Abstract
A dematiaceous hyphomycete with erect unbranched conidiophore stipes and Cladosporium-like branched conidial chains arranged at nodes along the stipe was found on living leaves of wild banana (Musa itinerans) in Taiwan. Scanning electron microscopy indicated a type of conidiogenous loci differing from that of Cladosporium by lacking the cleft between raised center and margin. Molecular phylogenetic analyses of several loci (LSU, ITS, TEF, RPB2) indicated a relationship with cercosporoid fungi (Mycosphaerellaceae). Since there is no other known lineage with similar morphology or DNA sequences, the new genus and species Cladocillium musae is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Within the last 100 years, the most important epidemic banana disease fungi were Mycosphaerella musicola (yellow sigatoka disease) followed by Pseudocercospora fijiensis (also known under the teleomorph name Mycosphaerella fijiensis) (black sigatoka disease), and Fusarium oxysporum f. sp. cubense (FOC). These diseases reduce the fruit production and cause premature ripening of fruits. Ca. 30% production costs are spent to prevention and control of sigatoka diseases (Marín et al. 2003). The impact of FOC leads to the replacement of the banana cultivar Gros Michel by the less tasty cultivar Cavendish worldwide in the 1960s (Ploetz 1994). Presently, two further major emerging fungal pathogens pose a risk to and loss in the global banana market, viz., Fusarium oxysporum f. sp. cubense (FOC) Tropical Race 4 (TR4) and Phyllosticta species (Ploetz 1994).
Different species, subspecies, cultivars, and hybrids of Musaceae are native to Taiwan or have been introduced as ornamental or food plants. Banana cultivars were introduced from Fujian Province of mainland China to Taiwan during the seventeenth century. During the Japanese rule in Taiwan between 1895 and 1945, banana growing gained economic importance (Pan 2014). Wild bananas in Taiwan are mainly different varieties of Musa itinerans Cheesman (Chiu et al. 2011). Breeding of new hybrids and cultivars is focusing more on disease resistance than on the food qualities (Ploetz 1994). This focus demonstrates the importance of fungi on banana. In contrast to this fact, however, comparatively few research has been done on fungi on wild and cultivated bananas, although further hitherto unknown pathogens are likely to emerge in the future, or other fungi may have antagonistic potential against pathogens. After random collections of fungi on wild and cultivated banana plants in Taiwan, two potentially plant pathogenic species of Mycosphaerellaceae were newly recorded for Taiwan (Kirschner and Piepenbring 2014). Some further investigations of fungi on banana leaves in Taiwan revealed another mycosphaerellaceous fungus which did not match with any known species so that it was studied in more detail.
Materials and methods
Specimens and isolation of strains
Leaf specimens of cultivated and wild bananas with disease symptoms were collected in the lowland hills of northern Taiwan, brought to the laboratory, and investigated with a dissecting microscope for the presence of conidiophores. With a flamed acupuncture needle, conidia were transferred from the conidiophores on the leaves to corn meal agar (CMA, Fluka) with 0.2% chloramphenicol. Online Auction Color Charts (Online Auction Color Chart Co., 2004 Palo Alto, CA, USA) were used for indicating colony colors.
Dried specimens were deposited at the herbarium of the National Museum of Natural Science, Taichung, Taiwan (TNM). Two of the three living strains relevant for this publication were deposited at the Bioresource Collection and Research Centre, Hsinchu City, Taiwan (BCRC).
Microscopy
For light microscopy, specimens from living banana leaves were mounted in 10% (w/v) aqueous KOH. Statistical treatment of measurements was based on n replicates and given as mean value ± standard deviation with extreme values given in brackets. For scanning electron microscopy, material from culture was fixed in 2% (w/v) aqueous osmium tetroxide for 25 h at 4 °C in the dark, washed three times in distilled water, dehydrated in a 10% grade ethanol series from 10 to 90% ethanol, followed by ethanol:acetone mixtures of 2:1, 1:1, and 1:2. Final dehydration was done with absolute acetone in three rounds each for 15 min, and critical point drying in liquid CO2 with a Hitachi HCP-2 critical point dryer. Critical point–dried material was mounted and coated with gold in a Hitachi E-1045 Ion Sputter Coater and investigated with a Hitachi S-4700 scanning electron microscope at 20 kV.
DNA sequence analysis
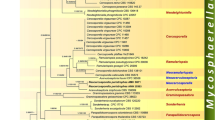
For DNA isolation, fresh material from a culture was crushed with a bead beater and the Genomic DNA Spin Kit for plants (Bioman Scientific Co., Ltd., Taiwan) used according to the manufacturer’s protocol. For PCR, primers ITS1F and ITS4 (Gardes and Bruns 1993; White et al. 1990) were used for amplifying the internal transcribed spacer region of the nuclear ribosomal RNA genes (ITS, including ITS1, 5.8S ribosomal rDNA, ITS2, and fragments of the small and large ribosomal subunit), primers NL1 and NL4 (Kurtzman and Robnett 1997) for the partial nuclear ribosomal large subunit gene (LSU), primers EF1-728F and EF1-986R (Carbone and Kohn 1999) for the translation elongation factor 1 alpha gene (TEF), and primers RPB2-5F and RPB2-414R for the gene for the DNA-directed RNA polymerase II second largest subunit (RPB2) (Quaedvlieg et al. 2013). The PCR conditions were the same as in the literature cited for the primers. PCR products were sequenced by Mission Biotech (Nankang, Taipei) with the same primers used for the PCR. The DNA sequences were edited with CodonCode Aligner version 4.0.1 (CodonCode Corporation, USA) and deposited at GenBank and the DNA Data Bank of Japan. The sequences were submitted to BLAST searches at GenBank (https://blast.ncbi.nlm.nih.gov/). The most similar species from the BLAST searches were selected for aligning the sequences from the three isolates from banana with similar sequences from GenBank. Only strains were included for which ITS, LSU, RPB2, and TEF were all available. Most sequences were from Quaedvlieg et al. (2014), since the four DNA regions were all covered for the same respective strain. Capnodium coffeae (Capnodiaceae) and species of Teratosphaeriaceae were added as more distantly related groups. BLAST searches were also done specifically for sequences of Cladosporium in GenBank and two most similar sequences included into the datasets. Table 1 shows the sequence accessions. Alignments done for the separate datasets with MUSCLE in MEGA 7 were compared with each other with respect to topology by running neighbor joining (NJ) analyses. Since the TEF sequence topology differed from the other ones, the TEF dataset was aligned with MAFFT with the G-INS-i and E-INS-I strategies; the NJ topology resulting from the G-INS-I produced a topology which was most similar to those in the other datasets so that this alignment was used further. Manual manipulation within the alignments was avoided. After trimming the uneven ends, the datasets were concatenated and formed a block along 1478 positions. The individual datasets and the concatenated one are provided as electronic supplementary materials (ESM1–5). As best model for the concatenated dataset (as well as the separate ITS and RPB2 datasets), Kimura’s two-parameter model with Gamma distribution and invariant sites was chosen, which was the second-best model for the separate datasets of LSU (Tamura 1992 model with Gamma distribution and invariant sites) and TEF (Kimura’s two-parameter model with Gamma distribution best model). The result of the maximum likelihood analysis run with 1000 bootstrap replicates is shown in Fig. 1.
Results
Three specimens of the same fungus on leaves from wild Musa itinerans were collected, cultivated, and used in phylogenetic estimates. Light microscopy was mainly based on specimen CCH05A from a leaf and SEM on the culture derived from it. When comparing the BLAST search results among sequences from the four loci (LSU, ITS, TEF, RPB2), the highest matches were all sequences of Mycosphaerellaceae. The highest matches were found with LSU sequences exceeding a length of 576 bp; the fragments of our isolates (CCH02B, CCH03D, CCH05A) and of several Pseudocercospora species showed an identity of up to 96%; for RPB2, Pseudocercospora spp. also showed the highest identities up to 76%. The ITS sequences had up to 94% identity with several sequences from Cercosporella in GenBank. For TEF sequences, the highest identities were sequences of Septoria spp. up to 82%. BLAST searches for Cladosporium sequences in GenBank retrieved up to 88% identity for the LSU, 90% for the ITS, no significant results for TEF, and up to 71% identity for RPB2. The three ITS sequences differed from each other for 3 bp (0.18%) or 7 bp (1.27%) of 545 bp. No chimeras were detected in the graphical representation of BLAST results with the ITS sequences. In the individual and the combined datasets, the three strains clustered with 100% bootstrap support in a distinct clade separate from the clades of Pseudocercospora and other mycosphaerellaceous fungi. Since the three isolates represent an unknown lineage within the cercosporoid fungi (Mycosphaerellaceae), a new genus and a new species are proposed.
Taxonomy
Cladocillium Chun-Hao Chen & R. Kirschner, gen. nov.MycoBank MB 835863
Etymology: Name composed of the elements—cillium for referring to the Verticillium-like conidiophore stipe and clado—for referring to the Cladosporium-like branched conidial chains.
Typus generis: Cladocillium musae Chun-Hao Chen & R. Kirschner in opere ipso.
Conidiophores macronematous, single, erect, unbranched or rarely branched, but with conidial chains arising in verticils along the conidiophore stipe, pigmented, conidia in branched acropetal chains, pigmented, rough, conidiogenous cells intercalary and terminal. Differing from Cladosporium by conidiogenous loci being crowded at nodes below septa, not conspicuously darkened in light microscopy, in SEM appearing flat or with a raised center, without cleft between center and margin.
Cladocillium musae Chun-Hao Chen & R. Kirschner, sp. nov. Figs. 2 and 3
Cladocillium musae (CCH05A) from leaf of Musa itinerans. a Young conidiophore with only two apical conidiogenous cells. b Typical mature conidiophore showing arrangement of branched conidial chains. c Apex of mature conidiophore showing the subterminal conidiogenous loci of the intercalary conidiogenous cells after conidium dehiscence. d Conidia. Scale bars: a–c, 20 μm; d, 5 μm.
MycoBank MB 835864
Etymology: Name referring to the host plant genus Musa.
Colonies not associated with distinct disease symptoms on the leaf, but with those caused by other fungi, olive-green to brown. Hyphae subhyaline to pale brown, smooth to finely verruculose, 1–2 μm wide. Conidiophores single, erect, with a non-rhizoidal single-celled foot distinguished from vegetative hyphae by deeper pigmentation and broader width, ca. 12–30 μm long and 5–10 μm high, with unbranched stipe (exceptionally with a single short lateral branch), appearing smooth in light microscopy, occasionally rough in SEM, (43–)87–222.5(–350) × (2.5–)3–5(–6) μm (n = 30). Distances between septa 14–28 μm, becoming shorter (ca. 10 μm) towards the apex of conidiophore, and cells becoming narrower, 1.5–2 μm, cells of the apical ca. 100–200 μm long part of stipe serving as intercalary conidiogenous cells (plus a terminal conidiogenous cell). Two to six conidiogenous loci crowded at distal end of conidiogenous cell and ramoconidia, not or slightly thickened, when thickened then refractive, but not darkened, minimally raised above the cell surface to slightly denticulate and less than 1 μm long, ca. 0.5 μm wide, in SEM flat or showing a raised center, without cleft between center and margin. Conidia in short branched acropetal chains, ovoid, fusiform, aseptate, subhyaline to light brown, verruculose, base truncated, apex broadly rounded, proximal ones (3–)3.5–5(–6) × (2–)2–3(–4) μm (n = 30), distal ones (1.5–)2.5–3.5(–3.5) × (2–)2–2.5(–3) μm (n = 30), hila with the same characteristics as conidiogenous loci.
Colonies on CMA pale vinaceous-pink (oac564), becoming ochre (oac646) by ageing, flat, appearing powdery by the production of conidia. Conidiophores and conidia identical to those on leaves.
Specimens examined: On Musa itinerans leaf, Taiwan, Yilan County, Datong Township, Jiuliao River, 200 m, 04 Oct., R. Kirschner, CCH02B (dried culture, TNM, paratype), living strain BCRC FU30629; on Musa itinerans leaf, Taiwan, Taoyuan City, Dasi District, Shimen reservoir, 15 Oct. 2014, R. Kirschner & Chun-Hao Chen, CCH03D (dried culture, TNM, paratype); ibid., 15 Nov. 2014, R. Kirschner, CCH05A (dried culture, TNM, holotype), ex-type strain BCRC FU30634.
Discussion
Light microscopy was based on material from leaves, complemented with some details from cultivated conidiophores and conidia which looked identical to those from leaves; SEM and DNA analysis were based on material in culture. The ITS sequences of the three strains had 98.72 to 99.82% identity, which is within the range of identity for the same species, even if the 97% threshold is considered too low (Hofstetter et al. 2019) or the variability can be exceptionally higher than 3% even within the same genome (Stadler et al. 2020).
The results either of morphology or DNA sequence analysis do not conform to a known genus. The DNA sequence analysis of the concatenated dataset (LSU, ITS, TEF, RPB2) strongly supported a close relationship with other cercosporoid fungi, but could not resolve a clear sister relationship with a single genus of the Mycosphaerellaceae. The four loci used in this study were proven useful for cercosporoid fungi in previous studies (Quaedvlieg et al. 2014; Verkley et al. 2013). The BLAST and preliminary phylogenetic analyses from the different loci yielded a distinct clade for the three strains of Cladocillium, but with different relationships within the Mycosphaerellaceae. The genera outside of Cladocillium were also less supported in the individual datasets than in the combined one. These data suggest a distinct lineage rather than a sister or subclade of a known genus. The morphology was strongly similar to that of Cladosporium, particularly with respect to pigmented blastocatenate verruculose conidia (Zalar et al. 2007; Zhan et al. 2014).
Cladosporium and similar genera were recently revised so that most of them can be considered comparatively well known (Bensch et al. 2012). The erect unbranched conidiophores of Cladocillium with conidial chains arising at nodes below the septa are similar to those of C. oxysporum Berk. & M.A. Curtis (Bensch et al. 2012). The typical conidiogenous locus in Cladosporium when seen from above shows a raised center which is clearly separated from the coronate margin by a cleft (Bensch et al. 2012; Zalar et al. 2007; Zhan et al. 2014). In light microscopy, the conidiogenous loci and hila of Cladosporium appear conspicuously darkened. This cleft could not be seen in the SEM of Cladocillium musae, although the center of the conidiogenous locus in some cases was raised. In light microscopy, the conidiogenous loci and hila were not darkened in Cladocillium. The distinctly macronematous and unbranched conidiophores with arrangement of conidial chains in whorls at intercalary conidiogenous cells and lack of septa in the conidia further distinguish Cladocillium form the majority of Cladosporium and Cladosporium-like fungi (Bensch et al. 2012). As far as DNA data are available from these fungi, they appear only distantly related to Cladocillium. Among these Cladosporium-like fungi, Metulocladosporiella with relationship to Chaetothyriales was segregated from Cladosporium. The species of Metulocladosporiella appear to be geographically widespread and specific to leaves of Musaceae where leaf speckle symptoms are caused (Jones 2019).
Cladocillium musae was only detected by chance with the dissecting and light microscope when searching for more conspicuous fungi associated with leaf spots. Because of the delicate nature of the tiny hyphae and conidiophores which were hardly discernable on the leaf even in the fresh state, it was not possible to clarify whether they grew exclusively superficially or emerged from the plant tissues. Further studies may clarify whether Cladocillium musae belongs either to the external commensalistic mycoflora on healthy or senescent leaves of bananas or is involved in leaf pathogenesis.
References
Bensch K, Braun U, Groenewald JZ, Crous PW (2012) The genus Cladosporium. Stud Mycol 72(1):1–401
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Chiu H-L, Shii C-T, Yang TYA (2011) A new variety of Musa itinerans (Musaceae) in Taiwan. Novon 21:405–412
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for Basidiomycetes: application to identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Hofstetter V, Buyck B, Eyssartier G, Schnee S, Gindro K (2019) The unbearable lightness of sequenced-based identification. Fungal Divers 96:243–284
Jones DR (2019) Handbook of diseases of banana, abacá and enset. CAB International, Boston
Kirschner R, Piepenbring M (2014) New records of three Ramichloridium species on banana leaves in Panama and Taiwan. Mycoscience 55:260–267
Kurtzman CP, Robnett CJ (1997) Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5'end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35(5):1216–1223
Marín DH, Romero RA, Guzmán M, Sutton TB (2003) Black sigatoka: An increasing threat to banana cultivation. Plant Dis 87(3):208–222
Pan F-J (2014) Formosan Plant Book, 2nd edn. Yuan-Liou Publishing Co., Ltd., Taipei 潘富俊2014福爾摩莎植物記.二版.遠流出版公司,臺北
Ploetz RC (1994) Panama disease: Return of the first banana menace. Int J Pest Manag 40(4):326–336
Quaedvlieg W, Verkley GJM, Shin HD, Barretto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW (2013) Sizing up Septoria. Stud Mycol 75:307–390
Quaedvlieg W, Binder M, Groenewald JZ, Summerell BA, Carnegie AJ, Burgess TI, Crous PW (2014) Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33:1–40
Stadler M, Lambert C, Wibberg D, Kalinowski J, Cox RJ, Kolařík M, Kuhnert E (2020) Intragenomic polymorphisms in the ITS region of high-quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycol Prog 19:235–245
Verkley GJ, Quaedvlieg W, Shin HD, Crous PW (2013) A new approach to species delimitation in Septoria. Stud Mycol 75(1):213–305
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego, pp 315–322
Zalar P, de Hoog GS, Schroers HJ, Crous PW, Groenewald JZ, Gunde-Cimerman N (2007) Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol 58:157–183
Zhan G, Tian Y, Wang F, Chen X, Guo J, Jiao M, Huang L, Kang Z (2014) A novel fungal hyperparasite of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One 9(11):e111484
Funding
The study was supported by the Ministry of Science & Technology, Taiwan (NSC 102–2621–B–008–001–MY3 and MOST 108-2621-B-002-007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Gerhard Rambold
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, CH., Hsieh, SY., Yeh, YH. et al. Cladocillium musae, a new genus and species of cercosporoid fungi (Mycosphaerellaceae) on wild banana in Taiwan. Mycol Progress 19, 837–843 (2020). https://doi.org/10.1007/s11557-020-01595-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-020-01595-3