Abstract
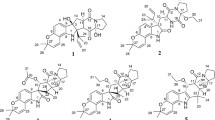
Two new indole alkaloids, 2-(3,3-dimethylprop-1-ene)-costaclavine (1) and 2-(3,3-dimethylprop-1-ene)-epicostaclavine (2), together with the known compounds costaclavine (3), fumgaclavine A (4) and C (5), were isolated from the marine-derived fungus Aspergillus fumigatus. The planar structures of the two new compounds were elucidated on the basis of chemical and physicochemical evidence including MS, UV, IR and NMR spectra. Their stereochemistry was studied by NOESY, 1H–1H coupling constant and CD spectra. The compounds 1, 2, 3 and 5 showed weak cytotoxicity against a mouse leukemia cell line (P388).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The marine environment is now recognized as an important resource of unusual and biologically active secondary metabolites [1]. Marine fungi are interesting sources of natural products. To avoid depletion of marine resources and to enable access to large quantities of interesting compounds, there has been a particular interest in marine microorganisms which are cultivable [2]. During our study of the symbiotic relationship between zoanthus and its related microorganisms, a fungus was isolated from the inside of a body of zoanthus and identified as Aspergillus fumigatus. Even though this genus of fungus has been studied for over 50 years, it continues to provide new bioactive metabolites [3–9]. During our study on its secondary metabolites, two new cytotoxic indole alkaloids, 2-(3,3-dimethylprop-1-ene)-costaclavine (1) and 2-(3,3-dimethylprop-1-ene)-epicostaclavine (2), together with the known compounds costaclavine (3) [3], fumgaclavine A (4) [7] and C (5) [8] (Fig. 1), were isolated. Here, we report the isolation, structure elucidation, and cytotoxic activity of two new costaclavines.
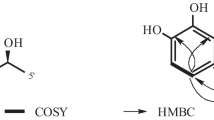
2-(3,3-Dimethylprop-1-ene)-costaclavine (1) was isolated as a pale yellow oil. The molecular formula of 1 was established as C21H28N2 (9 unsaturations) by HRFABMS and 13C NMR. The IR absorption spectrum of 1 showed the characteristic bands of the amino group (3299 cm−1) and indole (1673 cm−1) functionalities. The 13C NMR spectrum showed all the expected 21 signals, which could be assigned to four methyl groups including one methyl group attached to nitrogen (δ H = 3.02 ppm and δ C = 42.3 ppm, respectively), four methylene groups with one olefinic methylene (δ H = 5.12, 5.15 ppm and δ C = 112.2 ppm, respectively), and seven methine groups (Table 1). The planar structure of 1 was determined by detailed analyses of 2-D NMR spectra including COSY and HMBC. The 1H–1H COSY spectrum of 1 showed the connectivity of carbons 4-5-10-9-8(-18)-7 and 12-13-14, and the COSY correlation between H-22 and H-23 was also detected. The analysis of HMBC data of 1 connected all carbons, assigning the skeletal structure (Fig. 2). Positions of amino groups were determined by HMBC correlations from the amino proton to C-2 and C-15, and from the N-bearing methyl (δ C = 42.3) to C-5 and C-7. The general features of UV, IR and NMR spectra of 1 closely resembled those of compound 3 [3]. The NMR signals at C-2 were changed from sp 2-methine [7.04, d, J = 1.4 Hz, 120.4 (C-2)] in 3 to sp 2-quaternary carbon [137.9 (C-2)] in 1 and a 1,3-dimethylprop-1-ene group was connected to C-2. Detailed analyses of the 1H and 13C NMR spectra of 1, including the results from DEPT, HSQC and HMBC experiments, suggest that 1 is a 2-(1,3-dimethylprop-1-ene) derivative compound of 3. On the basis of all the evidence, we elucidated that the structure of 1 is 2-(1,3-dimethylprop-1-ene)-6,8-dimethylergoline.
The stereochemistry for 1 was elucidated using NOESY, 1H–1H coupling constant and CD spectra. The NOESY data of 1 showed correlations between Hβ-9 and H-12, and did not give a correlation between Hα-9 and H-12, thus it can be confirmed that Hβ-9 and Hα-9 are on different sides in the compound 1. At the same time, the NOESY signals between 18-Me and H-10 and Hβ-9, between Hβ-9 and H-10, and between H-5 and H-10, suggested that H-5, 18-Me, Hβ-9 and H-10 are on the same side. The NOESY signals between Hα-9 and H-8 and Hα-7, and between Hα-7 and H-8 and Hα-9, showed that Hα-7, H-8 and Hα-9 are on the other side. A cis relationship of both protons at C-5 and C-10 was further supported by comparing the coupling constant between H-5 and H-10 in 1 (J H5–H10 = 5.4 Hz and J H10–H5 = 5.4 Hz) with values reported for the cis stereoisomers of costaclavine (J = 3.1 Hz) and epicostaclavine (J = 3.9 Hz) and the trans stereoisomers of festuclavine (J = 9.5 Hz) [10].
Based on the CD spectra, the compound 1 showed negative cotton effect at 204 (−3.1) and 216 (−5.7) and positive cotton effect at 235 (+1.9), 273 (+1.0) and 294 (+0.8). By comparison of the CD data between 3 and 1, the CD spectrum of 1 was similar to that of 3 which showed negative cotton effect at 208 (−3.4) and 214 (−3.3) and positive cotton effect at 229 (+1.6), 273 (+0.6) and 294 (+0.5). It can be deduced that both compounds shared the same configuration. Based on the evidence described above, the stereostructure of 1 was determined to be (5R,8R,10S)-2-(1,3-dimethylprop-1-ene)-6,8-dimethylergoline.
2-(3,3-Dimethylprop-1-ene)-epicostaclavine (2) was a pale yellow oil which was isolated from the mycelium extract. A molecular formula of C21H28N2 gave nine degrees of unsaturation, which was established by HRFABMS and 13C NMR methods. The IR absorption spectrum of 2 showed the characteristic bands of the amino groups (3319 cm−1) and indole (1674 cm−1) functionalities. The 1H NMR and 13C NMR data of 2 were similar to those of compound 1 except for signals of positions 7, 8, 9, 10 and 18. The planar structure of 2 was established by analysis of 2-D NMR spectra including COSY and HMBC. Although the 1H and 13C chemical shift of positions 7, 8, 9, 10 and 18 were different from those of compound 1, it still has the same planar structure as 1 (Fig. 2). On the basis of all the foregoing evidence, the structure of 2 was suggested as 2-(1,3-dimethylprop-1-ene)-6,8-dimethylergoline.
The NOESY data of 2 showed correlations between Hβ-9 and H-12, and did not give a correlation between Hα-9 and H-12, thus it can be confirmed that Hβ-9 and Hα-9 are on opposite sides in compound 2. At the same time, the NOESY signals between H-8 and H-5, H-10 and Hβ-9, Hβ-9 and H-10, and H-5 and H-10, suggested that H-5, H-8, Hβ-9 and H-10 are on the same side. The NOESY signals between Hα-9 and 18-Me showed that 18-Me and Hα-9 are on the other side. Using the same method as compound 1, a cis relationship of both protons at C-5 and C-10 was further confirmed by comparing the coupling constant between H-5 and H-10.
Based on the optical rotation of compounds 2 and 1, it is difficult to judge the configuration at C-5, C-8 and C-10. However, the chemical shift of C-5 in 2 [3.01, ddd, J = 4.8, 5.8, 11.8 Hz, 69.0] is similar to 1 [3.02, ddd, J = 3.7, 5.4, 11.0 Hz, 69.0]. This suggested that the two compounds have the same configuration in C-5. Furthermore, by comparison of the CD data between 2 and 1, the CD spectrum of 2 was different to that of 1 and showed negative cotton effect at 208 (−3.4) and 224 (−8.9), and positive cotton effect at 260 (+3.7) and 293 (+2.0), indicating that the two compounds had a different configuration in C-8 (Fig. 3). Based on the evidence described above, the stereostructure of 2 was determined to be (5R,8S,10S)-2-(1,3-dimethylprop-1-ene)-6,8-dimethylergoline (2).
The known compounds 3, 4 and 5 were identified by spectroscopic analysis (1H and 13C NMR, LRFABMS, and [a] D ) and comparison to literature data [3, 7, 8].
The compounds of 1, 2, 3 and 5 showed weak cytotoxicity against mouse leukemia cell line (P388) with IC50 of 64.9, 259.7, 333.3 and 218.8 μM, respectively; 4 did not show this bioactivity.
Experimental section
General experimental procedures
Optical rotation was determined on a JASCO model P-1030 polarimeter. CD spectra were taken on a JASCO J-820 spectropolarimeter. UV/visible spectra were measured on a JASCO V-550 UV/vis spectrometer. IR spectra were recorded on a JASCO FT-IR model 420 spectrometer. 1H (400 MHz) and 13C NMR (100 MHz) spectra were obtained on a JEOL ECX400 NMR spectrometer, using solvent peaks as reference standard. 1H (500 MHz) and DQF COSY spectra were obtained on a JEOL ECA 500 NMR. MS spectra were obtained on a JEOL JMS-700P FAB MS spectrometer.
Fungal isolation and culture
The fungal strain was isolated from a zoanthid, Zoanthus sp., collected in Ayamaru Cape, Amami Island, Kagoshima Prefecture, Japan, in April 2003. This microorganism was identified as Aspergillus fumigatus by partial sequence analysis of both of the 28S rDNA-D1/D2 and the ITS-5.8S rDNA, followed by GenBank/DDBCJ/EMBL, which resulted in a closest match to Aspergillus fumigatus (100%), as judged by the fungal classification company Techno Suruga Co., Ltd. A voucher specimen is deposited at Tokyo University with the code ZTF001. The fungus was cultured (16 L) for 2 weeks (static) at 37°C in Malt medium consisting of soluble starch (0.4%), yeast extract (0.4%), malt extract (1.0%) and distilled water (100%).
Extraction and isolation
The mycelium and broth were separated by filtration using gauze. The mycelium was extracted with acetone–MeOH (v/v 1/1) to afford mycelium extract (0.57 g), which was subjected to silica gel flash chromatography. Elution was performed with n-hexane–EtOAc (stepwise, 0–100% EtOAc) to yield six fractions. Fraction 6 was separated into two fractions by GL-pak Carbograph using MeOH–EtOAc (v/v 1/1) and MeOH, respectively. The fraction 6.1 was further separated by PTLC on CH2Cl2–MeOH (10-1) with 0.1% TFA to afford four fractions. Fractions 1–3 were identified as crude compounds 5, 1, and 2, respectively, which were further purified by HPLC (YMC, ODS-C18) utilizing a 30-min gradient program of 50–100% MeOH in H2O to furnish 5 (30.0 mg), 1 (4.1 mg) and 2 (3.3 mg), respectively. Fraction 4 was purified by HPLC (YMC, ODS-C18) utilizing a 30-min gradient program of 50–75% MeOH in H2O to furnish 3 (1.9 mg) and 4 (2.7 mg).
Cytotoxicity test against P388 cells [11]
Mouse leukemia cell line P388 cells were grown and maintained in culture medium RPMI1640 (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 5% penicillin/streptomycin solution (Sigma) at 37°C in a humidified 5% CO2 incubator (Yamato, IT43) using EasyFlasks (Nunc). To maintain exponential growth, cells were subcultured every 96 h. P388 cells subcultured three times were used for the cytotoxicity test. Compounds were dissolved in methanol to obtain a concentration of 1 mg/mL. The solutions were diluted in the medium at concentrations ranging from 1 mg/mL to 1 μg/mL. The cell suspension (10,000 cells) was then blended in a 96-well plate (Nunc), each experimental group consisting of 5 wells. After 96 h culture in the incubator, 50 mL MTT solution was added to each well. After 4 h incubation, the 96-well plate was centrifuged and the supernatant discarded. Then, 100 mL DMSO was added to each well, and the absorbance of each well was measured at 570 nm using a micro-plate reader (Bio-Rad, Model 450).
2-(3,3-Dimethylprop-1-ene)-costaclavine (1) was obtained as a pale yellow oil: \( \left[ \alpha \right]_{D}^{25} \)-21 (c 0.29, MeOH); CD (MeOH) λ max (∆ε) 204 (−3.1), 216 (−5.7), 235 (+1.9), 273 (+1.0), 294 (+0.8) nm; UV (MeOH) λ max (log ε) 231 (4.2), 281 (4.0) nm; IR (film) λ max 3299, 2976, 1673, 1474, 1407, 1201, 1192, 1137 cm−1; 1H and 13C NMR, see Table 1; FAB MS m/z 308 [M]+, 309 [M+H]+; HRFAB MS m/z 309.2327 (calcd. for C21H29N2, 309.2331, ∆mmu 0.4).
2-(3,3-Dimethylprop-1-ene)-epicostaclavine (2) was obtained as a pale yellow oil: \( \left[ \alpha \right]_{D}^{25} \)-13 (c 0.34, MeOH); CD (MeOH) λ max (∆ε) 208 (−3.4), 224 (−8.9), 260 (+3.7), 293 (+2.0) nm; UV (MeOH) λ max (log ε) 212 (4.2), 226 (4.4), 281 (4.0) nm; IR (film) λ max 3319, 2975, 1674, 1464, 1202, 1189, 1139 cm−1; 1H and 13C NMR, see Table 1; FAB MS m/z 309 [M+H]+; HRFAB MS m/z 309.2328 (calcd. for C21H29N2, 309.2331, ∆mmu 0.4).
References
Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR (2009) Marine natural products. Nat Prod Rep 22:170–244
Li XF, Kim SK, Kang JS, Choi HD, Son BW (2006) Radical scavenging hydroxyphenyl ethanoic acid derivatives from a marine-derived fungus. J Microbiol Biotechnol 16:637–638
Abe M, Yamatodani S, Yamano T, Kusumoto M (1956) Isolation of two new water-soluble alkaloids, pyroclavine and costaclavine. Bull Agr Chem Soc Jpn 20:59–60
George B, Kin CL, Brunhilde K, James MT (1977) Four new mycotoxins of Aspergillus clavatus related to tryptoquivaline. J Org Chem 42(2):244–246
Atsushi N, Chika T, Tomochika M, Tamie M, Kenzo K, Yoshihide U, Eiko M, Masatoshi I, Hirofumi O, Tetsuro S (1992) Fumiquinazolines, novel metabolites of a fungus isolated from a saltfish. Tetrahedron Lett 33:1621–1624
Wang FZ, Fang YC, Zhu TJ, Zhang M, Lin AQ, Gu QQ, Zhu WM (2008) Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus Aspergillus fumigatus. Tetrahedron 64:7986–7991
Spilsbury JF, Wilkinson S (1961) The isolation of festuclavine and two new clavinet alkaloids from Aspergillus fumigatus. Fres J Chem Soc 2085–2091
Richard JC, Jerry WK, Joe WD, David MW, Joe CJJ, Johnson AN, David MB, James PS, Kuldip KC, Richard HC (1977) Mycotoxins produced by Aspergillus fumigatus species isolated from molded silage. J Agric Food Chem 25(4):826–830
Cui CB, Hideaki K, Hiroyuki O (1996) Novel mammalian cell cycle inhibitors, spirotryprostatins A and B, produced by Aspergillus fumigatus, which inhibit mammalian cell cycle at G2/M phase. Tetrahedron 52(39):12651–12666
Sakharovskii VG, Kozlovskii AG (1983) 1H NMR study of the structure of costaclavine and epicostaclavine. Zhurnal Strukturnoi Khimii 24:100–105
Norimasa S, Ryu-ichi F, Tamotsu M, Tetsuro M, Kiyotomo S, Ryozo S, Shinichi A (1992) Potentiation of the vincristine effect on P388 mouse leukemia cells by a newly synthesized dihydropyridine analogue, PAK-200. J Cancer Res Jpn 83:1011–1017
Acknowledgment
This work was financed by the Global COE Program for Chemistry Innovation, the University of Tokyo.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, D., Satake, M., Fukuzawa, S. et al. Two new indole alkaloids, 2-(3,3-dimethylprop-1-ene)-costaclavine and 2-(3,3-dimethylprop-1-ene)-epicostaclavine, from the marine-derived fungus Aspergillus fumigatus . J Nat Med 66, 222–226 (2012). https://doi.org/10.1007/s11418-011-0565-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11418-011-0565-3