Abstract
Fritillaria cirrhosa is a remarkably representative endangered species on the plateau, and its phenotype has undergone dramatic alterations due to global climate change and habitat destruction. However, the mechanism behind the phenotypic change associated with color variation has not been characterized, and subsequent physiological responses are still unknown. We investigated different phenotypes of cultivated F. cirrhosa and conducted a comprehensive transcriptomic analysis. Their agronomic traits, photosynthetic parameters, and the content of pharmaceutical ingredients were also compared. In the transcriptomic profiling, the purple phenotype had 754 up-regulated and 980 down-regulated genes compared with the green F. cirrhosa, in which a total of 37 significant differential expression genes (DEGs) regulated the anthocyanin biosynthesis by coding 6 vital enzymes (C4H, F3′H, ANS, DFR, DFT, and BA1). These DEGs were key genes responsible for the form of the purple phenotype of F. cirrhosa. Moreover, 10 DEGs were observed to be related to biotic and abiotic stress responses, such as regulation of defense response to bacterium and UV regulation in the actual unshaded field environment. The results of agronomic traits indicated that the purple phenotype exhibited a multitude of merits in plant height and stem diameter (p < 0.05), and produced more high-quality fruit and seeds, which demonstrated that the purple phenotype has high regeneration ability and potential resistance to cultivation conditions. Importantly, the content of total alkaloids as bioactive ingredients in medicinal bulbs of purple F. cirrhosa was significantly higher than that in the green phenotype by 57.14%. Overall, the present study not only reveals the potential mechanisms of phenotypic variation in F. cirrhosa but also contributes to a better understand adaptation of highland species related to ecological changes, as well as paves the way for the further breeding and large-scale cultivation of F. cirrhosa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant adaptation is an evolution strategy that allows organisms to respond to ecological fluctuations through various mechanisms such as gene families (Zhang et al. 2019), chromosome evolution (Ramsey 2011), anatomical traits physiological innovation (Taylor et al. 2012), and germination behavior (Donohue et al. 2010). Highland ecosystems are particularly vulnerable to global climate changes resulting in smaller biomass and slower growth of plants compared to those at lower elevations (Cunningham et al. 2018). Within these ecosystems, plants often develop adaptative traits for population expansion though through farmers’ selection (Beyene et al. 2005) and convergent phenotypic evolution (Wang et al. 2021a). Fritillaria cirrhosa D. Don, an endangered plant species found in the Himalayan-Hengduan Mountains (Chen et al. 2020; Wang et al. 2021b), serves as a valuable model for investigating speciation and ecological adaptation in extreme environmental conditions (Zhang et al. 2019). The dried bulbs of F. cirrhosa have been utilized for thousands of years to moisten dryness and clear lung hear (Quan et al. 2022). However, due to increasing demand and unsustainable excavation, the species is facing accelerated extinction. Consequently, its protection status was upgraded from Class III to Class II in 2021, announced by the National Forestry and Grassland Administration and National Park Administration in China. In recent years, the phenotype of F. cirrhosa, a representative rare plateau species, has undergone dramatic changes. Purple plants now dominate the cultivation field while green phenotypes remain prevalent in the wild environment. Despite these observations, there have been no reports focusing on the mechanism underlying this color variation or analyzing its correlation with adaptation strategies.
The wild F. cirrhosa primarily grows in the shade under the plant community of other bush fallow, such as Sibiraea angustata community and the Rhododendron litangense community (Chen et al. 2003). During our field investigation, we observed that the green aerial parts of the plant turned purple, especially during the seeding stage, under strong ultraviolet lights in cultivated condition. Previous studies have confirmed that plants can adapt to environmental fluctuations by switching phenotypes (Kussell and Leibler 2005). Color variation is a crucial adaptation mechanism in many plants under stressful conditions (Dwivedi et al. 2016; Kochian et al. 2015), such as high light intensity and ultraviolet stress. The formation of plant color was determined by pigment content, which consists of three types of pigments: flavonoids, carotenoids, and alkaloids (Qiao et al. 2022).
Flavonoids are major classes of pigments that participate in plant developmental stages and growth conditions, serving various physiological functions including ultraviolet protection, antioxidant activity, and defense against phytopathogens, addressing biotic and abiotic stresses (Petrussa et al. 2013). The implementation of multi-omics approaches for strategic improvement has accelerated plant breeding with recent technological advances in multiple disciplines, facilitating genetic modification or germplasm selection (Shen et al. 2022). By analyzing the combination of transcriptome sequencing and flavonoid metabolism in three cultivars of Acer truncatum, it was demonstrated that F3′H, BZ1, and ANS are crucial genes for breeding red phenotype, while ANR is the key gene for product high-content flavonoids varieties (Qiao et al. 2022). Unfortunately, few studies have focused on the mechanism of phenotype variation in plateau plants due to the harsh environment. Therefore, studying the phenotypic color variation mechanism of F. cirrhosa will not only enhance our understanding of the impact of global climate change on the genetic diversity of plateau species but also provide a research foundation for early molecularly assisted breeding of F. cirrhosa.
A comparative genomics study was conducted on two different phenotypes of F. cirrhosa to reveal the mechanism of color variation. The study also investigated the physiological consequences of adaptation to the harsh environment in which F. cirrhosa survives. The research facilitates the examination of the impact of global climate change on endangered plants on the plateau, which can contribute to a better understanding of the active adaptation of F. cirrhosa to changes in the ecological environment of natural habitats. Furthermore, the data obtained in this study can enhance our understanding of molecular breeding and large-scale cultivation of F. cirrhosa.
Materials and methods
Plant materials
The planting site of F. cirrhosa was located in Tu Autonomous County of Huzhu (36° 59′ E, 101° 59′ N, altitude: 3050 m), which is in Haidong City of Qinghai Province, China. The annual mean temperature is 0 °C; mean diurnal range is 13 °C; isothermality is 37; annual precipitation is 466 mm; max temperature of the warmest month is 18 °C; min temperature of the coldest month is − 17 °C. Fresh leaves were harvested without any damage by insects or diseases at the seedling stage when these plants didn’t grow anymore. All the materials were stored in a freezer at − 80 °C, awaiting metabolic measurement and RNA extraction. Six batches of leaf samples from two different phenotypes were conducted in the biological repetition experiments. Finally, mature bulbs were collected to determine the total alkaloid content and evaluate the medicinal quality.
Authentication of experimental samples
The fresh bulbs (0.1 g) and leaves (0.1 g) of two different phenotypes were used for DNA extraction followed by the manufacture of a Novel Plant Genome Extraction Kit (Beijing, China) and a modified CTAB method (Shi-lin Chen et al. 2013). The forward and reverse primers for ITS2 were 5′-ATGCGATACTTGGTGTGAAT-3′ and 5′-GACGCTTCTCCAGACTACAAT-3′, respectively. The PCR reactions were carried out using a 1 μL DNA template, 1 μM forward and reverse primers, 9.5 μL ddH2O and 12.5 μL 2 × Taq Master Mix. The PCR amplification procedure involved pre-denaturation for 5 min at 94 °C, followed by 40 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 56 °C, extension for 45 s at 72 °C, extension at 72 °C for 10 min, and storage at 4 °C.
Transcriptome measurement
RNA extraction and library construction
The total RNA of fresh leaves from two phenotypes F. cirrhosa was extracted using RNAprep Pure Plant Hit (Beijing, China) according to the manufacturer’s protocol. Agarose gel electrophoresis was used to assess the quality of the total RNA. The accurate RNA concentration and RNA integrity were determined using a 2100 Bioanalyzer (Agilent Technologies, CA, USA). The isolation and enrichment of messenger RNA (mRNA) were finished using Oligo (dT) and cut into short fragments after adding a fragmentation buffer. These short fragments were purified using AMPure XP heads and further synthesized the cDNA. The cDNA was then subject to polyA tails and ligated with adapters for library construction before fragment selection and PCR enrichment. The library quality was analyzed based on an accurate quality using the Q-PCR approach (effective concentration > 2 nM). A total of 9 cDNA libraries were sequenced using Illumina HiSeqTM 2000 (NEB, USA) after pooling.
Transcriptome assembly and annotation of differentially expressed genes
High-quality reads were obtained from raw sequences by removing low-quality transcripts and adapter sequences. The low-quality transcripts contained paired reads with N content exceeding 10% of the read base number, as well as paired reads in which the number of low-quality (Q ≤ 20) bases exceeded 50% of the total bases. These reads were then mapped to the assembled transcriptome for analysis of the gene expression quantity, due to the absence of reference genomes. The reference sequence was assembled using Trinity (Version v2.6.6) after obtaining clean reads for subsequent analysis (Grabherr et al. 2011).
Furthermore, DESeq2 software was used to obtain a gene database of differentially expressed genes (DEGs) between the two phenotypes, using reads without normalization (Love et al. 2014; Varet et al. 2016). The selection criteria for DEGs were |log2Fold Change| ≥ 1 and false discovery rate (FDR) < 0.05. A visualization method called M-versus-A plot (MA plot) was used to analyze data distribution, using M (log2 (Fold Change)) and mean/average (A) values. Gene expression levels were compared using the kilobase of transcript per million fragments mapped (FPKM) after normalizing the amounts of mapped reads and the transcript length. The function information of DEGs was analyzed using public database, such as the NCBI non-redundant protein sequences (Nr) for species authentication, the Kyoto Encyclopedia of Genes and Genomes (KEGG) for investigating the potential function of DEGs, and the Gene Ontology (GO) database, which contains extensive functional information on DEGs in biological process, molecular function, and cellular component. Finally, the unigenes sequences of DEGs were aligned with the PinTFDB and PlantTFDB databases to identify transcription factor families.
Validation of DEGs regulating anthocyanin biosynthesis using real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from two phenotypes of F. cirrhosa for three biological repetitions, and the extracted RNA was reversed transcribed to synthesize cDNA samples. Five DEGs related to anthocyanin biosynthesis were selected from the transcriptome sequencing results to verify the accuracy of the transcriptome data. GAPHD was used as the internal reference gene. The PCR reaction followed this protocol: 95 °C for 2 min, 95 °C for 15 s, 60 °C for 30 s, 72 °C for 30 s, after 40 cycles, 65 °C for 5 s. The relative gene expression was calculated using the 2−△△t method. The primer sequences for the targeted genes and internal reference gene are listed in Table S1.
Measurement of photosynthetic parameters
Fritillaria cirrhosa has a long flowering stage that lasts about 30 days. Those unfolded leaves at the top of the plants were used as the experimental object for obtaining parameters. The measurement time was controlled within the section of 8:30–11:30 in the forenoon when the weather was cloudless, avoiding the influence of photosynthesis. Each leaf was detected triple times equally, and 10 individuals of each phenotype were measured.
The Li-6800 portable photosynthesizer equipped with a 6800-01F fluorescent leaf chamber (Li-Cor, Lincoln, NE, United States) and CO2 small cylinders were used to control a statable environment around the detected leaf. The flow speed and fan revolutions were set at 500 μmol/s and 10,000 r/min, respectively. The humidity and CO2 concentrations were set at 55% and 400 μmol/mol, respectively. The Qin values were set at 2400, 2100, 1800, 1500, 1200, 1000, 800, 600, 500, 400, 300, 200, 150, 120, 90, 60, 40, 20, and 0 µmol/(m2·s). The obtained net photosynthetic rate combined with light intensity values were calculated by photosynthesis fitting software to form the photosynthetic light response curve.
Based on the abovementioned instrument parameters, the setpoint value of light was 1,500 μmol·m−2 s−1, and the match options under the leaf chamber of red and blue light were always open. After the leaf underwent detection of gas exchange parameters, it was subjected to 20 min of dark adaptation using silver paper. The leaf, after a dark adaption, was detected by the Li-6800 portable photosynthesizer equipped with a 6800-01F fluorescent leaf chamber to obtain fluorescence parameters of chlorophyll.
Detection of total alkaloid content and anthocyanin
The detection of total alkaloid content followed the specifications of the Chinese Pharmacopoeia and our published study (Wang et al. 2022b). In brief, dried bulb powers (2.0 g) were soaked using 3 mL ammonia solution (Batch No. 20210601) for 1 h before reflux extraction using a 40 mL chloroform (batch No. 20210104): methanol (Batch No. 20210104, 4:1, v/v) mixture. The filtered solution was then mixed with bromocresol green (Batch No. CS6218) liquor to obtain the underlying liquid after being divided by a separating funnel. The final extraction was detected using an ultraviolet spectrophotometer (752, Beijing, China). The alkaloid content was calculated based on the standard curve (Y = 25.374X − 0.0059, R2 = 0.9945) obtained from difference concentration of imperialine (purity > 98%, Batch No. BP6122).
The absorbance of anthocyanin was determined as Chen’s (Chen et al. 2022) and Pu’s methods (Pu et al. 2021), which involved extracting it with 0.1 mol·L−1 hydrochloric acid alcohol at room temperature. The extraction liquid was scanned at 530 nm, 620 nm, 650 nm, respectively, using a microplate reader (BioTek Synergy 2, USA). The anthocyanin content was calculated after obtaining the absorbance of anthocyanin according to the two aforementioned references.
Bioinformation and statistical analysis
The DNA sequence of two phenotypes from two botanical parts was processed and aligned using CodonCode and DNAMAN software. The sequence alignment was conducted using the BLAST site (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Correlation analysis based on Pearson’s correlation coefficient was utilized to evaluate the association within the biological replicates based on the whole FPKM values. Principal component analysis (PCA), a non-parametric and data dimension reduction chemometrics method, was used for cluster pattern and sample classification, with limited principal component linearly related to the original sequencing data. This analysis was performed using MetaboAnalyst (version 5.0, an online software, https://www.metaboanalyst.ca/). Significant differences were determined using the pared-sample T-test in SPSS Statistic (version 21, IBM company, USA), and p < 0.05 was considered as statistically significant.
Results
Species authentication and agronomic traits between purple and green F. cirrhosa phenotypes
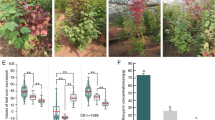
During the period of emergence, these young plants exhibited purple and green traits (Fig. 1A), Specifically, 50 individuals were selected and labeled for further examination of agronomic characteristics during the flower and fruit stages. The purple plants demonstrated a purple hue from the basal stem to the leaves (Fig. 1B), while the green phenotypes displayed green features from the middle stem to the leaves (Fig. 1C). Additionally, we observed that bulbs of the purple phenotype germinated the lilac-colored young stem tip before breaking through the soil (Fig. 1D), whereas bulbs of the green phenotype developed the aqua-colored stem tip before emerging (Fig. 1E). The anthocyanin content in the leaves of the purple phenotype measured 8.1566 ± 0.3371 nmol g−1, exhibiting an extremely significant difference at p < 0.01 (Fig. 1F, G). Ten F. cirrhosa samples, including leaves and bulbs parts, were subjected to DNA extraction. The purity of extracted DNA, as indicated by the A260/A280 ratio, was approximately 2.0, and most PCR amplifications produced clear bands with strong signals. The amplified fragments had a length of 500 bp (Figure S1). DNA barcoding revealed that both phenotypes displayed 100% similarity to the standard reference sequencing data of F. cirrhosa from the NCBI database.
The morphological characters of the aerial part and fresh bulbs of F. cirrhosa in the cultivated site. A The whole distribution in the large-scale area of two phenotypes. B Purple phenotype. C Green phenotype. D The fresh bulb of purple phenotype before breaking through the soil. E The fresh bulb of purple phenotype before breaking through the soil. F Fresh leaves of green and purple phenotypes. G Anthocyanin content of fresh leaves of two kinds of leaves
Six agronomic traits were measured at the flowering stage (30th May–5th June), including plant height, stem diameter, leaf number, leaf length, leaf width, and flower number (Figure S2A–S2F). The purple phenotype exhibited a significant advantage in terms of plant height and stem diameter (p < 0.05), with purple plants averaging height of 41.04 ± 7.99 cm compared to 34.36 ± 7.39 cm for the green phenotype. In other words, the purple phenotype showed strong growth vigor. Although there were no significant differences in the other four traits between purple and green F. cirrhosa, purple plants showed a higher tendency than green individuals, especially in flower number. Additionally, the agronomic traits of fruit were compared between purple and green phenotypes (Fig. 2A–F). The results indicated extremely significant differences in fruit diameter, fruit weight, and seed setting have between the two phenotypes (p < 0.01), while the length/diameter of the fruit and fruit shell weight showed significant variation (p < 0.05). There was no significant difference in fruit length. The purple phenotype exhibited a higher seed setting rate (average above 57.81%) compared to green plants, which was consistent with the fruit weight and fruit shell weight comparisons.
The agronomic traits of fruits F. cirrhosa. A Fruit length. B Fruit diameter. C Length/diameter of fruit. D Fruit weight. E Fruit shell weight. F Seed setting rate. * Means the significant difference with p < 0.05; ** means the extremely significant difference with p < 0.01. The significant test is calculated according to the T-test analyses
Transcriptomic results
Quality assessment of RNA extraction, assembly, and DEGs selection
The extracted RNA concentration ranged from 248 and 473 ng·μL−1, with a total amount of more than 8.68 μg and excellent integrity (more than 7.5) in Table S2. The RNA sequence and assembly are shown in Table S3, we obtained nearly 45,000,000 clean reads, with a Q20 value of over 97.68% and a Q30 value of over 93.57% after sequence and assembly. GC content was approximately 50%, with little AT and a clear separation of GC. Furthermore, the correlation heatmap (Fig. 3A) showed that the similarity within the same phenotype was higher than that between different groups, with a correlation coefficient of more than 0.94. Based on the selection criterion, we identified 1734 DEGs between the green and purple phenotypes. The MA plot (Fig. 3B) displayed the expression level and fold change. Herein, these DEGs were comprised of 754 up-regulated genes and 980 down-regulated genes. The detailed gene information of two phenotypes, including gene ID, averaged FPKM, log2Fold-change, value, padj (False discovery rate corrected for multiple hypothesis testing), and regulated status, is displayed in Table S4. PCA results indicated that the PC1, which explained 83.2% of DEGs information, could distinguish between purple and green phenotypes in Fig. 3C. These DEGs showed excellent classification in the clustering heatmap (Fig. 3D).
The analytical results of sequencing data of purple and green F. cirrhosa. A Correlation heatmap and the redder the color, the higher the correlation. B The MA plot (M-versus-A plot (MA plot) was used as a visualization method of data distribution using M (log2 (Fold Change)) and mean/average (A)), that red points mean up-regulated genes, green points mean down-regulated genes, and blue points false regulation. C PCA score plot that pink circle represents green phenotype while the green circle represents purple phenotype. D Cluster tree and heatmap of DEGs that red means high expression level and green means low expression level
Annotation results of DEGs and transcription factors
These DEGs were first annotated in the Nr database, with 201 DEGs annotated in F. cirrhosa and 335 genes annotated in congeneric species including F. agrestis, F. hupehensis, F. liliacea, F. taipaiensis, etc. Moreover, these DEGs were annotated into the public KEGG and GO databases. The KEGG annotation results showed most DEGs participated in the metabolic pathways and biosynthesis of secondary metabolites (Fig. 4). Some genes were related to the biosynthesis of phenylpropanoid, flavonoid and anthocyanin antenna. Two photosynthesis-relation pathways were also involved in the differential annotation results. The GO enrichment analysis indicated that 31 DEGs participated in the biosynthesis and metabolic process of anthocyanin-containing components (Fig. 5). The enrichment results reflect a clear difference in anthocyanidin 3-O-glucosyltransferase activity. Moreover, there also exist enrichment pathways in the photosynthesis-relation process, such as regulation of chlorophyll biosynthetic process, response to red light, photosystem, and photosystem II.
The circle plot of GO enrichment of DEGs in purple and green F. cirrhosa. GO:0016210: Naringenin-chalcone synthase activity; GO:0022625: Cytosolic large ribosomal subunit; GO:0022626: Cytosolic ribosome; GO:0030529: Extracellular ribonucleoprotein complex; GO:0042440: Pigment metabolic process; GO:0043043: Peptide biosynthetic process; GO:0043228: Non-membrane-bounded organelle; GO:0043603: Cellular amide metabolic process; GO:0043604: Amide biosynthetic process; GO:0044391: Ribosomal subunit; GO:0044444: Obsolete cytoplasmic part; GO:0044445: Obsolete cytosolic part; GO:0045552: Dihydrokaempferol 4-reductase activity; GO:0046148: Pigment biosynthetic process; GO:0046283: Anthocyanin-containing compound metabolic process; GO:1990904: Intracellular ribonucleoprotein complex. The first circle means three types of GO enrichment, biological process (orange), molecular function (blue), and cellular components (green). The second circle means the − log10 (p value) and the values increase with the deep color. The third circle means upregulated DEGs and downregulated DEGs. The fourth circle means the rich factor of each GO term
Significant DEGs of forty-nine participated in the activity of 25 transcription factors (TFs) and 7 transcriptional regulators (TRs) were listed in Table S5. Most of TFs and TRs simultaneously have up-and down-regulated genes, such as bHLH which was regulated by 2 up- and 2 down-regulated DEGs. Those DEGs coded the bZIP, C2H2, CSD, MADS-MIKC, S1Fa-like, and WRKY were down-regulated while these DEGs coded LUG and TCP were up-regulated.
The DEGs involved in anthocyanin biosynthesis between purple and green F. cirrhosa phenotypes
The predictive biosynthesis pathway of anthocyanin is displayed in Fig. 6, along with a heatmap of the DEGs related to the enzyme involved in the process. A total of 23 DEGs were found to regulate the anthocyanin biosynthesis, starting from phenylalanine. Six vital enzymes were identified to participate in the biosynthesis process. Among these DEG, 14 genes coding C4H have higher expression levels in the purple phenotype compared to the green individuals. Similarly, most of the DEGs related to CHS displayed low expression levels in the green phenotypes. Additionally, nine DEGs regulating four key enzymes (F3′H, ANS, DFR, and BA1) showed higher expression levels in the purple F. cirrhosa compared to the green phenotype. The F3′H determined the biosynthesis of dihydrokaemoferol from naringenin and ANS intervened in the formation of leucopelargonidin from dihydrokaemoferol. DFR and BA1 affect anthocyanin biosynthesis by regulating the route of dihydromyricetin-leucodelphinidin-delphinidin-anthocyanin.
The predicted pathway of anthocyanin biosynthesis, DEGs heatmap, RT-qPCR analysis of anthocyanin-relation in purple and green F. cirrhosa phenotypes. C4H is cinnamate 4-hydroxylase; CHS is chalcone synthase; F3′H is flavanone 3′-hydroxylase; ANS: anthocyanidin synthase; DFR: dihydroflavonol 4-reductase; BZ1: anthocyanidin 3-O-glucosyltransferase. The first three lines are green phenotype and the later three lines are purple phenotype in the heatmap. The green indicates low genes expression and red show high gene expression. Error bars of column plots indicate the mean ± SD after three duplicates
Furthermore, RT-qPCR was performed to performed to verify the reliability and expression level of the RNA sequencing data. Five DEGs related to anthocyanin biosynthesis, including CHS, F3′H, ANS, DFR and BA1, were selected for analysis (Fig. 6). The results showed consistent patterns between RNA sequencing data and RT-qPCR analysis results, indicating the reliability of the transcriptome sequencing data. The combined results suggested that the color variation in F. cirrhosa was determined by the upregulation of structural genes regulating anthocyanin biosynthesis.
The DEGs involved in the growth and potential stress between purple and green F. cirrhosa phenotypes
Several DEGs related to plant growth and potential stress was identified based on the GO annotation results for further investigation of the difference in agronomic traits. Herein, 28 DEGs were upregulated in the purple phenotype compared to the green plants, and they were in the regulation of plant growth and potential resistance (Table S6). Further analysis revealed that five genes were related to the regulation and formation of the cell wall, seven genes were involved in meristem growth, cell differentiation, and cell proliferation, and 13 genes were associated with the regulation of organ growth, including maintenance of floral organ identity, regulation of leaf development, inflorescence morphogenesis and development. Ten genes were found to be related to the biotic stimulus and abiotic stress, such as regulation of defense response to bacterium (GO:1900425, GO:1900424, GO:0016045) and virus (GO:0051607, GO:0009615, GO:0098586) in biotic response and UV regulation (GO:0010224, GO:0043478, GO:0043479, GO:0043481) in abiotic stress. Interestingly, we found two genes (Cluster-2539.154075, Cluster-2539.154070) have annotation information of cellular response to nitrogen levels (GO:0043562).
Comparison of photosynthetic parameters between purple and green F. cirrhosa phenotypes
Photosynthetic light response curves of purple and green phenotypes of F. cirrhosa are shown in Figure S3. The curve indicated that the photosynthetic rate of both phenotypes displayed an increasing tendency with the photon flux density when the value was less than 850 μmol/(m2 s). The curve became smooth between 850 and 1500 μmol/(m2 s), and the photosynthetic rate showed a descending trend after 1500 μmol/(m2 s). We found that the purple phenotype always displayed a higher photosynthesis rate compared with that of the green phenotype. After curve fitting, we obtained five characteristic parameters of the photosynthetic light response curve in Table S7. The comparison results indicated that purple phenotype has the higher light saturation point (853.19 ± 96.35 μmol/(m2·s)), light compensation point (8.4 ± 2.4 μmol/(m2·s)), dark respiration rate (− 0.65 ± 0.1 μmol/(m2 s)), maximum photosynthetic rate (16.91 ± 2.48 μmol/(m2 s)) but lower apparent quantum efficiency (0.09 ± 0.02) compared with green phenotype (749.99 ± 139.74, 7.2 ± 4.8, − 0.71 ± 0.44, 15.17 ± 2.27, 0.1 ± 0.01, respectively).
There were 15 photosynthetic parameters were detected between purple and green F. cirrhosa during the flowering stage (Table S8). The results indicated that there was a minor difference in these parameters. Compared with green plants, the purple phenotype has a higher net photosynthetic rate, stomatal conductance to boundary layer water vapor, net photosynthetic rate at steady-state fluorescence, electron transport rate, and the quantum efficiency of CO2 assimilation, and non-photochemical quenching coefficient. Moreover, green individuals showed lower values of the other 9 parameters, such as transpiration rate and non-photochemical quenching coefficient. In addition, purple plants displayed higher chlorophyll fluorescence values including minimum initial fluorescence, maximum fluorescence after a dark adaption, the difference between the maximum and minimum fluorescence under dark adaption, and maximum photochemical efficiency of PSII reaction centers when fully open under dark adaptation, whereas lower net photosynthetic rate under dark adaptation compared with the green phenotype (Table S9).
Content of total alkaloid between purple and green F. cirrhosa phenotypes
The validation results of the detection method were reported in our previous research (Wang et al. 2022b). The performance indicated that accuracy, repeatability, and stability were excellent within 12 h. The standard curve equation obtained via imperialine was Y = 25.374X − 0.0059 with R2 = 0.9945, and the linearity range was between 0.0006 and 0.0111 mg·mL−1. The comparison result of total alkaloid is displayed in Figure S4, which indicated that the purple phenotype has higher content (0.11% ± 0.02%) compared with the content (0.08% ± 0.02%) of green F. cirrhosa. The content of alkaloids in both phenotypes exceeded the standard of Chinese Pharmacopoeia.
Discussion
As a typical plateau plant, F. cirrhosa not only survives in the extreme environments of the Himalayan-Hengduan Mountains but also provides precious medicinal materials. Unfortunately, fewer studies have been conducted to investigate the underlying mechanisms of phenotypic variations and ecological adaptations of F. cirrhosa from wild environment to cultivated conditions, primarily due to global climate change and deterioration of the ecological environment.
Fritillaria cirrhosa is a high-altitude species and sexual propagation using mature seeds is the primary method of expanding propagation (Qu et al. 2022; Wang et al. 2021b). Therefore, it is increasingly important for plant enterprises to select phenotypes with high seed yields. Interestingly, in our current study, we discovered a purple phenotype of F. cirrhosa that produces high-quality fruit containing numerous seeds. The high-yield advantage can be attributed to the fact that these purple leaves can absorb more energy to withstand the low temperature in high-altitude areas. The temperature during the emergence period of F. cirrhosa in May remains extremely low for plant growth. The advantages of the purple phenotype may enhance plant growth conditions, which is supported by our comparison results indicating that purple plants have tall and sturdy stems. The dominant plant traits are beneficial for flower and fruit formation, thereby helping plants withstand harsh windy weather. Additionally, in our research sites, the purple F. cirrhosa phenotype accounted for a high percentage (more than 90%) in the young stage, in contrast to the green individuals. The percentage differed from our previous research conducted in the Sichuan Province of China, where the green phenotype accounted for a high percentage (89.5%) (Zhang et al. 2010). The difference can be interpreted as the present provenance was collected from Tibet while previous samples were from local plants in Sichuan Province. We speculate that this difference may be attributed to the long-term, high-intensity ultraviolet irradiation in Tibet, which led to the emergence of this new phenotype in F. cirrhosa as an adaptation to cope with strong light stress. This interpretation aligns with the high expression of anthocyanin synthesis genes observed in our transcriptome results for the purple phenotype.
Plants with purple traits seem to possess unique advantages in promoting healthy growth, which is a common exceptional characteristic observed in most plants (Fan et al. 2021; Landi et al. 2013; Meng et al. 2022; Yan et al. 2020; Yuan et al. 2021). Tang et al. illustrated that BcTT8 is the candidate gene responsible for regulating anthocyanin synthesis, which resulted in two phenotypes of purple and green non-heading Chinese cabbage (Tang et al. 2022). Purple peppers exhibit favorable physiological tolerance to drought stresses and high temperatures (Meng et al. 2022). Purple ginseng had showed high resistance under high-light stress compared with green phenotypes (Chen et al. 2021). This is similar to our findings that purple plants have a higher light saturation point compared with green plants. Based on our comparison results, it appears that purple F. cirrhosa can utilize a higher level of photosynthetic active radiation to adapt to strong light intensities in high-altitude region (Gao et al. 2017). We have observed that upregulated genes in the purple phenotype response to UV light, which aligned with their performance. Furthermore, purple phenotypes exhibit upregulated genes that respond to biotic response, particularly against bacterium and virus. This consistency is observed in the purple leaf type of castor bean (Ricinus communis), which demonstrates stable resistance to leafminer, an insect pest that caused economic damage to the plant, compared with the green leaf type susceptible to the pest (Anjani et al. 2007). In a comparison study between purple and green-leafed Ocimum basilicum, the green cultivars were found to be more susceptible to boron toxicity and higher concentrations of iodine due to lower anthocyanins and phenolic content (Incrocci et al. 2019; Landi et al. 2013). Purple sesame had the ability to produce high-quality commercial production with a high antioxidant capacity (Landi et al. 2013). Additionally, exogenous application of anthocyanins has been found to enhance arsenic tolerance and improve plant growth and productivity (Ahammed and Yang 2022). We have also observed that the purple phenotype of F. cirrhosa demonstrates excellent growth performance after long-time drought stress and high temperature, allowing for the production of full fruits. Published literature has also shown that anthocyanins can protect photosynthetic tissue by absorbing light in the visible range, reducing light stress (Zhang et al. 2014). In our study, we found two genes (Cluster-2539.154075, Cluster-2539.154070) are associated with cellular response to nitrogen levels, indicating that the purple phenotype has high-efficiency nitrogen utilization. Our previous study has shown that nitrogen supply promotes the quality of fruits and seeds (Wang et al. 2022a). In our further study, we plan to investigate the tolerance of the two phenotypes to further validate the potential molecular mechanisms of the purple phenotype under conditions of long-time drought stress and high temperature.
Investigating the molecular mechanism of anthocyanin biosynthesis in purple and green F. cirrhosa can provide theoretical and technical support for expanding the gene resource of purple F. cirrhosa and promoting its molecular breeding. The clear KEGG enrichment analysis results presented in Fig. 4 indicate that leaf color differences may be caused by anthocyanin biosynthesis, as the regulated genes have a higher expression in the purple phenotypes. The GO enrichment analysis results displayed in Fig. 5 also showed DEGs between purple and green phenotypes participating in the photosystem. However, there were no significant differences in photosynthetic parameters, although the purple phenotype shows a slight advantage. This suggests that the expression of DEGs was not sufficient to lead to significant differences between the two phenotypes.
The predicted pathway of anthocyanin biosynthesis reveals the involvement of six key enzymes (C4H, CHS, F3′H, ANS, DFR, and BA1), leading to the formation of anthocyanin and the purple phenotype. Previous research has also indicated that ANS plays a key role in breeding red Acer truncatum (Qiao et al. 2022), while DFR and F3′H have been confirmed to be involved in anthocyanin biosynthesis in various plant species (Khusnutdinov et al. 2021). The comparison results of TFs and TRs identified down-regulated bZIP, C2H2, CSD, MADS-MIKC, S1Fa-like, and WRKY, while up-regulated these LUG and TCP genes were observed. Additionally, some genes that regulate bHLH and MYB also demonstrated high expression in the purple phenotype, which is consistent with the role of the MYB-bHLH-WD40 (MBW) complex in the color component biosynthesis (Chaves-Silva et al. 2018; Zhang et al. 2022).
Conclusions
In conclusion, F. cirrhosa demonstrates adaptation to artificial environments from its natural shade habitat through several characteristics, including color variation from green to purple, advantageous agronomic traits (strong aerial parts and high seed yield), and the presence of differentially expressed genes related to potential resistance. The comparative transcriptome analysis indicated 37 significant DEGs regulating anthocyanin biosynthesis, which are key genes responsible for the formation of the purple phenotype in F. cirrhosa. Additionally, 10 DEGs were found to be related to the biotic and abiotic stress, indicating that the purple phenotype is an adaptation of F. cirrhosa to the change in its ecological environment. Agronomic traits comparison also supported the advantage of the purple phenotype is an in reproduction and population evolution. Furthermore, a significant difference in total alkaloid was observed, and DEGs coding F3′H, ANS, DFR, and BA1 can be considered as candidate genes for the molecular breeding of F. cirrhosa. Overall, this comprehensive study provides a research basis for understanding the adaptation of plateau species.
Data availability
We have uploaded the RNA sequencing data to NCBI and the link is https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881243. The data sets supporting the results of this article are included within the article and supplementary table.
Abbreviations
- ANS:
-
Anthocyanidin synthase
- BA1:
-
Anthocyanidin 3-O-glucosyltransferase
- bHLH:
-
Basic helix-loop-helix
- bZIP:
-
Basic-leucine zipper
- C4H:
-
Cinnamate 4-hydroxylase
- CHS:
-
Chalcone synthase
- DEGs:
-
Differentially expressed genes
- DFR:
-
Dihydroflavonol 4-reductase
- F3′5′H:
-
Flavonoid 3050-hydroxylase
- F3′H:
-
Flavanone 3′-hydroxylase
- FDR:
-
False discovery rate
- FPKM:
-
Kilobase of transcript per million fragments mapped
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- Nr:
-
NCBI non-redundant protein sequences
- PCA:
-
Principal component analysis
- TFs:
-
Transcription factors
- TRs:
-
Transcriptional regulators
References
Ahammed GJ, Yang Y (2022) Anthocyanin-mediated arsenic tolerance in plants. Environ Pollut 292:118475. https://doi.org/10.1016/j.envpol.2021.118475
Anjani K, Pallavi M, Sudhakara Babu SN (2007) Uniparental inheritance of purple leaf and the associated resistance to leafminer in castor bean. Plant Breed 126(5):515–520. https://doi.org/10.1111/j.1439-0523.2007.01395.x
Beyene T, Botha AM, Myburg AA (2005) Phenotypic diversity for morphological and agronomic traits in traditional Ethiopian highland maize accessions. S Afr J Plant Soil 22(2):100–105. https://doi.org/10.1080/02571862.2005.10634689
Chaves-Silva S, Santos ALD, Chalfun-Júnior A, Zhao J, Peres LEP, Benedito VA (2018) Understanding the genetic regulation of anthocyanin biosynthesis in plants—tools for breeding purple varieties of fruits and vegetables. Phytochemistry 153:11–27. https://doi.org/10.1016/j.phytochem.2018.05.013
Chen S, Jia M, Wang Y, Gang X, Xiao P (2003) Study on the plant community of Fritillaria cirrhosa. China J Chin Mater Media 28:18–22
Chen SL, Hui Y, Jian-ping H, Tian-yi X, Xiao-hui P, Lin-chun S, Kun L, Jing-yuan S, Dian-yun H, Shang-mei S, Zhong-zhi Q (2013) Principles for molecular identification of traditional Chinese materia medica using DNA barcoding. China J Chin Materia Media 38:141–148. https://doi.org/10.4268/cjcmm20130201
Chen T, Zhong F, Yao C, Chen J, Xiang Y, Dong J, Yan Z, Ma Y (2020) A systematic review on traditional uses, sources, phytochemistry, pharmacology, pharmacokinetics, and toxicity of Fritillariae Cirrhosae Bulbus. Evid Based Complement Altern Med 2020:1–26. https://doi.org/10.1155/2020/1536534
Chen JY, Wang Y, Xu FR, Li XW (2021) Resistance evaluation of different phenotypes of farmland Ginseng Radix et Rhizoma germplasm under high light stress. Chin J Exp Tradit Med Formulae 27:121–129. https://doi.org/10.13422/j.cnki.syfjx.20211349
Chen T, Fu W, Yu J, Feng B, Li G, Fu G, Tao L (2022) The photosynthesis characteristics of colored rice leaves and its relation with antioxidant capacity and anthocyanin content. Sci Agric Sin 55:467–478
Cunningham AB, Brinckmann JA, Pei SJ, Luo P, Schippmann U, Long X, Bi YF (2018) High altitude species, high profits: can the trade in wild harvested Fritillaria cirrhosa (Liliaceae) be sustained? J Ethnopharmacol 223:142–151. https://doi.org/10.1016/j.jep.2018.05.004
Donohue K, Rubio De Casas R, Burghardt L, Kovach K, Willis CG (2010) Germination, postgermination adaptation, and species ecological ranges. Annu Rev Ecol Evol Syst 41(1):293–319. https://doi.org/10.1146/annurev-ecolsys-102209-144715
Dwivedi SL, Ceccarelli S, Blair MW, Upadhyaya HD, Are AK, Ortiz R (2016) Landrace germplasm for improving yield and abiotic stress adaptation. Trends Plant Sci 21(1):31–42. https://doi.org/10.1016/j.tplants.2015.10.012
Fan K, Shi Y, Luo D, Qian W, Shen J, Ding S, Ding Z, Wang Y (2021) Comparative transcriptome and hormone analysis of mature leaves and new shoots in tea cuttings (Camellia sinensis) among three cultivars with different rooting abilities. J Plant Growth Regul. https://doi.org/10.1007/s00344-021-10478-0
Gao S, Yan Q, Chen L, Song Y, Li J, Fu C, Dong M (2017) Effects of ploidy level and haplotype on variation of photosynthetic traits: novel evidence from two Fragaria species. PLoS ONE 12(6):e179899. https://doi.org/10.1371/journal.pone.0179899
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652. https://doi.org/10.1038/nbt.1883
Incrocci L, Carmassi G, Maggini R, Poli C, Saidov D, Tamburini C, Kiferle C, Perata P, Pardossi A (2019) Iodine accumulation and tolerance in sweet basil (Ocimum basilicum L.) with green or purple leaves grown in floating system technique. Front Plant Sci 10:1494. https://doi.org/10.3389/fpls.2019.01494
Khusnutdinov E, Sukhareva A, Panfilova M, Mikhaylova E (2021) Anthocyanin biosynthesis genes as model genes for genome editing in plants. Int J Mol Sci 22(16):8752. https://doi.org/10.3390/ijms22168752
Kochian LV, Piñeros MA, Liu J, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 66(1):571–598. https://doi.org/10.1146/annurev-arplant-043014-114822
Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309(5743):2075–2078. https://doi.org/10.1126/science.1114383
Landi M, Remorini D, Pardossi A, Guidi L (2013) Purple versus green-leafed Ocimum basilicum: which differences occur with regard to photosynthesis under boron toxicity? J Plant Nutr Soil Sci 176(6):942–951. https://doi.org/10.1002/jpln.201200626
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Meng Y, Zhang H, Fan Y, Yan L (2022) Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum). BMC Plant Biol 22:258. https://doi.org/10.1186/s12870-022-03746-y
Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, Vianello A (2013) Plant flavonoids—biosynthesis, transport and involvement in stress responses. Int J Mol Sci 14(7):14950–14973. https://doi.org/10.3390/ijms140714950
Pu Q, Yang P, Yong L, Deng Y, He Z, Lin B, Shi S, Xiang C, Fang F (2021) Studies on pigment content and photosyntheic characteristics of purple-red leaf color mutant in radish. J Agric Sci Technol 23:45–54. https://doi.org/10.13304/j.nykjdb.2020.0788
Qiao Q, Si F, Wu C, Wang J, Zhang A, Tao J, Zhang L, Liu Y, Feng Z (2022) Transcriptome sequencing and flavonoid metabolism analysis in the leaves of three different cultivars of Acer truncatum. Plant Physiol Biochem 171:1–13. https://doi.org/10.1016/j.plaphy.2021.12.027
Qu A, Wu Q, Su J, Li C, Yang L, Wang ZA, Wang Z, Li Z, Ruan X, Zhao Y, Wang Q (2022) A review on the composition and biosynthesis of alkaloids and on the taxonomy, domestication, and cultivation of medicinal Fritillaria species. Agronomy 12(8):1844. https://doi.org/10.3390/agronomy12081844
Quan Y, Li L, Yin Z, Chen S, Yi J, Lang J, Zhang L, Yue Q, Zhao J (2022) Bulbus Fritillariae Cirrhosae as a respiratory medicine: is there a potential drug in the treatment of COVID-19? Front Pharmacol 12:784335. https://doi.org/10.3389/fphar.2021.784335
Ramsey J (2011) Polyploidy and ecological adaptation in wild yarrow. Proc Natl Acad Sci 108(17):7096–7101. https://doi.org/10.1073/pnas.1016631108
Shen Y, Zhou G, Liang C, Tian Z (2022) Omics-based interdisciplinarity is accelerating plant breeding. Curr Opin Plant Biol 66:102167. https://doi.org/10.1016/j.pbi.2021.102167
Tang L, Xiao D, Yin Y, Wang H, Wang J, Liu T, Hou X, Li Y (2022) Comparative transcriptome analysis of purple and green non-heading Chinese cabbage and function analyses of BcTT8 gene. Genes 13(6):988. https://doi.org/10.3390/genes13060988
Taylor SH, Franks PJ, Hulme SP, Spriggs E, Christin PA, Edwards EJ, Woodward FI, Osborne CP (2012) Photosynthetic pathway and ecological adaptation explain stomatal trait diversity amongst grasses. New Phytol 193(2):387–396. https://doi.org/10.1111/j.1469-8137.2011.03935.x
Varet H, Brillet-Guéguen L, Coppée J, Dillies M (2016) SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS ONE 11(6):e157022. https://doi.org/10.1371/journal.pone.0157022
Wang L, Josephs EB, Lee KM, Roberts LM, Rellán-Álvarez R, Ross-Ibarra J, Hufford MB (2021a) Molecular parallelism underlies convergent highland adaptation of maize landraces. Mol Biol Evol 38(9):3567–3580. https://doi.org/10.1093/molbev/msab119
Wang Y, Hou H, Ren Q, Hu H, Yang T, Li X (2021b) Natural drug sources for respiratory diseases from Fritillaria: chemical and biological analyses. Chin Med 16(1):40. https://doi.org/10.1186/s13020-021-00450-1
Wang X, Wang Y, Wu J, Feng X, Gao D, Fu S, Li X, Chen S (2022a) Effects of topdressing in the flowering stage on yield and quality of Fritillaria cirrhosa D. Don. World Chin Med 17:1789–1796
Wang Y, Feng X, Gao D, Xie H, Fu S, Li X (2022b) Comparative analysis of physiological indexes and alkaloid content of Fritillaria cirrhosa D. Don of different bulb sizes. World Chin Med 17:1833–1839
Yan J, Qian L, Zhu W, Qiu J, Lu Q, Wang X, Wu Q, Ruan S, Huang Y (2020) Integrated analysis of the transcriptome and metabolome of purple and green leaves of Tetrastigma hemsleyanum reveals gene expression patterns involved in anthocyanin biosynthesis. PLoS ONE 15(3):e230154. https://doi.org/10.1371/journal.pone.0230154
Yuan L, Zhang L, Wu Y, Zheng Y, Nie L, Zhang S, Lan T, Zhao Y, Zhu S, Hou J, Chen G, Tang X, Wang C (2021) Comparative transcriptome analysis reveals that chlorophyll metabolism contributes to leaf color changes in wucai (Brassica campestris L.) in response to cold. BMC Plant Biol 21(1):438. https://doi.org/10.1186/s12870-021-03218-9
Zhang SF, Jianhe W, Shilin C, Yong D, Xiwen L (2010) Floral dynamic and pollination habit of Fritillaria cirrhosa. China J Chin Mater Med 35:27–29. https://doi.org/10.4268/cjcmm20100105
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19(4):81–90. https://doi.org/10.1016/j.pbi.2014.05.011
Zhang T, Qiao Q, Novikova PY, Wang Q, Yue J, Guan Y, Ming S, Liu T, De J, Liu Y, Al-Shehbaz IA, Sun H, Van Montagu M, Huang J, Van de Peer Y, Qiong L (2019) Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc Natl Acad Sci 116(14):7137–7146. https://doi.org/10.1073/pnas.1817580116
Zhang X, Li B, Duan R, Han C, Wang L, Yang J, Wang L, Wang S, Su Y, Xue H (2022) Transcriptome analysis reveals roles of sucrose in anthocyanin accumulation in “Kuerle Xiangli” (Pyrus sinkiangensis Yü). Genes 13(6):1064. https://doi.org/10.3390/genes13061064
Funding
The research was funded by the National Key Research and Development Program of China (2019YFC1710601), Scientific and technological innovation project of China Academy of Chinese Medical Sciences (CI2021A03910). Fundamental Research Funds for the Central public welfare research institutes (ZXKT22061, L2022053, L2023022, ZZ16-XRZ-067, Z2023014 and L2022038).
Author information
Authors and Affiliations
Contributions
Experiment, Z.Y., X.W. and Z.L.; writing—original draft preparation, Y.W. and D.G.; writing—review and editing, X.L.; funding acquisition, G.X., S.F. and X.L. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Informed consent
Not applicable.
Additional information
Communicated by J. Huang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Yang, Z., Wang, X. et al. Transcriptomic profiling reveals color variation mechanism of Fritillaria cirrhosa for the molecular plant breeding. Acta Physiol Plant 46, 64 (2024). https://doi.org/10.1007/s11738-024-03688-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-024-03688-y