Abstract
The electrochemical property of dinuclear copper(II) complex containing mixed ligands of N-hydroxyethylaminoethyl oxamido and 2,2′-bipyridine [Cu2(bpy)2(HAO) 2+2 ] was studied with cyclic voltammetry. Cu2(bpy)2(HAO) 2+2 had irreversible oxidation peaks in 0.1 mol/L NaCl solution at the stearic acid-modified carbon paste electrode. Cyclic voltammetry and absorption spectra measurements were used to study the interaction between Cu2(bpy)2(HAO) 2+2 and herring fish sperm DNA. All the experimental results showed that Cu2(bpy)2(HAO) 2+2 interacted with DNA mainly through electrostatic affinity to make tiny difference between Cu2(bpy)2(HAO) 2+2 –ssDNA and Cu2(bpy)2(HAO) 2+2 –dsDNA. The binding ratio and the binding constant of DNA–Cu2(bpy)2(HAO) 2+2 were calculated as 1:1 and 6.41 × 104, respectively. The redox peak current of Cu2(bpy)2(HAO) 2+2 decreased markedly after its interaction with DNA. This was used to detect the concentration of DNA quantitatively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There is continuing interest in small molecules, especially metal complexes which can interact with DNA [1–4]. A precise understanding of the DNA-binding affinity and the cleavage mechanism would provide insight into the nature and structure of the reactive intermediate and would be potentially useful in the design of new site-specific scission reagents and chemical probes. Copper is a bioessential element with relevant oxidation states. Copper complexes are involved in a variety of important biochemical processes [5, 6]. Some copper complexes with different types of ligands have been synthesized and studied. Electrochemical techniques are used to characterize their electrochemical behaviors since the biochemical reactions related to the redox processes. Among the copper complexes explored so far, attention has been primarily focused on the copper complexes with rigid bidentate ligands, such as 2,2′-bipyridine [1, 3] or 1,10-phenanthroline and their derivatives [7–10] due to their high nucleolytic efficiency and their potential application in the recognition of the DNA sequence. Herein, we characterized the electrochemical property of a new dinuclear copper(II) complex containing mixed ligands of N-hydroxyethylaminoethyl oxamido and 2,2′-bipyridine [Cu2(bpy)2(HAO) 2+2 ]. Its interaction with herring fish sperm DNA was also investigated with cyclic voltammetry and spectroscopy.
Experimental
Materials
Double-strand herring fish sperm DNA from Sigma was used as received. The stock solution of DNA (pH 8.1) was prepared by dissolving appropriate amount of DNA in 10 mM Tris containing 1 mM EDTA and stored at 4 °C. The ratio of the absorbance at 260 and 280 nm (A 260/A 280) was checked to be 1.89, indicating that the DNA was sufficiently free from protein. Its concentration was determined by ultraviolet absorption at 260 nm (ε = 6,600 L mol−1 cm−1). Denatured single-strand DNA (ssDNA) was obtained by heating double-stranded DNA (dsDNA) in a water bath at 100 °C for 10 min, followed by rapidly cooling in an ice bath. Cu2(bpy)2(HAO) 2+2 was friendly supplied by Professor Yantuan Li, who works in Marine Drug and Food Institute, Ocean University of China. Stearic acid, carbon powder, and other reagents were all purchased from Shanghai Reagent Company and used as received.
Procedure
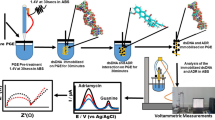
A CHI 832 electrochemical analyzer (Shanghai CH Instrument Company, China), which was in connection with a homemade carbon paste-modified working electrode (Φ 4 mm), a saturated calomel reference electrode, and a platinum wire auxiliary electrode, was used for the electrochemical measurement. Absorption spectra were recorded on a Varian 300 spectrophotometer. Spectrometric titrations were performed by adding successive aliquots of DNA to Cu2(bpy)2(HAO) 2+2 solution, in a 10-mm quartz cell, and recording the spectrum after each addition.
Results and discussion
The electrochemical properties of Cu2(bpy)2(HAO) 2+2
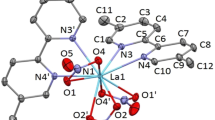
The structure of Cu2(bpy)2(HAO) 2+2 was illustrated in Fig. 1. The carbon paste electrodes were prepared according to our previous works [11].
As shown in Fig. 2, the peak currents of 2.0 × 10−4 mol/L Cu2(bpy)2(HAO) 2+2 at the carbon glassy electrode (GCE, Φ 3 mm) were very small. Modified carbon paste electrodes (CPEs) were made to enhance the electrochemical signals of Cu2(bpy)2(HAO) 2+2 . Different mass ratio of graphite powder, stearic acid, and solid paraffin was heated at 80 °C for 2 h and ground intensively to obtain uniform carbon paste. The hot carbon paste was tightly packed into the cave of a glass tube (Φ 4 mm) and a copper wire was introduced for electrical contact. And then, the carbon paste was allowed to cool down to the ambient temperature under certain pressure. The obtained electrode was smoothed on a weighing paper before use.
Cyclic voltammograms of 2.0 × 10−4 mol/L Cu2(bpy)2(HAO) 2+2 in 0.1 mol/L NaCl solution at (a) GCE and (b–e) CPE. The mass ratio of solid paraffin, stearic acid, and carbon powder was 1∶0∶4; 0∶1∶4; 0∶1∶3; and 1∶3∶12, respectively. The starting potential was 0.2 V and the direction of scan was negative
The electrochemical signals of Cu2(bpy)2(HAO) 2+2 were a little bigger at pure carbon paste electrode (the mass ratio of carbon powder to solid paraffin was 4:1), but still too small to detect exactly. Considering the positively charged Cu2(bpy)2(HAO) 2+2 and the hydrophilic ligands of N-hydroxyethylaminoethyl oxamide, stearic acid was added to the carbon paste to improve the hydrophilicity and the charged property of the electrode surface. The peak currents increased evidently at the stearic acid-modified CPE. The reversibility of the electrochemical reaction became worse when the percentage of stearic acid exceeded 20 %. On the other hand, when the percentage of stearic acid was too small, the obdurability of the electrodes decreased and Cu2(bpy)2(HAO) 2+2 was easy to penetrate into the carbon paste, which made it difficult to eliminate the remaining signal of Cu2(bpy)2(HAO) 2+2 at the test electrode. Considering both the obdurability of the electrodes and the reversibility of the reaction of Cu2(bpy)2(HAO) 2+2 , the mass ratio of carbon powder to stearic acid employed in all the subsequent work was 4∶1.
The multi-sweep cyclic voltammograms of 1.5 × 10−4 mol/L Cu2(bpy)2(HAO) 2+2 in 0.1 mol/L NaCl solution were recorded after the CPE soaking in the test solution for 5 min (Fig. 3). With the increase of the scan cycle (a→f), the cathodic peak potential experienced a positive shift and the cathodic peak current decreased markedly. By contrast, the anodic peak currents increased slightly and the anodic peak potentials experienced slightly negative shift up to the third scan cycle and then tended to keep constant. A new cathodic peak appeared at the third scan cycle between the potential of −0.6 to approximately −0.7 V and the peak current increased with the increase of the scan cycle. All the above characteristics indicated that the electroreduction of Cu2(bpy)2(HAO) 2+2 at the CPE is irreversible. The new cathodic peak increased markedly when the terminal scan potential negatively shifted to −1.4 V (the inset of Fig. 3). In other words, the more negative terminal potential favored the new substance.
When the scan rate changed in the range from 20 to 250 mV/s, the i pc and i pa of 2.0 × 10−4 mol/L Cu2(bpy)2(HAO) 2+2 were directly proportional to the scan rate. The linear regression equations of i pc and i pa were i pa = 10.695 v + 1.3569 (r = 0.9971) and i pc = 11.28 v + 4.0179 (r = 0.9957), respectively, where i pa was in 10−5 A and v in volts per second. Thus, the electrochemical reaction of Cu2(bpy)2(HAO) 2+2 at CPE was an adsorption-controlled process.
The dissociation process of Cu2(bpy)2(HAO) 2+2 from the CPE was also discussed(Fig. 4). After the CPE was inserted into the Cu2(bpy)2(HAO) 2+2 solution for 5 min, it was transferred into 0.1 mol/L NaCl solution immediately and subsequently scanned in the potential range of 0.2 to approximately −0.8 V. The peak currents of Cu2(bpy)2(HAO) 2+2 decreased gradually with the prolonging of the dissociation time. A linear relation between the logarithmic value of the i pa of Cu2(bpy)2(HAO) 2+2 and the dissociation time was obtained with the corresponding regression equation lgi pa = −0.0028 t − 4.697 (r = 0.993), which suggested that the dissociation of Cu2(bpy)2(HAO) 2+2 from the CPE obeyed the model of first-order kinetic process. The new cathodic peak above also appeared and increased with the scan cycle. Some Cu2(bpy)2(HAO) 2+2 being translated into the new substance in addition to the dissociation of Cu2(bpy)2(HAO) 2+2 led to its i pc decreasing more quickly than the i pa and the regression curve of i pc was bended down.
Electrochemical study of the interaction between Cu2(bpy)2(HAO) 2+2 and DNA
Typical CV behaviors of 2.0 × 10−4 mol/L Cu2(bpy)2(HAO) 2+2 in the absence and presence of DNA were shown in Fig. 5. When 3.2 × 10−5 mol/L ssDNA or dsDNA was added, the i ps of Cu2(bpy)2(HAO) 2+2 decreased along with the widening of the ΔE p(E pa − E pc), and no other new peak appeared. On the other hand, the formal potential [E f = 1/2(E pa + E pc)] of Cu2(bpy)2(HAO) 2+2 exhibited ∼8 mV (mean value of five measurements with the standard deviation of 1.91 mV) negative shift after adding ssDNA or dsDNA, which indicated that Cu2(bpy)2(HAO) 2+2 was associated with DNA through electrostatic binding [12]. The ratio of the equilibrium constants, K 2+/K + for the binding of the Cu(II) and Cu(I) forms of complexes to DNA could be estimated from the net shift in E 1/2, using the equation:
where E 0f ′ and E 0b ′ are the formal potentials of the Cu(II)/Cu(I) couple in the free and bound species, respectively. For both of ssDNA and dsDNA, K 2+ was higher than K +. DNA tended to stabilize the Cu(II) over the Cu(I) state by electrostatic interaction. The i pa/i pc ratio of Cu2(bpy)2(HAO) 2+2 both increased after adding ssDNA or dsDNA suggesting that the adsorption of the Cu(I) tended to be suppressed in the presence of DNA. Cu2(bpy)2(HAO) 2+2 cannot be used to distinguish ssDNA from dsDNA because there was no obvious difference between the i ps of dsDNA + Cu2(bpy)2(HAO) 2+2 and ssDNA + Cu2(bpy)2(HAO) 2+2 .
According to reference [13], the binding ratio and binding constant of Cu2(bpy)2(HAO) 2+2 –DNA complex can be determined. It is assumed that Cu2(bpy)2(HAO) 2+2 and DNA produce only a single complex DNA–mCu2(bpy)2(HAO) 2+2 :
The binding constant is
Because of: \( \left[ {\text{DNA}} \right] = {C_{{(DNA)}}} - \left[ {{\text{DNA}} - {\text{mC}}{{\text{u}}_{{2}}}{{\left( {\text{bpy}} \right)}_{{2}}}{{\left( {\text{HAO}} \right)}_{{2}}}^{{{2} + }}} \right] \)
Therefore, \( \Delta {i_{{{\text{p}},{ \max }}}} - \Delta {i_{\text{p}}} = {\text K}\left[ {\text{DNA}} \right] \)
and the following equations can be deduced:
where △i p,max represents the maximum difference of the peak currents before and after the addition of DNA. If DNA and Cu2(bpy)2(HAO) 2+2 form a single complex, the plot of lg [△i p / (△i p,max − △i p)] versus lg [Cu2(bpy)2(HAO) 2+2 ] is linear. In Fig. 6, curve a showed the relationship between the current i p and the concentration of Cu2(bpy)2(HAO) 2+2 . Curve b typically represented i p change at C (DNA) 3.2 × 10−5 mol/L on varying the concentration of Cu2(bpy)2(HAO) 2+2 . Curve c was the relationship between △i p, which was i p(a) − i p(b) and the concentration of Cu2(bpy)2(HAO) 2+2 . When the concentration of Cu2(bpy)2(HAO) 2+2 was over 1.6 × 10−4 mol/L, △i p tended to be a stable value, which indicated that here was almost no free DNA in the solution. Figure 7 showed the dependence of lg[△i p/(△i p,max − △i p)]on lg[Cu2(bpy)2(HAO) 2+2 ], by showing a fine straight line (y = 1.0243x + 4.8071, r = 0.9945). From the slope and the intercept of the straight line, the values of m and β were calculated as 1 and 6.41 × 104, respectively. A tentative idea was that the binding ratio of DNA and Cu2(bpy)2(HAO) 2+2 is 1:1. In Figs. 6 and 7, C(Cu2(bpy)2(HAO) 2+2 ) and [Cu2(bpy)2(HAO) 2+2 ] represent the concentration added and the equilibrium concentration of Cu2(bpy)2(HAO) 2+2 , respectively. [Cu2(bpy)2(HAO) 2+2 ] in Fig. 7 can be obtained from the data in Fig. 6.
Electronic spectral study of the interaction between Cu2(bpy)2(HAO) 2+2 and DNA
Cu2(bpy)2(HAO) 2+2 absorbed in the ultraviolet range with maxima at 310 and 299 nm, respectively. With the addition of DNA, Cu2(bpy)2(HAO) 2+2 showed a decrease in molar adsorptivity (hypochromism) as well as a slightly blue shift (1∼2 nm). The hypochromism induced by ssDNA was almost the same as dsDNA to indicate that the interaction between Cu2(bpy)2(HAO) 2+2 and ssDNA made no obvious difference with that between Cu2(bpy)2(HAO) 2+2 and dsDNA. The intrinsic binding constant K b of Cu2(bpy)2(HAO) 2+2 for dsDNA had been determined from the spectral titration data using the equation [14]:
where ε a, ε f, and ε b corresponded to A obsd/[Cu], the extinction coefficient for the free copper complex, and the extinction coefficient for the copper complex in the fully bound form, respectively. In the plot of [DNA]/(ε a − ε f) Vs [DNA], K b was given by the ratio of the slope to intercept. The K b value was calculated to be 5.24 × 104, which was closed to the result of electrochemical method (6.41 × 104), but lower than those observed for typical classical intercalators. Cu2(bpy)2(HAO) 2+2 should interact with DNA mainly by electrostatic interaction (Fig. 8).
Analytical application
Keeping the concentration of DNA as 4.0 × 10−6 mol/L, the influence of 20 times of substances such as l-lysine, l-glutamate, l-arginine, l-leucine, l-cysteine, citric acid, glucose, and 1.0 × 10−5 mol/L of cations such as Mg2+, Ni2+, Zn2+, Ca2+, Cu2+, and Ba2+, on the Δi p of DNA−Cu2(bpy)2(HAO) 2+2 system was tested according to the standard procedure. The relative errors of the results were smaller than ±8.7 %, which illustrates that these usual coexisting substances have little effects on the determination of DNA. It shows that the interaction of Cu2(bpy)2(HAO) 2+2 with DNA provides a possibility for detecting the content of DNA. The decrease of i pc (Δi pc) was proportional to the added concentration of dsDNA over the range from 1.2 × 10−6 to 1.1 × 10−5 mol/L, with the regression equation Δi p = 1.5628 C − 0.0487 (Δi p, 10−5 A; C, 10−5 mol/L, r = 0.9982). The limit of detection was given by the equation limit of detection (LOD) = 3α, where α is the standard deviation of the blank measurements (n = 11); here, the LOD was 4.8 × 10−7 mol/L.
To test this new method, the DNA contents in two synthetic samples, constructed based on the effects of foreign substances, were analyzed under the optimum conditions, namely, 2.0 × 10−4 mol/L Cu2(bpy)2(HAO) 2+2 in 0.10 mol/L NaCl solution. As shown in Table 1, the results are reproducible and reliable.
Conclusion
Cu2(bpy)2(HAO) 2+2 had irreversible oxidation peaks in 0.1 mol/L NaCl buffer solution at the stearic acid-modified carbon paste electrode. A new substance with more negative cathodic potential was produced during the reductive process. All the experimental results from cyclic voltammetry and absorption spectra measurements showed that Cu2(bpy)2(HAO) 2+2 interacted with DNA mainly through electrostatic affinity. There was only a tiny difference between Cu2(bpy)2(HAO) 2+2 –ssDNA and Cu2(bpy)2(HAO) 2+2 –dsDNA. The decreases of the cathodic peak current of Cu2(bpy)2(HAO) 2+2 were proportional to the added concentration of dsDNA over the range from 1.2 × 10−6 to 1.1 × 10−5 mol/L.
References
Laine M, Richard F, Tarnaud E, Bied CC, Verchere BC (2004) J Biol Inorg Chem 9:550
Ju HX, Ye YK, Zhao JH, Zhu YL (2003) Anal Biochem 313:255
Liu FQ, Wang QX, Jiao K, Jian FF, Liu GY, Li RX (2006) Inorganica Chimica Acta 359:1524
Humphreys KJ, Karlin KD, Rokita SE (2002) J Am Chem Soc 124:8055
Holm RH (1995) Pure & Appl Chem 67:217
Tamas G, Yvonne D, Ibolya T (2001) Inorg Chem 40:4918
Carter MT, Rodriguez M, Bard AJ (1989) J Am Chem Soc 111:8901
Sethuraman M, Mallayan P (1998) Inorg Chem 37:693
Li XM, Ju HQ, Ding CF (2007) Analytica Chimica Acta 582:158
Macías B, Villa MV, Salgado M (2006) Inorganica Chimica Acta 359:1465
Ren Y, Jiao K, Xu GY, Sun W, Gao HW (2005) Electroanalysis 17:2182
Sethuraman M, Mallayan P (1998) Inorg Chem 37:3927
Jiao K, Li QJ, Sun W, Wang ZJ (2005) Electroanalysis 17(11):997
Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK (1989) J Am Chem Soc 111:3051
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, G., Tang, L. & Liu, H. Electrochemical behavior and DNA-binding properties of binuclear copper(II) complex containing mixed ligands of N-hydroxyethylaminoethyl oxamido and 2,2′-bipyridine. Ionics 19, 309–314 (2013). https://doi.org/10.1007/s11581-012-0721-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11581-012-0721-0