•] and gene network analyses [22], indicating that researchers may be shifting from identifying methylation within individual genes to exploring how these modifications affect larger systems (e.g., regulatory pathways, gene networks, etc.) in disease. Methods are also being developed to employ epigenetic data as potential biomarkers. For example, Irvin et al. observed that epigenetic age acceleration (estimated from epigenetic clock algorithms) was positively associated with inflammatory markers and TG and negatively associated with HDL in the GOLDN study [31]. More studies, however, are needed before these tools are clinically translatable. Overall, the methods for understanding DNA methylation in the context of lipids are vast, as the field is shifting from candidate gene and EWAS-based approaches to more integrated “omic” analytical methods.
Widely Replicated Genes
CPT1A
Carnitine palmitoyltransferase 1A (CPT1A) codes for a key enzyme that initiates long-chain fatty acid beta-oxidation by the mitochondria, thus playing an important role in lipid metabolism. Multiple cross-sectional EWAS in whole blood have identified methylation variations of CpG sites annotated to CPT1A in association with blood lipid levels. In a population-based cohort study of ~ 1500 Dutch adults aged 45 years and older, Braun et al. identified differential methylation of two CpG sites (cg00574958 and cg17058475) in association with TG levels [7]. Methylation of these CpG sites were inversely associated with TG and very low-density lipoprotein (VLDL) cholesterol [32, 33]. GOLDN investigators also found that methylation of cg00574958, specifically, explained ~ 12% of the variation in TG [32]. Further study showed that decreased methylation of cg00574958 was linked to elevated plasma adiponectin levels [8]. Adiponectin is a hormone primarily secreted from adipose tissue, and it plays multiple roles in fatty acid oxidation, insulin resistance, and atherosclerosis [34]. This relationship not only remained significant after accounting for body mass index (BMI) and cigarette smoking, but also was replicated in a cohort of Amish adults (n ~ 500), as well as white, but not black, adults in the Bogalusa Heart Study (n ~ 850).
Furthermore, CPT1A methylation (specifically cg00574958) has been linked to other cardiometabolic traits and pathologies: hypertriglyceridemic waist phenotype, a potential marker of type 2 diabetes (T2D) [35]; familial hypercholesterolemia [36]; BMI [37]; adiposity [38]; carbohydrate and fat intake [33]; and metabolic syndrome [39]. A longitudinal study in rats suggested that high fat diet may increase CPT1A expression in blood [40], but human studies are lacking. Considering these findings, some investigators have sought to elucidate the causal relationship between CPT1A methylation and lipid traits through Mendelian randomization analyses. While these studies are not yet abundant, the current findings suggest that the lipid levels are causal for methylation variations rather than methylation being causal for the lipid trait variation. Sayols-Baixeras et al. identified causal effects of fasting TG levels on the methylation of CpG sites annotated to CPT1A in a cohort of ~ 1000 adults [41]. Similarly, Dekkers et al. observed causal effects of TG on the same CpG site (cg00574958) in the BIOS Consortium (n ~ 2000) [42]. Moreover, a metabolomics study (n ~ 360) identified metabolite levels as causal on methylation of multiple CpG loci, including those annotated to CPT1A [43]. These findings suggest that CPT1A methylation plays an important role in cardiometabolic diseases but that the mechanisms are complicated and could be environmental.
ABCG1
ABCG1 codes for a protein involved in cholesterol transport in macrophages and lipid homeostasis. ABCG1 methylation has previously been linked to T2D and glycemic traits, including as a potential mediator between statin use and T2D risk [44, 45]. EWAS have identified increased methylation at CpG sites annotated to ABCG1 in association with elevated TG and lower HDL levels: cg06500161 and cg27243685 [7, 23, 24••]. Other studies have evaluated associations between these CpG sites and hypertriglyceridemic waist [35], metabolic syndrome [46], prior myocardial infarction [23], insulin resistance [47•], and adiposity [48]. Additionally, an EWAS of ~ 650 German adults in a population-based study (KORA F4) showed that methylation of cg06500161 was inversely associated with ABCG1 expression in whole blood; this association was marginally significant after correction for multiple comparisons [24••]. These findings were supported by an earlier study in the KORA F4 cohort also reporting the inverse association between ABCG1 methylation and corresponding mRNA transcripts as well as HDL levels; the investigators further identified a positive association between ABCG1 mRNA and HDL levels, suggesting that the relationship between ABCG1 methylation and HDL may be mediated by ABCG1 expression [23]. Another study showed that methylation at cg07397296 partially mediated the relationship between in utero famine exposure and adult TG levels, and these methylation variations were associated with gene expression in an external dataset [49]. Overall, the directional relationship between ABCG1 methylation and lipids has been difficult to assess from cross-sectional research, although a Mendelian randomization analysis showed a causal effect of HDL on methylation of ABCG1 CpG sites [42]. Candidate gene and case–control studies have also linked increased methylation of ABCG1 to atherosclerotic markers and elevated LDL [30•, 50]. In sum, differential methylation of ABCG1 has been widely associated with lipids and related traits. Still, further analyses are needed to characterize the relationship between gene expression, environmental factors, methylation, and lipid levels.
SREBF1
SREBF1 binds a motif in the promoter region of the LDL receptor gene (LDLR) to activate its transcription for cholesterol metabolism. EWAS in whole blood have repeatedly found that increased methylation at cg11024682 and cg20544516 (located in the promoter region of SREBF1) is associated with a worsening lipid profile. Increased methylation of cg11024682 was also linked to elevated TG levels and postprandial lipemia in GOLDN [7, 33], as well as decreased HDL levels in the Registre Gironí del Cor (REGICOR), Framingham Offspring Study, and GOLDN cohorts (n ~ 3300) [51]. Moreover, a meta-analysis of European and Indian adults (n ~ 5500) showed that cg11024682 and cg20544516 methylation was positively associated with VLDL and TG levels [24••]. In secondary analyses (n ~ 1700), Gomez-Alonzo et al. also found that cg20544516 methylation was inversely related to expression of SREBF1 cis-transcripts in subcutaneous fat. These findings are consistent with previous analyses showing that differential methylation of cg11024682 and cg20544516 in relation to lipid traits persist in both whole blood and adipose tissue. In a cohort of ~ 1800 adults, not only did methylation of these CpG sites in blood each explain ~ 3% of the variation in TG level, but they were differentially methylated in skin and adipose tissue samples from an external cohort [23]. Like CPT1A and ABCG1, Mendelian randomization has showed a causal effect of TG levels on methylation of this gene [42]. Moreover, increased methylation at SREBF1 has been associated with central adiposity [52]; BMI [47•, 53,54,55]; childhood and adult obesity [30•, 56]; and T2D [57, 58]. These findings have been replicated in diverse cohorts, increasing their generalizability. Given the relationship between these CpG sites and several traits, SREBF1 methylation may serve as a potential “multipurpose” biomarker for cardiometabolic dysfunction, not exclusive to lipids.
Overall, candidate gene and EWAS analyses have identified and repeatedly validated that CPT1A, ABCG1, and SREBF1 are targets of DNA methylation in lipid metabolism and associated disease. Furthermore, Pfeiffer et al. have demonstrated how these genes are interrelated to regulate cholesterol homeostasis and fatty acid metabolism: MIR33a/b is co-transcribed with SREBF1, and the intronic miRNA acts as a negative regulator of ABCG1 and CPT1A [23]. While current findings suggest that the relationships between lipid traits and methylation of these genes in blood are concordant with those in adipose tissue, more studies are needed to evaluate how these methylation variations affect gene transcript levels in target tissue. Other genes (e.g., DHCR24, ABCA1) may also be of interest, but they have not been validated as extensively as those discussed above. More studies are needed to fully describe the directionality of the environment, methylation, and lipid traits.
Environmental Considerations
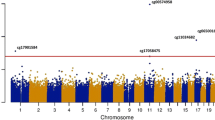
While Mendelian randomization studies of methylation and lipids are limited, current analyses suggest that methylation changes are the consequence of lipid traits rather than the cause, supporting an important role for exposures which induce methylation changes [42]. Prospective epigenetic studies of environmental factors also consistently show that external exposures may induce methylation changes relevant to lipids [59] (Fig. 1). Importantly, environmental epigenetic study findings have shown overlap with putative metabolic and lipid-related pathways, but the findings are mostly independent of previous lipid EWAS discoveries discussed above. These analyses have primarily focused in the areas of pollution and diet.
The interrelation of the environment, epigenome, and lipids are not fully understood. Environmental factors may induce methylation changes at genes that are associated with lipid traits. Conversely, Mendelian randomization studies suggest that lipid traits are causal for methylation. Further studies (via a diverse set of analytical methods) are needed clarify the directionality of these relationships. Abbreviations: microRNA (miRNA); long non-coding RNA (lncRNA); low-density lipoprotein cholesterol (LDL); triglycerides (TG); total cholesterol (TC); high-density lipoprotein cholesterol (HDL); coronary artery disease (CAD); myocardial infarction (MI); epigenome-wide association study (EWAS); methylation quantitative trait loci (meQTL); expression quantitative trait loci (eQTL)
Pollution
Air pollution is a risk factor for exposure to particulate matter: hazardous, microscopic particles that are suspended in the atmosphere [60]. An EWAS meta-analysis in the Women’s Health Initiative (WHI) and the Atherosclerosis Risk in Communities (ARIC) studies identified significant methylation changes in association with particulate matter exposure (PM2.5–10, PM10), including at a CpG site annotated to miR128-2 [61]. This miRNA has been posited as an inhibitor of ABCG1 [62], and its methylation could result in ABCG1 upregulation. However, as previously discussed, ABCG1 silencing (via increased methylation) has been associated with elevated lipid levels in EWAS studies. Studies are lacking on the association between miR128-2 and lipids, but these findings suggest that there may be complex and potentially multidirectional relationships between pollution, methylation, and lipids. Furthermore, in a randomized crossover study of healthy young adults in China (n = 36), investigators observed increased methylation in the promoter regions of additional sex comb-like 2 (ASXL2) and lamin A/C (LMNA) following short-term, high exposure to particulate matter (PM2.5) [63]. ASXL2 encodes a transcriptional regulator that facilitates lipid homeostasis via the PPAR-γ pathway [64]. LMNA encodes lamins A and C, structural nuclear proteins that contribute to lipid metabolism and storage. Mutations in LMNA can cause laminopathies, of which hyperlipidemia and atherosclerosis are clinical presentations [65]. More prospective studies in larger cohorts are needed, but preliminary analyses suggest that, at the very least, there is a connection between exposure to particulate matter and lipid-related genes via DNA methylation.
Other studies have explored the role of endocrine disrupting chemicals (EDCs), such as phthalates and parabens, in methylation changes. Among a cohort of Dutch adults, an EWAS identified differentially methylated CpG sites related to urinary concentration of EDCs. Multiple CpG sites were annotated to genes that are functionally related to TG and HDL levels [66]. Another EDC, di(2-ethylhexyl)phthalate (DEHP), is added to plastics to make them flexible, and chronic DEHP exposure may cause adverse cardiovascular effects [67, 68]. In both animal models and epidemiologic studies, DEHP exposure was associated with global hypermethylation, elevated cholesterol levels, and carotid intima-media thickness (CIMT, a marker of subclinical atherosclerosis) [69, 70].
Heavy metal exposure, thought to affect cardiovascular tissues through oxidative stress pathways, has similarly been linked to both atherosclerosis and DNA methylation [71]. In a study of children and young adults (n ~ 700), investigators observed that urinary concentrations of lead and cadmium were positively associated with both CIMT and global DNA methylation [72]. Similarly, in a pilot study of epigenetic changes in a cohort of middle-aged men (n = 23), 46% of DMRs associated with exposure to metals overlapped with atherosclerosis-related DMRs [73]. Pathway analyses showed that these genes were involved in inflammatory and metabolic processes. In a human cell line, treatment with arsenic exposure upregulated the transcription of DNA (cytosine-5)-methyltransferase 1 (DNMT1) via reactive oxygen species. The DNMT1 enzyme in turn methylated the ABCA1 promoter, induced global hypomethylation, and inhibited cholesterol efflux in macrophages [74]. ABCA1 is a member of the superfamily of ATP-binding cassette transporters that includes ABCG1, and its encoded protein plays a role in cholesterol efflux and HDL formation [75]. While investigators of these studies have proposed that DNA methylation may mediate the relationship between pollutants and lipids, further studies, such as Mendelian randomization, are needed to establish causality.
Diet
The relationship between methylation, diet, and lipids have been extensively explored. In the GOLDN study (n ~ 1000), CpG sites annotated to multiple genes, including CPT1A, were associated with postprandial lipemia—elevation of TG levels after eating a high-fat meal [33]. The association remained significant after adjusting for baseline TG levels for cg005794958 and cg1705847 (both annotated to CPT1A). Another EWAS in GOLDN found that increased methylation of the ABCA1 promoter was associated with lower circulating omega-3 fatty acid and HDL levels [10]. Furthermore, a cross-sectional analysis of Japanese adults showed inverse relationships between ABCA1 methylation and dietary vitamin intake. Investigators suggested that ABCA1 methylation may mediate the effect of vitamin intake on HDL [76].
In a randomized controlled diet intervention trial, researchers evaluated the effect of a Mediterranean diet on methylation over a 5-year period [77]. Components of the Mediterranean diet induced methylation changes in genes associated with metabolism, diabetes, inflammation, and signal transduction. Specifically, Arpón et al. identified a CpG site (cg01081346) annotated to CPT1B—a paralog of CPT1A that encodes the rate-limiting enzyme of fatty acid oxidation in skeletal muscle—that was associated with polyunsaturated fatty acid intake among study participants. In another randomized, placebo-controlled trial of a dietary intervention, Lima et al. found that hazelnut oil consumption was associated with decreased methylation at ADRB3 and an increase in HDL [78]. In a later EWAS, the research group found that ADRB3 methylation was associated with higher fat intake and LDL [79]. ADRB3 encodes the adrenoreceptor beta 3, which is involved in regulating lipolysis.
In both prospective and cross-sectional analyses across multiple populations, exposures to pollutants and endocrine disruptors (e.g., phthalates, heavy metals) have been linked to differential DNA methylation. Many of these genes are involved in lipid metabolism and homeostasis. Additionally, dietary factors, particularly the consumption of different types of fats, have also been associated with the methylation of these linked to lipid-related genes. Further investigation is needed to explain the complex relationship between environmental exposures, DNA methylation, and lipids.
Conclusion and Future Directions
In summary, studies suggest that DNA methylation, variation of plasma lipids, and pathogenesis of dyslipidemias are entwined. Associations have been validated across multiple cohorts (particularly for CPT1A, ABCG1, and SREBF1). Candidate gene and EWAS approaches are still broadly applied, but investigators are expanding their analyses to multi-omics approaches that include genomic, transcriptomic, and metabolomic data. Intervention trials that explore the effects of environmental factors, such as diet, have shown methylation variations in association with lipids. Still, large epigenomic studies are predominantly cross-sectional in design.
Future analyses should continue to increase the diversity of study populations, as European ancestry populations are disproportionately overrepresented in the current literature. Additionally, explanations of the causal effects of methylation on lipids (or vice versa) are lacking. More studies that apply approaches such as Mendelian randomization or that capture prospective data are needed to clarify the relationship between the environment, DNA methylation, and lipids. Studies should also continue to assess how these relationships alter gene expression across relevant tissues. Other considerations include the evaluation of methylation variation over the lifespan, or in the context of medication responses (i.e., pharmacoepigenetics). Overall, while there has been significant progress in our understanding of DNA methylation and lipids, considerable research is still needed to translate this information into clinically applicable tools.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Muro E, Atilla-Gokcumen GE, Eggert US. Lipids in cell biology: how can we understand them better? Mol Biol Cell. 2014;25(12):1819–23. https://doi.org/10.1091/mbc.E13-09-0516.
Hegele RA. Lipoprotein and lipid metabolism. In: Rimoin D, Pyeritz R, Korf B, editors. Emery and Rimoin’s principles and practice of medical genetics, 6th ed. Academic Press; 2013. https://doi.org/10.1016/B978-0-12-383834-6.00100-2.
Nelson RH. Hyperlipidemia as a risk factor for cardiovascular disease. Prim Care. 2013;40(1):195–211. https://doi.org/10.1016/j.pop.2012.11.003.
Tada H, Won HH, Melander O, Yang J, Peloso GM, Kathiresan S. Multiple associated variants increase the heritability explained for plasma lipids and coronary artery disease. Circ Cardiovasc Genet. 2014;7(5):583–7. https://doi.org/10.1161/CIRCGENETICS.113.000420.
Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–83. https://doi.org/10.1038/ng.2797.
Natarajan P, Peloso GM, Zekavat SM, Montasser M, Ganna A, Chaffin M, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat Commun. 2018;9(1):3391. https://doi.org/10.1038/s41467-018-05747-8.
Braun KVE, Dhana K, de Vries PS, Voortman T, van Meurs JBJ, Uitterlinden AG, BIOS consortium, Hofman A, Hu FB, Franco OH, Dehghan A. Epigenome-wide association study (EWAS) on lipids: the Rotterdam Study. Clin Epigenetics. 2017;9:15. https://doi.org/10.1186/s13148-016-0304-4.
Aslibekyan S, Do AN, Xu H, Li S, Irvin MR, Zhi D, Tiwari HK, Absher DM, Shuldiner AR, Zhang T, Chen W, Tanner K, Hong C, Mitchell BD, Berenson G, Arnett DK. CPT1A methylation is associated with plasma adiponectin. Nutr Metab Cardiovasc Dis. 2017;27(3):225–33. https://doi.org/10.1016/j.numecd.2016.11.004.
Hedman ÅK, Mendelson MM, Marioni RE, Gustafsson S, Joehanes R, Irvin MR, et al. Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circ Cardiovasc Genet. 2017;10(1):e001487. https://doi.org/10.1161/CIRCGENETICS.116.001487.
Ma Y, Follis JL, Smith CE, Tanaka T, Manichaikul AW, Chu AY, et al. Interaction of methylation-related genetic variants with circulating fatty acids on plasma lipids: a meta-analysis of 7 studies and methylation analysis of 3 studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Am J Clin Nutr. 2016;103(2):567–78. https://doi.org/10.3945/ajcn.115.112987.
Xie T, Gorenjak V, Stathopoulou GM, Dadé S, Marouli E, Masson C, et al. Epigenome-wide association study (EWAS) of blood lipids in healthy population from STANISLAS Family Study (SFS). Int J Mol Sci. 2019;20(5):1014. https://doi.org/10.3390/ijms20051014.
Jones GT, Marsman J, Bhat B, Phillips VL, Chatterjee A, Rodger EJ, et al. DNA methylation profiling identifies a high effect genetic variant for lipoprotein(a) levels. Epigenetics. 2020;15(9):949–58. https://doi.org/10.1080/15592294.2020.1739797.
Shen Y, Peng C, Bai Q, Ding Y, Yi X, Du H, et al. Epigenome-wide association study indicates hypomethylation of MTRNR2L8 in large-artery atherosclerosis stroke. Stroke. 2019;50(6):1330–8. https://doi.org/10.1161/STROKEAHA.118.023436.
Wright ML, Ware EB, Smith JA, Kardia SLR, Taylor JY. Joint influence of SNPs and DNA methylation on lipids in African Americans from hypertensive sibships. Biol Res Nurs. 2018;20(2):161–7. https://doi.org/10.1177/1099800417752246.
Bandesh K, Prasad G, Giri AK, Kauser Y, Upadhyay M, INDICO, et al. Genome-wide association study of blood lipids in Indians confirms universality of established variants. J Hum Genet. 2019;64(6):573–87. https://doi.org/10.1038/s10038-019-0591-7.
Ghanbari M, Peters MJ, de Vries PS, Boer CG, van Rooij JGJ, Lee YC, et al. A systematic analysis highlights multiple long non-coding RNAs associated with cardiometabolic disorders. J Hum Genet. 2018;63(4):431–46. https://doi.org/10.1038/s10038-017-0403-x.
Manning AK, Goustin AS, Kleinbrink EL, Thepsuwan P, Cai J, Ju D, et al. A long non-coding RNA, LOC157273, is an effector transcript at the chromosome 8p23.1-PPP1R3B metabolic traits and type 2 diabetes risk locus. Front Genet. 2020;11:615. https://doi.org/10.3389/fgene.2020.00615.
Li WJ, Yin RX, Huang JH, Bin Y, Chen WX, Cao XL. Association between the PPP1R3B polymorphisms and serum lipid traits, the risk of coronary artery disease and ischemic stroke in a southern Chinese Han population. Nutr Metabo (Lond). 2018;15:27. https://doi.org/10.1186/s12986-018-0266-y.
Love-Gregory L, Kraja AT, Allum F, Aslibekyan S, Hedman ÅK, Duan Y, et al. Higher chylomicron remnants and LDL particle numbers associate with CD36 SNPs and DNA methylation sites that reduce CD36. J Lipid Res. 2016;57(12):2176–84. https://doi.org/10.1194/jlr.P065250.
Chien KL, Hsu HC, Liu PH, Lin HJ, Chen MF. Common sequence variants in CD36 gene and the levels of triglyceride and high-density lipoprotein cholesterol among ethnic Chinese in Taiwan. Lipids Health Dis. 2012;11:174. https://doi.org/10.1186/1476-511X-11-174.
Pietka TA, Schappe T, Conte C, Fabbrini E, Patterson BW, Klein S, et al. Adipose and muscle tissue profile of CD36 transcripts in obese subjects highlights the role of CD36 in fatty acid homeostasis and insulin resistance. Diabetes Care. 2014;37(7):1990–7. https://doi.org/10.2337/dc13-2835.
Tremblay BL, Guénard F, Lamarche B, Pérusse L, Vohl MC. Network analysis of the potential role of DNA methylation in the relationship between plasma carotenoids and lipid profile. Nutrients. 2019;11(6):1265. https://doi.org/10.3390/nu11061265.
Pfeiffer L, Wahl S, Pilling LC, Reischl E, Sandling JK, Kunze S, et al. DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet. 2015;8(2):334–42. https://doi.org/10.1161/CIRCGENETICS.114.000804.
•• Gomez-Alonso MDC, Kretschmer A, Wilson R, Pfeiffer L, Karhunen V, Seppälä I, et al. DNA methylation and lipid metabolism: an EWAS of 226 metabolic measures. Clin Epigenetics. 2021;13(1):7. https://doi.org/10.1186/s13148-020-00957-8. Findings from this study indicate that differential methylation of multiple CpG sites in relation to lipid levels persist in both whole blood and adipose tissue.
Allum F, Hedman ÅK, Shao X, Cheung WA, Vijay J, Guénard F, et al. Dissecting features of epigenetic variants underlying cardiometabolic risk using full-resolution epigenome profiling in regulatory elements. Nat Commun. 2019;10(1):1209. https://doi.org/10.1038/s41467-019-09184-z.
Lacey M, Baribault C, Ehrlich KC, Ehrlich M. Atherosclerosis-associated differentially methylated regions can reflect the disease phenotype and are often at enhancers. Atherosclerosis. 2019;280:183–91. https://doi.org/10.1016/j.atherosclerosis.2018.11.031.
Ehrlich KC, Lacey M, Ehrlich M. Tissue-specific epigenetics of atherosclerosis-related ANGPT and ANGPTL genes. Epigenomics. 2019;11(2):169–86. https://doi.org/10.2217/epi-2018-0150.
Wang X, Liu AH, Jia ZW, Pu K, Chen KY, Guo H. Genome-wide DNA methylation patterns in coronary heart disease. Herz. 2018;43(7):656–62. https://doi.org/10.1007/s00059-017-4616-8.
Tristán-Flores FE, Guzmán P, Ortega-Kermedy MS, Cruz-Torres G, de la Rocha C, Silva-Martínez GA, et al. Liver X receptor-binding DNA motif associated with atherosclerosis-specific DNA methylation profiles of Alu elements and neighboring CpG islands. J Am Heart Assoc. 2018;7(3):e007686. https://doi.org/10.1161/JAHA.117.007686.
• Płatek T, Polus A, Góralska J, Raźny U, Gruca A, Kieć-Wilk B, et al. DNA methylation microarrays identify epigenetically regulated lipid related genes in obese patients with hypercholesterolemia. Mol Med. 2020;26(1):93. https://doi.org/10.1186/s10020-020-00220-z. Although a preliminary study, these findings suggest that hypercholesterolemia is accompanied by a distinct DNA methylation profile in lipid-related genes.
Irvin MR, Aslibekyan S, Do A, Zhi D, Hidalgo B, Claas SA, et al. Metabolic and inflammatory biomarkers are associated with epigenetic aging acceleration estimates in the GOLDN study. Clin Epigenetics. 2018;10:56. https://doi.org/10.1186/s13148-018-0481-4.
Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, et al. Epigenome-wide association study of fasting blood lipids in the genetics of lipid-lowering drugs and diet network study. Circulation. 2014;130(7):565–72. https://doi.org/10.1161/CIRCULATIONAHA.114.009158.
Lai CQ, Wojczynski MK, Parnell LD, Hidalgo BA, Irvin MR, Aslibekyan S, et al. Epigenome-wide association study of triglyceride postprandial responses to a high-fat dietary challenge. J Lipid Res. 2016;57(12):2200–7. https://doi.org/10.1194/jlr.M069948.
Maeda N, Funahashi T, Matsuzawa Y, Shimomura I. Adiponectin, a unique adipocyte-derived factor beyond hormones. Atherosclerosis. 2020;292:1–9. https://doi.org/10.1016/j.atherosclerosis.2019.10.021.
Mamtani M, Kulkarni H, Dyer TD, Göring HH, Neary JL, Cole SA, et al. Genome- and epigenome-wide association study of hypertriglyceridemic waist in Mexican American families. Clin Epigenetics. 2016;20(8):6. https://doi.org/10.1186/s13148-016-0173-x.
Reeskamp LF, Venema A, Pereira JPB, Levin E, Nieuwdorp M, Groen AK, et al. Differential DNA methylation in familial hypercholesterolemia. EBioMedicine. 2020;61:103079. https://doi.org/10.1016/j.ebiom.2020.103079.
Karlsson IK, Ericsson M, Wang Y, Jylhävä J, Hägg S, Pedersen NL, et al. Replicating associations between DNA methylation and body mass index in a longitudinal sample of older twins. Int J Obes (Lond). 2020;44(6):1397–405. https://doi.org/10.1038/s41366-019-0498-6.
Meeks KAC, Henneman P, Venema A, Burr T, Galbete C, Danquah I, et al. An epigenome-wide association study in whole blood of measures of adiposity among Ghanaians: the RODAM study. Clin Epigenetics. 2017;9:103. https://doi.org/10.1186/s13148-017-0403-x.
Das M, Sha J, Hidalgo B, Aslibekyan S, Do AN, Zhi D, et al. Association of DNA methylation at CPT1A locus with metabolic syndrome in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. PLoS ONE. 2016;11(1):e0145789. https://doi.org/10.1371/journal.pone.0145789.
Díaz-Rúa R, Palou A, Oliver P. Cpt1a gene expression in peripheral blood mononuclear cells as an early biomarker of diet-related metabolic alterations. Food Nutr Res. 2016;60:33554. https://doi.org/10.3402/fnr.v60.33554.
Sayols-Baixeras S, Tiwari HK, Aslibekyan SW. Disentangling associations between DNA methylation and blood lipids: a Mendelian randomization approach. BMC Proc. 2018;12(Suppl 9):23. https://doi.org/10.1186/s12919-018-0119-8.
Dekkers KF, van Iterson M, Slieker RC, Moed MH, Bonder MJ, van Galen M, et al. Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17(1):138. https://doi.org/10.1186/s13059-016-1000-6.
Zaghlool SB, Mook-Kanamori DO, Kader S, Stephan N, Halama A, Engelke R, et al. Deep molecular phenotypes link complex disorders and physiological insult to CpG methylation. Hum Mol Genet. 2018;27(6):1106–21. https://doi.org/10.1093/hmg/ddy006.
Ling C. Epigenetic regulation of insulin action and secretion - role in the pathogenesis of type 2 diabetes. J Intern Med. 2020;288(2):158–67. https://doi.org/10.1111/joim.13049.
Ochoa-Rosales C, Portilla-Fernandez E, Nano J, Wilson R, Lehne B, Mishra PP, et al. Epigenetic link between statin therapy and type 2 diabetes. Diabetes Care. 2020;43(4):875–84. https://doi.org/10.2337/dc19-1828.
Nuotio ML, Pervjakova N, Joensuu A, Karhunen V, Hiekkalinna T, Milani L, et al. An epigenome-wide association study of metabolic syndrome and its components. Sci Rep. 2020;10(1):20567. https://doi.org/10.1038/s41598-020-77506-z.
• Krause C, Sievert H, Geißler C, Grohs M, El Gammal AT, Wolter S, et al. Critical evaluation of the DNA-methylation markers ABCG1 and SREBF1 for Type 2 diabetes stratification. Epigenomics. 2019;11(8):885–97. https://doi.org/10.2217/epi-2018-0159. This study found that differentially methylated CpG sites that have been widely validated in studies of lipid traits were also associated with insulin resistance and BMI in blood, liver, and adipose tissues. These findings suggest that these CpGs may have some utility as multipurpose biomarkers for cardiometabolic diseases.
Campanella G, Gunter MJ, Polidoro S, Krogh V, Palli D, Panico S, et al. Epigenome-wide association study of adiposity and future risk of obesity-related diseases. Int J Obes (Lond). 2018;42(12):2022–35. https://doi.org/10.1038/s41366-018-0064-7.
Tobi EW, Slieker RC, Luijk R, Dekkers KF, Stein AD, Xu KM, et al. DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci Adv. 2018;4(1):eaao4364. https://doi.org/10.1126/sciadv.aao4364.
Qin X, Li J, Wu T, Wu Y, Tang X, Gao P, et al. Overall and sex-specific associations between methylation of the ABCG1 and APOE genes and ischemic stroke or other atherosclerosis-related traits in a sibling study of Chinese population. Clin Epigenetics. 2019;11(1):189. https://doi.org/10.1186/s13148-019-0784-0.
Sayols-Baixeras S, Subirana I, Lluis-Ganella C, Civeira F, Roquer J, Do AN, et al. Identification and validation of seven new loci showing differential DNA methylation related to serum lipid profile: an epigenome-wide approach. The REGICOR study. Hum Mol Genet. 2016;25(20):4556–65. https://doi.org/10.1093/hmg/ddw285.
Justice AE, Chittoor G, Gondalia R, Melton PE, Lim E, Grove ML, et al. Methylome-wide association study of central adiposity implicates genes involved in immune and endocrine systems. Epigenomics. 2020;12(17):1483–99. https://doi.org/10.2217/epi-2019-0276.
Dragic D, Ennour-Idrissi K, Michaud A, Chang SL, Durocher F, Diorio C. Association between BMI and DNA methylation in blood or normal adult breast tissue: a systematic review. Anticancer Res. 2020;40(4):1797–808. https://doi.org/10.21873/anticanres.14134.
Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman ÅK, Aslibekyan S, et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 2017;14(1):e1002215. https://doi.org/10.1371/journal.pmed.1002215.
Al Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai PC, Kumar P, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenetics. 2016;8:13. https://doi.org/10.1186/s13148-016-0177-6.
Kochmanski J, Goodrich JM, Peterson KE, Lumeng JC, Dolinoy DC. Neonatal bloodspot DNA methylation patterns are associated with childhood weight status in the Healthy Families Project. Pediatr Res. 2019;85(6):848–55. https://doi.org/10.1038/s41390-018-0227-1.
Cardona A, Day FR, Perry JRB, Loh M, Chu AY, Lehne B, et al. Epigenome-wide association study of incident type 2 diabetes in a British Population: EPIC-Norfolk Study. Diabetes. 2019;68(12):2315–26. https://doi.org/10.2337/db18-0290.
Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H, et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1clevels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia. 2018;61(2):354–68. https://doi.org/10.1007/s00125-017-4497-7.
Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–8. https://doi.org/10.1016/j.tig.2004.06.009.
“Particulate Matter (PM) Basics.” EPA, Environmental Protection Agency, 1 Oct. 2020, https://www.epa.gov/pm-pollution/particulate-matter-pm-basics#PM.
Gondalia R, Baldassari A, Holliday KM, Justice AE, Méndez-Giráldez R, Stewart JD, et al. Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environ Int. 2019;132:104723. https://doi.org/10.1016/j.envint.2019.03.071.
Adlakha YK, Khanna S, Singh R, Singh VP, Agrawal A, Saini N. Pro-apoptotic miRNA-128–2 modulates ABCA1, ABCG1, and RXRa expression and cholesterol homeostasis. Cell Death Dis. 2013;4:e780.
Li H, Chen R, Cai J, Cui X, Huang N, Kan H. Short-term exposure to fine particulate air pollution and genome-wide DNA methylation: a randomized, double-blind, crossover trial. Environ Int. 2018;120:130–6. https://doi.org/10.1016/j.envint.2018.07.041.
Izawa T, Rohatgi N, Fukunaga T, Wang QT, Silva MJ, Gardner MJ, et al. ASXL2 Regulates Glucose, Lipid, and Skeletal Homeostasis. Cell Rep. 2015;11(10):1625–37. https://doi.org/10.1016/j.celrep.2015.05.019.
Di Pasquale E, Condorelli G. Endoplasmic reticulum stress at the crossroads of progeria and atherosclerosis. EMBO Mol Med. 2019;11(4):e10360. https://doi.org/10.15252/emmm.201910360.
Lu X, Fraszczyk E, van der Meer TP, van Faassen M, Bloks VW, Kema IP, et al. An epigenome-wide association study identifies multiple DNA methylation markers of exposure to endocrine disruptors. Environ Int. 2020;144:106016. https://doi.org/10.1016/j.envint.2020.106016.
Jaimes R 3rd, Swiercz A, Sherman M, Muselimyan N, Marvar PJ, Posnack NG. Plastics and cardiovascular health: phthalates may disrupt heart rate variability and cardiovascular reactivity. Am J Physiol Heart Circ Physiol. 2017;313(5):H1044–53. https://doi.org/10.1152/ajpheart.00364.2017.
Posnack NG. The adverse cardiac effects of Di(2-ethylhexyl)phthalate and Bisphenol A. Cardiovasc Toxicol. 2014;14(4):339–57. https://doi.org/10.1007/s12012-014-9258-y.
Lin CY, Lee HL, Hwang YT, Wang C, Hsieh CJ, Wu C, et al. The association between urine di-(2-ethylhexyl) phthalate metabolites, global DNA methylation, and subclinical atherosclerosis in a young Taiwanese population. Environ Pollut. 2020;265(Pt B):114912. https://doi.org/10.1016/j.envpol.2020.114912.
Xu Q, Qi W, Zhang Y, Wang Q, Ding S, Han X, et al. DNA methylation of JAK3/STAT5/PPARγ regulated the changes of lipid levels induced by di (2-ethylhexyl) phthalate and high-fat diet in adolescent rats. Environ Sci Pollut Res Int. 2020;27(24):30232–42. https://doi.org/10.1007/s11356-020-08976-x.
Bimonte VM, Besharat ZM, Antonioni A, Cella V, Lenzi A, Ferretti E, et al. The endocrine disruptor cadmium: a new player in the pathophysiology of metabolic diseases. J Endocrinol Invest. 2021. https://doi.org/10.1007/s40618-021-01502-x. Online ahead of print.
Lin CY, Lee HL, Hwang YT, Huang PC, Wang C, Sung FC, et al. Urinary heavy metals, DNA methylation, and subclinical atherosclerosis. Ecotoxicol Environ Saf. 2020;204:111039. https://doi.org/10.1016/j.ecoenv.2020.111039.
Riffo-Campos AL, Fuentes-Trillo A, Tang WY, Soriano Z, De Marco G, Rentero-Garrido P, et al. In silico epigenetics of metal exposure and subclinical atherosclerosis in middle aged men: pilot results from the Aragon Workers Health Study. Philos Trans R Soc Lond B Biol Sci. 2018;373(1748):20170084. https://doi.org/10.1098/rstb.2017.0084.
Song Y, Zhou T, Zong Y, Gu B, Tan X, Yang L. Arsenic inhibited cholesterol efflux of THP-1 macrophages via ROS-mediated ABCA1 hypermethylation. Toxicology. 2019;424:152225. https://doi.org/10.1016/j.tox.2019.05.012.
Frambach SJCM, de Haas R, Smeitink JAM, Rongen GA, Russel FGM, Schirris TJJ. Brothers in arms: ABCA1- and ABCG1-mediated cholesterol efflux as promising targets in cardiovascular disease treatment. Pharmacol Rev. 2020;72(1):152–90. https://doi.org/10.1124/pr.119.017897.
Fujii R, Yamada H, Munetsuna E, Yamazaki M, Ando Y, Mizuno G, et al. Associations between dietary vitamin intake, ABCA1 gene promoter DNA methylation, and lipid profiles in a Japanese population. Am J Clin Nutr. 2019;110(5):1213–9. https://doi.org/10.1093/ajcn/nqz181.
Arpón A, Milagro FI, Razquin C, Corella D, Estruch R, Fitó M, et al. Impact of consuming extra-virgin olive oil or nuts within a Mediterranean diet on DNA methylation in peripheral white blood cells within the PREDIMED-Navarra randomized controlled trial: a role for dietary lipids. Nutrients. 2017;10(1):15. https://doi.org/10.3390/nu10010015.
Lima RPA, do Nascimento RAF, Luna RCP, Persuhn DC, da Silva AS, da Conceição Rodrigues Gonçalves M, et al. Effect of a diet containing folate and hazelnut oil capsule on the methylation level of the ADRB3 gene, lipid profile and oxidative stress in overweight or obese women. Clin Epigenetics. 2017;9:110. https://doi.org/10.1186/s13148-017-0407-6.
Lima RPA, Ribeiro MR, de Farias Lima KQ, Sena EA, de Oliveira CD, Luna RCP, et al. Methylation profile of the ADRB3 gene and its association with lipid profile and nutritional status in adults. Biol Res. 2019;52(1):21. https://doi.org/10.1186/s40659-019-0226-7.
Funding
This work was supported by the National Institutes of Health (NIH)—National Heart, Lung, and Blood Institute (NHLBI) R01HL091357 and the National Institute of Diabetes and Digestive Kidney Diseases (NIDDK) T32DK116672.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Genetics and Genomics
Rights and permissions
About this article
Cite this article
Jones, A.C., Irvin, M.R., Claas, S.A. et al. Lipid Phenotypes and DNA Methylation: a Review of the Literature. Curr Atheroscler Rep 23, 71 (2021). https://doi.org/10.1007/s11883-021-00965-w
Accepted:
Published:
DOI: https://doi.org/10.1007/s11883-021-00965-w