Abstract
Purpose
The coastal tidal flat is one of the most dynamic environments in the biosphere and provides an important habitat for marine organisms. Pollution by excessive nutrients and potentially toxic elements (PTEs) can threaten microbial processes. The objective of this study was to investigate the effects of environmental properties on microbial diversity and functional profiling, and uncover the correlations between communities, nutrients, and PTEs, laying theoretical support for ecological studies of coastal tidal flat.
Materials and methods
Surface sediments from the typical coastal tidal flat zones (including Longshan Wharf (LS), Gaobeipu (GBP), and Shuiyunpu (SYP)) at the South Bank of Hangzhou Bay in Zhejiang Province, China, were investigated using Illumina sequencing and metagenome sequencing for the diversity and function of the microbial communities. Water quality and sediment monitoring were performed to assess the environment of the coastal tidal flat.
Results and discussion
The concentrations of water chemical oxygen demand (COD), available phosphorus (AP), dissolved inorganic nitrogen (DIN), and sediment Cr indicated that the coastal tidal flat was mainly polluted by nutrients and PTEs. The dominant microorganisms of the sediment samples were bacteria, of which 32 known phyla and 26 dominant classes were detected. Cr was positively correlated with Desulfuromonadia and Verrucomicrobiae, whereas it was negatively correlated with Gemmatimonadetes, Dadabacteriia, Gammaproteobacteria, and Dehalococcoidia, which showed opposite trends to Pb and AP. Metabolic functional genes were analyzed and differences in both N- and P-cycling processes were observed. Furthermore, multidrug resistance genes were the most abundant in sediment samples, whereas macB and tetA genes were more abundant than the whole antibiotic resistance genes. Network analysis revealed that Plactomycetia was the major bacterial host.
Conclusions
This study characterized the microbial diversity and functional profiling in the sediments of the coastal tidal flat, and revealed the effects of available nutrients and PTE pollution that may prompt the selection of dominant microbial members, which provides the theoretical support for future studies on monitoring and managing the coastal tidal flat ecosystems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The intertidal region, especially the coastal tidal flat, is an important area that consists of various types of ecosystems and provides an important habitat for marine organisms (Nordlund et al. 2014; Li et al. 2022). Coastal tidal flat has a variety of sediment types, complex hydrodynamic conditions, rich biological resources, and biodiversity (Boehm et al. 2014; Li et al. 2016), which is an active zone in the geochemical cycles of nutrients. However, the emergence of pollution sources such as land industry and agriculture, domestic wastewater, and pollutants has accelerated the degradation of tidal flat wetlands (Alahmed et al. 2022). It is necessary to conduct ecological studies on coastal tidal flats, which can provide theoretical support for the scientific utilization and high-quality development of coastal aquaculture.
Accumulation of nutrients has become one of the key environmental concerns in coastal tidal flats (Hu et al. 2014), as it has significant effects on microbial processes and directly or indirectly affects the biogeochemical cycle (Deng et al. 2020). The large consumption of dissolved oxygen stimulates the microbe-mediated degradation of organic matters, in which metabolites of ammonium and nitrite are biotoxic (Hu et al. 2014; Jin et al. 2017) and released gases of CH4 and N2O are also considered significant emission sources of the greenhouse effect (Groffman et al. 2000; Yang et al. 2015). During the sulfur cycle mediated by sulfate-reducing bacteria and hydrogen sulfide-oxidizing bacteria, the sediments are enriched with high concentrations of hydrogen sulfide, causing the mudflat mud to become black and smelly, which leads to the deterioration of water quality and affects the growth of cultured species (Yang et al. 2017). Furthermore, the excessive nutrition causes mass multiplication of pathogenic microorganisms (Defoirdt 2014), which leads to a decline in the productivity of aquaculture and further destroys the ecological balance of the coastal tidal flat (Li et al. 2017). Meanwhile, the biodeposits of aquatic products accelerate the sedimentation rate of nutrients in tidal flats, which promote the growth of benthic algae (Gibbs et al. 2004), further changing the microbial community (Jordan and Valiela 1982). Therefore, sediment microorganisms have a great metabolic potential for nutrient transformation and pollutant degradation in coastal tidal flats. Their community structure and metabolic activities can be used as important indicators for assessing nutrient dynamics in tidal flat ecosystems (Huang et al. 2016).
Pollution by potentially toxic elements (PTEs) is a key problem in aquatic environments (Capangpangan et al. 2016). Higher concentrations of metal contaminants adversely affect the composition of the microbial communities (Shi et al. 2018). PTEs change the microbial community structure by destroying cells to a certain extent (Yan et al. 2020), whereas microorganisms can convert PTEs into organometallic compounds, thereby enhancing their toxicity and increasing the risk of water pollution. Bacteroides had a significant negative correlation with Cd, Cu, and Zn in highly polluted sediments (Gillan et al. 2005), and Nitrospirae was significantly negatively affected by PTEs except Cr, while Cr was significantly positively correlated with the Planctomycoetes-Verrucomicrobia-Chlamydia super-phylum in nearshore sediments (Zhuang et al. 2019). Deltaproteobacteria, Acidobacteria, Gemmatimonadetes, and Nitrospira may be indicators of Zn and Cu pollution in the eutrophic water environments (Wang et al. 2019). With the enhancement of the tolerance of species to PTE pollution, the abundance of sensitive species susceptible to metal pollution gradually decreases, which then changes the microbial diversity (Tipayno et al. 2018). Hence, exploring the impact of PTE pollution on the microbial community can reflect the biological indicators of metal contaminant status, which is helpful for monitoring and managing coastal tidal flat ecosystems.
Antibiotic resistance is regarded as an increasingly global public health threat (Cassini et al. 2019), and emerging pollutants of antimicrobial resistance genes (ARGs) and resistant bacteria (ARB) persist in the dissemination of resistance in aquatic environments (Yang et al. 2018). The wide use of antibiotics means that the discharged pollutants circulate around the tidal flats, which may cause potential risks for long retention times to the ecosystem and cause pollution control in coastal tidal flat. Sediment is considered an important area for ARG accumulation and transmission (Kümmerer 2009), while ARB is repositories of ARGs in the sediments (Martínez 2008). Bacterial hosts acquire ARGs through integrons, which further accelerate the spread of ARGs in the environment (Zhang et al. 2020). Moreover, long-term nutrient input promoted the reproduction of ARGs (Zhao et al. 2017). Previous studies have indicated that carbon, nitrogen, and phosphorus affect the distribution and abundance of ARGs by changing the composition of the microbial communities (Zhou et al. 2017; Pan et al. 2020). Therefore, it is of great significance to study the relationships between ARGs, microbial communities, and environmental factors in the sediments of coastal tidal flats.
In the present study, using 16S rRNA sequencing and metagenome sequencing, three typical coastal tidal flat zones located at the South Bank of Hangzhou Bay, China, were chosen to investigate the community dynamics, potential function, and adaption mechanism of the microbiome in surface sediments. It is hypothesized that microbial composition and functional genes would respond to pollution of nutrient and PTEs in coastal tidal flat. The main objectives of this study were to (1) determine the distribution and diversity of the microbial community, (2) evaluate the impact of PTE contaminant on sediment microbial structures, and (3) examine the responses of functional genes with biogeochemical cycles to the accumulation of nutrients in coastal tidal flat.
2 Materials and methods
2.1 Site description and sample collection
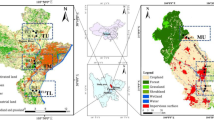
Cixi City is located at the South Bank of Hangzhou Bay in Zhejiang Province, China, between 30°2' and 30°19' north latitude and 121°2' and 121°36' east longitude. Its coastline is 78.5 km long and has the largest contiguous beach south of the Yangtze River. Using abundant beach resources to develop mariculture, Cixi City gradually formed a large-scale coastal aquaculture production base with regional characteristics. The Longshan Wharf (LS), Gaobeipu (GBP), and Shuiyunpu (SYP) are three typical offshore aquaculture zones. The LS is a mass fishing port wharf with a berthing grade of 100 tons. The SYP is one of the main flood gates in the Zhonghe District of the Yaojiang River Basin. Using a Van Veen grab sampler (35 cm × 25 cm × 30 cm), surface sediment samples (0–5 cm) were collected from three offshore sites in May 2021 (Fig. 1). Three parallel samples were collected and pooled into one sample mixing for each site to ensure that the sampling truly represented the coastal tidal flat as much as possible (Yuan et al. 2003; Shen et al. 2022). Each sample was then divided into two parts: one was refrigerated in a sterile tube at 4 °C for physicochemical parameter measurements, and the other was stored at − 80 °C until molecular analysis.
2.2 Physicochemical analyses
Water quality and sediment monitoring were performed at LS, GBP, and SYP sampling sites in the aquaculture zones. The pH, dissolved oxygen (DO), and chemical oxygen demand (COD) of the water were monitored in situ using a multi-parameter water quality analyzer (UPW-T700N, China). Available phosphorus (AP) was measured using a spectrophotometer (UV2550, Shimadzu, Japan). The dissolved inorganic nitrogen (DIN) was determined using a Discrete Chemistry Analyzer (CleverChem380, Dechem-Tech, Germany). The oil content was measured using a water–oil analyzer (InfraCal 2 ATR-SP, Spectro Scientific, USA). Sediments Cu, Pb, Cd, Zn, and Cr were analyzed using inductively coupled plasma mass spectrometry (ICP-MS, 7900, Agilent, USA) after digestion with a microwave digester (Ethos A, Milestone, Italy). Physicochemical characteristics of each sampling site are presented in Table 1.
2.3 DNA extraction
Genomic DNA was extracted using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Southern California, USA) from 0.5-g fresh sediment of each sample. The final DNA quality was confirmed by electrophoresis on 1% agarose gel. The DNA concentration and purification were determined using Qubit 3.0 (Thermo Fisher Scientific, Waltham, MA, USA). Three parallel samples of DNA were extracted from each site, manually pooled into one sample, and stored at − 80 °C.
Plasmid standards were constructed by inserting the bacterial 16S rRNA gene fragment amplified by PCR using the universal primers 341F/534R (Muyzer et al. 1993). qPCR was performed in duplicate using a Bio-Rad CFX384 Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). The gene copy numbers of the sediment samples were calculated from the cycle threshold (CT) value and are presented as copies per gram of wet sediment using the absolute standard curve. The average amplification efficiency of these genes was 87.5%.
2.4 16S rRNA sequencing and data analysis
The V3–V4 region of the bacterial 16S rRNA gene was amplified using the universal primer 338F/806R with adaptor sequences to link to the barcodes using a thermocycler PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA) (Xu et al. 2016). The amplification conditions included initial denaturation at 95 °C for 3 min, 25 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 min, and elongation at 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. Each sample was amplified in triplicate, containing 5 × FastPfu Buffer, 2.5 mM dNTPs, 50 mM of Mg2+, 0.1 µM of each primer, 1 unit of TransStart® FastPfu DNA Polymerase (TransGen, Beijing, China), and 1 ng of DNA template. The PCR products were analyzed by 2% agarose gel electrophoresis, purified using Agencourt AMPure XP (Beckman, USA), and quantified using Qubit 3.0 (Thermo Fisher Scientific, Waltham, MA, USA). After purification, three parallel amplicons from each site were manually mixed into a single DNA library. According to standard protocols (http://www.illumina.com/), sequencing was performed using the Illumina MiSeq PE300 platform (Illumina, San Diego, CA, USA).
The raw fastq sequencing data were quality-filtered, and the paired reads were merged into long sequences using qiime2 (http://qiime.org/scripts/assign_taxonomy.html). The optimized sequences were then compared with the Silva (Release138, http://www.arb-silva.de) 16S rRNA database to remove eukaryotic and archaeal sequences. The screened bacterial sequences were classified into operational taxonomic units (OTUs) with a 97% similarity cutoff. The sequencing depth for each sample was 30,000.
2.5 Metagenome sequencing and Gene Catalogue Construction
The extracted genomic DNA was sheared into approximately 300-bp fragments and connected with the Y-shaped linker. A PE library was constructed using PCR amplification and library enrichment. One segment of DNA fragment was fixed on the chip, and the other was randomly complementary of the primer, forming a “bridge.” Subsequently, DNA clusters were generated by bridge PCR amplification and linearized into single DNA strands. According to standard protocols (http://www.illumina.com/), sequencing was performed at Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China) using NovaSeq 6000 platform (Illumina, San Diego, CA, USA).
After the raw sequences obtained by sequencing were split, quality cut, and decontaminated, the optimized sequences were assembled, and genes were predicted by MetaGene (http://metagene.cb.k.u-tokyo.ac.jp/). Genes with nucleic acid length of ≥ 100 bp were selected and translated into amino acid sequences (Noguchi et al. 2006). The high-quality reads of each sample were compared with the non-redundant gene catalogue using SOAPaligner (Version 2.21), and the abundance information of predicted genes in the corresponding samples was obtained (http://soap.genomics.org.cn/). The resulting genes were annotated and classified by species and function using BLASTP (V2.2.2), including non-redundant (NR) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
2.6 Bioinformatics and statistical analysis
The sample alpha diversity index, including richness, evenness, diversity indices, and coverage, was determined using mothur software (version 1.30.2, https://www.mothur.org/wiki/Download_mothur). Principal coordinates analysis (PCoA) with Bray–Curtis distance was calculated to compare the similarities and differences in environmental samples. Redundancy analysis (RDA) with Monte Carlo permutation tests (999 unrestricted permutations, p < 0.05) was employed to analyze the relationships between the physicochemical parameters and microbial communities. Random forest analysis (RF) was conducted to evaluate the relative importance of each environmental factor in predicting the distribution of microbial communities. The contribution of environmental factors was assessed using “mean decrease accuracy” in the “randomForest” R package (Liaw and Wiener 2002). All analyses were visualized in R “vegan” package (Oksanen et al. 2019). Using SPSS 20.0 (IBM, Armonk, NY, USA), Pearson’s rank correlation and significance analyses were used to examine the potential relationships among diversity, community, and physicochemical parameters.
2.7 Accession numbers
The 16S rRNA gene sequences and metagenome sequences obtained from Illumina sequencing have been deposited in the public NCBI database (http://www.ncbi.nlm.nih.gov/) under the accession numbers PRJNA906945 and PRJNA908223.
3 Results
3.1 Physicochemical properties of the samples
At the time of sampling in May 2021, routine water quality and sediment monitoring were conducted for Longshan Wharf (LS), Gaobeipu (GBP) and Shuiyunpu (SYP) sampling sites in the offshore aquaculture zones (Table 1). No significant differences were noted among the three sampling sites in water pH (8.01, 7.92, and 7.91, respectively), dissolved oxygen (DO, 6.09, 6.80, and 7.73 mg L−1, respectively), inorganic nitrogen (DIN, 0.933, 1.118, and 0.832 mg L−1, respectively), and sediment Cu, Pb, Cd, Zn, and Cr (p > 0.05). However, the concentrations of chemical oxygen demand (COD), available phosphorus (AP), and oil in the samples were significantly different (p < 0.05). The concentrations of COD and oil were higher in LS and GBP than in SYP, whereas the concentrations of AP were highest in the LS sample. According to the Seawater Quality Standard of China (GB3097-1997), seawater quality is classified into four classes (I, II, III, and IV, corresponding to marine fishery water areas, aquaculture areas, industrial water areas, and marine port water areas, respectively). The pH and DO in the three sampling sites were classified as class I, whereas the concentrations of oil were classified as class I in SYP, and class II in LS and GBP. The concentrations of COD in LS and GBP (up to 1.042 times higher), AP in LS (up to 1.711 times higher), and DIN in three samples (up to 1.66 to 2.24 times higher) were considerably higher than the specified safety limits. According to the Marine Sediment Quality of China (GB18668-2002), marine sediment quality is classified into three classes (I, II, and III, corresponding to marine fishery water, industrial water, and marine port water areas, respectively). The concentrations of Cu, Pb, Cd, and Zn in the three sediment samples were classified as class I, while the concentration of Cr was classified as class II. Physicochemical characteristics indicated that GBP and LS were more hypernutrified and polluted than SYP.
3.2 Overview of 16S rRNA sequencing and metagenome sequencing
To investigate the diversity and function of the microbial community, 16S rRNA and metagenome sequencing were performed. After filtering and trimming the raw reads of 16S rRNA sequencing, 219,367 high-quality sequences were obtained from the three samples with an average length of 419.58 bp and were flattened by minimum sample sequence number (Table 2). Good’s coverage values ranged between 0.987 and 0.990 across samples, indicating that sequencing recovered most of the local species. A total of 770 bacterial OTUs in the sediments from the three sampling sites were classified with a threshold value of 97%.
Through metagenomic sequencing, 15.77 million clean reads (> 50 bp) were generated from the three samples (Table 3). After assembly, 1,404,224 contigs (> 300 bp) were obtained for prediction and annotation. The contigs_N50 and _N90 illustrated that the assembly efficiency was comparable with previous metagenome reports (Albertsen et al. 2012; Ye et al. 2012; Deng et al. 2020). A total of 565,459, 708,575, and 477,911 open reading frames (ORFs) (> 100 bp) were retrieved from GBP, LS, and SYP, respectively.
3.3 Abundance, diversity, and composition of the bacterial community in sediment samples
The copy number of the bacterial 16S rRNA gene ranged from 2.46 × 1010 to 6.05 × 1010 copies g−1 wet sediment in the three samples (Fig. S1). No significant spatial differences were noted in bacterial 16S rRNA gene abundance in the sediments of tidal flat aquaculture (p > 0.05). The richness and diversity indices based on the OTUs are shown in Table 2. The Ace and Chao1 indices of the three samples indicated that LS had the highest richness (Ace index of 3644 and Chao1 index of 3576), followed closely by SYP (3411 and 3400) and GBP (3321 and 3252). Similarly, the sediments of GBP had the lowest diversity, with a Shannon index of 6.38 and Simpson index of 0.00581, whereas the sediments of LS (6.64 and 0.00494) and SYP (6.62 and 0.00392) showed considerably greater diversity.
Circos showed the composition of the bacterial communities in the three sediment samples based on taxonomic assignment (Fig. 2). A total of 32 known bacterial phyla were detected in the samples, of which 17 were dominant phyla (relative abundance > 1%), including Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteriota, Bacteroidota, and Desulfobacterota (Fig. 2A). Proteobacteria was the predominant bacterial phylum in all samples, with a prevalence of 31.2%, 31.5%, and 28.7% in the GBP, LS, and SYP, respectively. The relative proportions of Bacteroidetes and Firmicutes, accounting for 10.8% and 3.90% in SYP, respectively, were the highest, followed closely by those in GBP and LS, whereas Actinobacteriota and Cyanobacteria in GBP were significantly higher than those in LS and SYP (p < 0.05).
Composition and abundance of the sediment samples from Longshan Wharf (LS), Gaobeipu (GBP), and Shuiyunpu (SYP) at different taxonomic levels. Circos visualizing the distribution of microbial community for each sample at phylum (A), class (B), and genus (C) levels. The width of the bars from each phylum indicates the relative abundance in the sample. Ternary plot representing the relative occurrence of individual OTU (D), class (E), and genus (F) that are members of the most abundant phyla in samples. Species enriched in different compartments are colored by taxonomy of the most abundant phyla. The size of the circles is proportional to the mean abundance in the community
At the class level, there were 26 dominant bacterial classes (relative abundance > 1%) in the three samples (Fig. 2B). Gammaproteobacteria was the most abundant class in all samples, representing 21.6%, 22.6%, and 15.2% of GBP, LS, and SYP, respectively. The second most dominant class varied among the three samples, Alphaproteobacteria (13.5%) in SYP and Acidimicrobiia belonging to Acidobacteriota in GBP (12.3%) and LS (9.55%). Cyanobacteria comprised a relatively higher proportion in GBP than in LS and SYP, whereas the relative abundance of Bacteroidia and Bacilli in SYP was three orders of magnitude higher than those in GBP and LS.
At the genus taxonomic level, a total of 30 dominant bacterial genera (relative abundance > 1%) were observed in sediments of which accounted for more than 51.8% of classified sequences (Fig. 2C). The predominant genera observed were Woeseia and Actinomarinales, which accounted for more than 12% of the classified sequences. The relative proportions of Woeseia and Actinomarinales in LS and GBP was significantly higher than in SYP (p < 0.05). Gaiella belonged to Actinobacteria, accounting for 3.95%, 3.04%, and 2.09% in GBP, LS, and SYP, respectively. Similar to Cyanobacteria, Chloroplast had a relatively higher proportion in GBP than in LS and SYP (p < 0.05).
3.4 Correlation between microbial community and environmental parameters
Correlations between the environmental parameters and the microbial communities were analyzed by redundancy analysis (RDA) (Fig. S2). Nutrients and PTEs were significantly influential factors in explaining the total variation (p < 0.001). The random forest analysis (RF) found that DIN was the most important predictor for microbial community, with COD, Pb, and oil also highly ranked (Fig. S3), indicating that compared with PTEs, nutrient was the most contribution of environmental factor to the variations of microbial communities in the coastal tidal flat.
The relationship between the microbial community and environmental factors was examined using a heatmap analysis (Fig. 3). At the phylum level, DIN and oil were positively correlated with Actinobacteria and Planctomycetota (p < 0.001), whereas they were negatively linked with Firmicutes, Bacteroidetes, and Desulfobacterota (p < 0.001). PTEs, including Zn, Cu, and Cd, positively contributed to Chloroflexi and Myxococcota (p < 0.001) (Fig. 3A). Interestingly, MBNT15, Latescibacterota, Dadabacteria, Gemmatimonadota, Proteobacteria, and Acidobacteria were significantly positively correlated with Pb, pH, and AP (p < 0.001), whereas these dominant phyla were strongly negatively correlated with Cr and DO (p < 0.001).
At the class level, DIN and oil contributed positively to Cyanobacteria, KD4-96, Thermoleophilia, Acidimicrobiia, and Vicinamibacteria (p < 0.001), whereas they contributed negatively to Desulfobulbia, Bacilli, Anaerolineae, Alphaproteobacteria, and Bacteroidia (p < 0.001) (Fig. 3B). DO and Cr were positively related to Desulfuromonadia and Verrucomicrobiae (p < 0.001), whereas they were negatively correlated to classes including Gemmatimonadetes, Subgroup_22, Subgroup_21, Dadabacteriia, Gammaproteobacteria, and Dehalococcoidia (p < 0.001), which showed opposite trends to the correlation between Pb, pH, and AP with those classes.
At the genus level, Woeseia, Subgroup_10, Dadabacteriales, and Anaerolineaceae were significantly positively correlated with Pb, pH, and AP (p < 0.001), but strongly negatively correlated with Cr and DO (p < 0.001) (Fig. 3C). DIN and oil contributed positively to Actinomarinales and Gaiella (p < 0.001), whereas they contributed negatively to Robiginitalea and Flavobacteriaceae (p < 0.001). Chloroplast was strongly negatively correlated with PTEs, including Zn, Cu, and Cd (p < 0.001).
3.5 Microbial processes in sediment samples
Based on the phylogenetic annotation of the predicted non-redundant ORFs, the overall taxonomic breakdown of the communities was assigned to bacteria, archaea, eukaryotes, viruses, and norank (Table 3). Domain bacteria dominated the genetic composition of the microbial communities, and matched metagenomic reads accounted for 96.34%, 96.54%, and 98.34% of all taxonomically assigned reads in GBP, LS, and SYP, respectively.
To determine the microbial contribution to specific functions, functional potentials were classified using Clusters of Orthologous Group (COG) analysis against the evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) databases (Fig. S4). The total number of predicted functional genes was significantly higher in GBP and LS than in SYP (p < 0.05). The functional genes in the three sediments were more closely associated with metabolism, such as “Amino acid transport and metabolism” (9.73%) and “Energy production and conversion” (8.94%). COG classification related to “Cytoskeleton” was less abundant in LS (p < 0.05). Among functions, Proteobacteria was the most contributing phylum (Fig. 4). Actinobacteria and Thaumarchaeota contributed more to GBP than LS and SYP, while Chloroflexi and Acidobacteria contributed more to LS and SYP than GBP. The contributions of [W] extracellular structures were the phyla Proteobacteria, Bacteroidetes, and Firmicutes, while a reduced contribution to [W] extracellular structures at the phylum level from GBP to SYP belonging to Bacteroidetes and Firmicutes was observed.
3.6 Functional genes involved in N- and P-cycling
Non-redundant genes focused on N- and P-cycling were searched against the Kyoto Encyclopedia of Genes and Genomes Orthologs (KO) databases. Seven and six key processes were involved in N- and P-cycling detected, respectively (Fig. S5). Genes nifD/H/K, nasA, nirA, nrtA/B/C, gltB/D, glnA, and ureB/C/A were more abundant in GBP than in LS and SYP, whereas norC, narG/H/I, napA, nrfA/H, narB, NRT, and gdhB in SYP were significantly higher than in GBP and LS (Fig. S5A). These results suggest that the processes of nitrogen fixation, assimilatory nitrate reduction (ANRA), nitrate assimilation, and organic N metabolism in GBP contributed more than those in LS and SYP, whereas denitrification and dissimilatory nitrate reduction to ammonia (DNRA) were the main N-cycling process in SYP. In the P-cycling processes, most genes related to P-cycling in SYP were distinct from those in GBP and LS (Fig. S5B). The relative abundance of regulatory and transporter genes in SYP was greater than that in GBP and LS. Moreover, the genes involved in organic P mineralization and polyphosphate degradation processes in GBP were more abundant than those in LS and SYP.
Heatmap analysis confirmed that functional genes, similar to microbial composition, are also affected by environmental properties (Fig. 5). The functional genes of nitrification, organic P mineralization, and polyphosphate degradation were positively correlated with COD, DIN, and oil contents in water. Some PTEs, such as Zn, Cu, and Cd, are related to denitrification, organic N metabolism, and polyphosphate synthesis. Whether N- or P-cycling, DO/Cr and Pb/Ph/AP showed opposite correlation trends with functional genes. Functional genes for dissimilatory nitrate reduction to ammonia (DNRA), regulatory, and transporters were positively correlated with DO and Cr, whereas functional genes for nitrification had positive correlations with Pb, pH, and AP. Therefore, the correlation analysis indicated the consistency of species and functions in coastal tidal flats.
3.7 Abundance and composition of antibiotic resistance genes in aquaculture-impacted tidal flat
To investigate the abundance and distribution of ARGs in tidal flat aquaculture, the top 20 ARGs categories and types were chosen by metagenome-assembly (Fig. 6). The highest abundance of multidrug resistance genes was detected in all sediment samples, followed by MLS, tetracycline, and glycopeptides (Fig. 6A). The distribution of ARGs is shown in Fig. 6B. The MacB and tetA genes were greater than the whole genes in the three sediment samples. Both msbA and novA gene abundance showed a similar pattern, which was higher in GBP than in LS and SYP. The tetA, oleC, and rpoB2 genes comprised a larger proportion of genes in GBP and SYP than in LS. Overall, ARG levels at different sample sites were not significantly different.
3.8 Co-occurrence of microbial community and antibiotic resistance genes in tidal flat sediments
The co-occurrence patterns between ARGs and microbial communities were analyzed using network analysis (Fig. 7). At the phylum level, most ARGs, including multidrugs, tetracycline, fluoroquinolone, and sulfonamide, were positively correlated with Bacteroidetes, and negatively correlated with Nitrospirae. Proteobacteria and Thaumarchaeota had a significant positive correlation with pleuromutilin and bicyclomycin, and showed a negative correlation with phenicol and aminoglycoside, which was opposite to Chloroflexi, Acidobacteria, and Candidatus_Eisenbacteria (Fig. 7A). Meanwhile, Euryarchaeota, Firmicutes, Planctomycetes, Gemmatimonadetes, and Cyanobacteria showed similar trends in the correlation of some ARGs, while Candidatus_Dadabacteria, Actinobacteria and Verrucomicrobia showed opposite trends in the correlation with these ARGs. At the class level, Plactomycetia displayed significant positive correlations with tetA, oleC, mtrA, macB, efrA, and arlR, suggesting that although Proteobacteria was the most abundant in the community composition, Plactomycetia was the major potential bacterial host (Fig. 7B). The bacterial hosts encoding cpxA, baeS, and bcrA were opposite to those encoding novA, msbA, and TaeA. At the genus level, the zinc tolerance gene baeS had a negative contribution pattern with Woeseia and Nitrosopumilus (Fig. 7C). Gemmatimonas and Enhygromyxa showed similar trends in the correlation of tetB and evgS. Interestingly, Nitrospira and Xanthomonadales showed opposite trends in the correlation with macB, tetA, oleC, mtrA, arlR, efrA, and MexW.
4 Discussion
4.1 Microbial community variation in response to nutrients availability
In this study, many common members of the sedimentary bacterial communities from the three typical coastal tidal flat zones were noted, which is consistent with previous studies (Omont et al. 2020). The dominant phyla in the coastal tidal flat were Proteobacteria, Actinobacteriota, Acidobacteriota, Chloroflexi, and Bacteroidetes. Previous studies have demonstrated that the dominance of Proteobacteria is involved in the availability of high nutrients, including the ability to degrade a large variety of organic compounds, fix atmospheric carbon, and transform nitrogen (Tomczyk-Żak and Zielenkiewicz 2015). Among Proteobacteria, Gammaproteobacteria was the most abundant and diverse class in the coastal tidal flat, which attested to the possibility of sulfidogenic contamination in the sediments of the tidal flat (Bowman et al. 2005). Alphaproteobacteria were related to photosynthesis, nitrogen fixation, ammonia oxidation, and methylotrophy (Williams et al. 2007; Dhami et al. 2018). High abundance of Alphaproteobacteria indicated that microorganisms in the coastal tidal flat depended on surface-derived carbon to maintain their growth. Compared with GBP and LS, Alphaproteobacteria had obvious advantages in SYP, suggesting that it had a high heterotrophic nitrogen fixation capacity in an environment with relatively low nutrient levels (Larouche et al. 2012).
Members of Actinobacteria have been reported to be involved in the decomposition of organic matter and carbon fixation (Gonzalez et al. 2019). Nitrogen metabolism is an important feature of the Actinobacteria, whose members participate in nitrogen fixation, dissimilatory nitrate reduction, and denitrification pathways (Zhang et al. 2019a). Actinomarinales was found to be relatively abundant in pesticide wastewater (Chen et al. 2022). In this study, a higher abundance of Actinobacteria in GBP and LS was observed than in SYP, which may be due to the accumulation of high inorganic nitrogen and oil in the former sediments. Members of Firmicutes have been reported to exist mainly in environments polluted by PTEs and organic compounds and participate in heterogenic degradation processes (Ettoumi et al. 2013; Lu et al. 2020). The relative abundances of Firmicutes and Bacilli in SYP were significantly higher than those in GBP and LS, implying high rates of accumulation and mineralization of organic matter in the sediment in SYP. Furthermore, the lower oil content in SYP confirms the decomposition role of Bacilli in PAH-contaminated sediments (Zhuang et al. 2002). Additionally, Bacteroidetes is considered to be an abundant phylum of sediments (Fan et al. 2018) and mainly plays a role in the degradation of organic particles, especially polysaccharides and proteins (Church 2008; Thomas et al. 2011). The reduction of oils and the increase in dissolved oxygen in SYP may be attributed to the higher abundance of Bacteroidetes. Cyanobacteria are mainly involved in the process of nitrogen fixation, nitrification, and nitrate assimilation in sediments, whose members are autotrophic by fixing nitrogen in the atmosphere and using inorganic nitrogen (ammonium, nitrate, and nitrite) (Zhao et al. 2016; Herrero and Flores 2019). The growth of Cyanobacteria in GBP was nearly 6–9 times higher than in LS and SYP, indicating nitrogen enrichment in GBP sediments. Thus, diversity studies have confirmed that the effects of available nutrients and PTEs may prompt the selection of dominant microbial members in the sediments of coastal tidal flats, which subsequently changes the diversity and composition of phylogenetic groups.
4.2 Nutrients, PTEs, and ARGs drive distribution of microbial communities
Sediments provide a series of niches for nutrient-cycling microbes, and the diversity and distribution of functional groups are the basis for the transformation of nutrient processes. The coastal tidal flat may have deposited amounts of nutrients due to the large amount of land industry and agriculture, domestic wastewater, and pollutant excretion, containing more than 80–88%, 52–95%, and 85% of residual carbon, nitrogen, and phosphorus, respectively (Xia et al. 2004; Yang et al. 2004). Based on physicochemical characteristics, the concentrations of COD, DIN, AP, and oil in the three sediments were clearly beyond the scope of class I water, and some exceeded class IV water, suggesting eutrophication of the coastal tidal flat in this study (Table 1). This was also consistent with the abundant metabolism of the functional potentials in the three tidal flat zones, such as [E] amino acids, [P] inorganic ions, and [I] lipid transport and metabolism (Fig. S4). Notably, abundant and diverse functional genes only reflect the resilience of microbial communities in the nutrient cycling process to environmental stresses, and transcriptomic or proteomic analyses are needed to demonstrate the expression of functional genes.
PTEs have attracted extensive attention owing to their toxicity, persistence, and bioaccumulation in ecosystems (Gan et al. 2017). The response of microbial communities to PTEs is mainly related to the toxicity of PTEs (Du et al. 2017). Previous studies comparing the toxicity of various metals to soil microbial activity and communities showed that Cr exhibited the strongest toxicity to microbes, followed by Pb, As, Co, Zn, Cd, and Cu (Wang et al. 2010). In this study, the concentrations of Cu, Pb, Cd, and Zn in the coastal tidal flat were uniformly classified as class I, whereas the concentration of Cr was classified as class II (Table 1), indicating that the coastal tidal flat was mainly polluted by Cr. This was in line with a previous study in which Zhejiang led to greatest discharge of Cr via wastewater discharge in nearshore sediments in China (Liu and Yu 2022). The toxicity of metals changes the microbial community structure through the interaction of metals with proteins and the inhibition of metabolic processes and decreases the diversity and activity of populations through their environmental stress (Gasic and Korban 2006; Khan et al. 2007). Cr was positively correlated with Desulfuromonadia and Verrucomicrobiae, whereas it inhibited the growth of Gemmatimonadetes, Subgroup_22, Subgroup_21, Dadabacteriia, Gammaproteobacteria, and Dehalococcoidia. In particular, Cr and Pb had opposite effects on microbial community composition. Desulfuromonadia and Verrucomicrobia play key roles in sulfur reduction and anaerobic fermentation of carbohydrates in sediments (Elshahed et al. 2007; Aoki et al. 2014). Gammaproteobacteria was the predominant bacterial class in the three sediments and was reported to be dominant in ecosystems with high nutrient pollution (Nogales et al. 2011; Custodio et al. 2022). Members of Gammaproteobacteria are important in the degradation of organic pollutants and the cycle of sulfur compounds (Quero et al. 2015). Recent surveys of 16S rRNA gene sequences have revealed that Woeseiales is a globally prominent group of bacteria in coastal sediments (Dyksma et al. 2016). Members of Woeseiales have the potential to facultative chemolithoautotrophy driven by the oxidation of hydrogen and inorganic sulfur compounds (Baker et al. 2015; Mußmann et al. 2017; Hoffmann et al. 2020). Dehalococodia is the only class of Chloroflexi known to participate in the degradation of organic halides. Previous studies have found that microorganisms may generate resistance and enable them to survive by increasing the transport and excretion of Cr ions (Li et al. 2020), and the activity of coenzyme F420, which is involved in methanogenesis, is affected by Cr concentration during anaerobic fermentation (Zhang et al. 2019b). Microbially mediated sulfate and metal reductions are the main drivers of the nutrient cycle in anaerobic environments (Hori et al. 2015; Otwell et al. 2016). Some functional microorganisms involved in sulfate and PTE reduction can reduce Cr (VI) to its less toxic and soluble form (Cr (III)) (Somenahally et al. 2013; Yu et al. 2016). The significant correlation between Cr and the dominant classes further indicates that these sediment microbial communities may have the ability to reduce Cr (VI).
Antibiotic analysis showed high abundances of multidrug, MLS, tetracycline, and glycopeptide in the sediment samples. MacB and tetA showed a similar tendency for MLS and tetracycline residues, suggesting that antibiotics may impose selective pressure on ARGs (Guan et al. 2019). Network analysis showed that the concentrations of multidrugs, MLS, and tetracycline in sediment samples were positively correlated with the abundance of Bacteroides, and negatively correlated with Nitrospirae. Actinobacteria and Verrucomicrobia were positively correlated with glycopeptides and aminocoumarins. The different responses of dominant microbes to antibiotic contamination may be attributed to the selective pressure of antibiotics on bacterial communities. Previous studies have shown that ARG transfer between bacterial species mainly occurs among Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria (Hu et al. 2016). Similarly, the relative abundance of tetA, oleC, mtrA, macB, efrA, and arlR in this study displayed significant positive correlations with Planctomycetia and negative correlations with Bacilli and Nitrospira, and novA, msbA, and TaeA were significantly positively correlated with Gammaproteobacteria and Alphaproteobacteria. Furthermore, ARGs are generally transferred and disseminated through mobile genetic elements (Gillings et al. 2015). However, the intI1 gene was not detected in the coastal tidal flat. This result implies that the integron was not the main mechanism of ARG propagation in the tidal flat (Zhao et al. 2017) and further research on the relationship between horizontal gene transfer vehicles and ARGs is needed.
5 Conclusions
Our results demonstrated that the long-term accumulation of nutrients and PTE contaminants could jointly contribute to the microbial communities in sediments impacted by the coastal tidal flat. The effects of available nutrients and PTEs may prompt the selection of dominant microbial members in the sediments of tidal flat zones, subsequently changing the diversity and composition of the phylogenetic groups. The abundance and diversity of functional genes reflect the resilience of microbial communities during the nutrient cycling process with environmental stresses. Furthermore, the emergence and dissemination of ARGs have been detected in sediment. However, because the integron was not the main mechanism of ARG propagation, aquaculture activities were less likely to cause serious ARG contamination in the tidal flat under the current conditions.
References
Alahmed S, Ross L, Smith SMC (2022) Coastal hydrodynamics and timescales in meso-macrotidal estuaries in the Gulf of Maine: a model study. Estuar Coast 45:1888–1908. https://doi.org/10.1007/s12237-022-01067-9
Albertsen M, Hansen L, Saunders AM, Nielsen PH, Nielsen KL (2012) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6(6):1094–1106. https://doi.org/10.1038/ismej.2011.176
Aoki M, Ehara M, Saito Y, Yoshioka H, MiyazakM I, Saito Y, Miyashita A, Kawakami S, Yamaguchi T, Ohashi A, Nunoura T, Takai K, Imachi H (2014) A long-term cultivation of an anaerobic methane-oxidizing microbial community from deep-sea methane-seep sediment using a continuous-flow bioreactor. PLoS One 9:e105356. https://doi.org/10.1371/journal.pone.0105356
Baker BJ, Lazar CS, Teske AP, Dick GJ (2015) Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3:14. https://doi.org/10.1186/s40168-015-0077-6
Boehm AB, Yamahara KM, Sassoubre LM (2014) Diversity and transport of microorganisms in intertidal sands of the California coast. Appl Environ Microbiol 80:3943–3951. https://doi.org/10.1128/AEM.00513-14
Bowman JP, Mccammon SA, Dann AL (2005) Biogeographic and quantitative analyses of abundant uncultivated gamma-proteobacterial clades from marine sediment. Microb Ecol 49:451–460. https://doi.org/10.1007/s00248-004-0070-2
Capangpangan RY, Pagapong NK, Pineda CP, Sanchez PB (2016) Evaluation of potential ecological risk and contamination assessment of heavy metals in sediment samples using different environmental quality indices–a case study in Agusan River. Caraga Philippines J Bio Env Sci 8(1):1–16
Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A et al (2019) Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19(1):56–66. https://doi.org/10.1016/S1473-3099(18)30605-4
Chen W, Wei J, Su Z, Wu L, Liu M, Huang X, Yao P, Wen D (2022) Deterministic mechanisms drive bacterial communities assembly in industrial wastewater treatment system. Environment International 168:107486. https://doi.org/10.1016/j.envint.2022.107486
Church MJ (2008) Resource control of bacterial dynamics in the sea. In: Kirchman DL (ed) Microbial ecology of the oceans, 2nd edn. Wiley, Hoboken, pp 335–382. https://doi.org/10.1002/9780470281840.ch10
Custodio M, Espinoza C, Peñaloza R et al (2022) Microbial diversity in intensively farmed lake sediment contaminated by heavy metals and identification of microbial taxa bioindicators of environmental quality. Sci Rep 12:80. https://doi.org/10.1038/s41598-021-03949-7
Defoirdt T (2014) Virulence mechanisms of bacterial aquaculture pathogens and antivirulence therapy for aquaculture. Rev Aquacult 6(2):100–114. https://doi.org/10.1111/raq.12030
Deng M, Hou J, Song K, Chen JY, Gou JW, Li DP, He XG (2020) Community metagenomic assembly reveals microbes that contribute to the vertical stratification of nitrogen cycling in an aquaculture pond. Aquaculture 520:734911. https://doi.org/10.1016/j.aquaculture.2019.734911
Dhami NK, Mukherjee A, Watkin ELJ (2018) Microbial diversity and mineralogical-mechanical properties of calcitic cave speleothems in natural and in vitro biomineralization conditions. Front Microbiol 9:40. https://doi.org/10.3389/fmicb.2018.00040
Du H, Harata N, Li F (2017) Responses of riverbed sediment bacteria to heavy metals: integrated evaluation based on bacterial density, activity and community structure under well-controlled sequencing batch incubation conditions. Water Res 130(1):115–126. https://doi.org/10.1016/j.watres.2017.10.070
Dyksma S, Bischof K, Fuchs BM, Hoffmann K, Meier D, Meyerdierks A et al (2016) Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J 10:1939–1953
Elshahed MS, Youssef NH, Luo Q, Najar FZ, Roe BA, Sisk TM, Buhring SI, Hinrichs KU, Krumholz LR (2007) Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich zodletone spring, Oklahoma. Appl Environ Microbiol 73:4707–4716. https://doi.org/10.1128/AEM.00591-07
Ettoumi B, Guesmi A, Brusetti L, Borin S, Najjari A, Boudabous A, Cherif A (2013) Microdiversity of deep-sea Bacillales isolated from Tyrrhenian ssediments as revealed by ARISA, 16S rRNA gene sequencing and BOX-PCR fingerprinting. Microbes Environ 28:361–369. https://doi.org/10.1264/jsme2.me13013
Fan L, Wang Z, Chen M, Qu Y, Li J, Zhou A et al (2018) Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci Total Environ 657(20):1194–1204. https://doi.org/10.1016/j.scitotenv.2018.12.069
Gan Y, Wang L, Yang G, Dai J, Wang R, Wang W (2017) Multiple factors impact the contents of heavy metals in vegetables in high natural background area of China. Chemosphere 184:1388–1395. https://doi.org/10.1016/j.chemosphere.2017.06.072
Gasic K, Korban SS (2006) Heavy metal stress. In: Rao KVM, Raghavendra AS, Reddy KJ (ed) Physiology and molecular biology of stress tolerance in plants, 1st edn. Springer, Dordrecht, pp 219–254. https://doi.org/10.1007/1-4020-4225-6_8
Gibbs M, Funnell G, Pickmere S, Norkko A, Hewitt J (2004) Benthic nutrient fluxes along an estuarine gradient: influence of the pinnid bivalve Atrina zelandica in summer. Mar Ecol Prog Ser 288:151–164. https://doi.org/10.3354/meps288151
Gillan DC, Danis B, Pernet P, Joly G, Dubois P (2005) Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl Environ Microbiol 71:679–690. https://doi.org/10.1128/AEM.71.2.679-690.2005
Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu YG (2015) Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J 9:1269–1279. https://doi.org/10.1038/ismej.2014.226
Gonzalez SV, Johnston E, Gribben PE, Dafforn K (2019) The application of bioturbators for aquatic bioremediation: review and meta-analysis. Environ Pollut 250:426–436. https://doi.org/10.1016/j.envpol.2019.04.023
Groffman PM, Gold AJ, Addy K (2000) Nitrous oxide production in riparian zones and its importance to national emission inventories. Chemosphere - Global Change Science 2(3–4):291–299. https://doi.org/10.1016/S1465-9972(00)00018-0
Guan YJ, Jia J, Wu L, Xue X, Zhang G, Wang ZZ (2019) Analysis of bacterial community characteristics, abundance of antibiotics and antibiotic resistance genes along a pollution gradient of Ba River in Xi’an. China Front Microbiol 9:3191. https://doi.org/10.3389/fmicb.2018.03191
Herrero A, Flores E (2019) Genetic responses to carbon and nitrogen availability in Anabaena. Environ Microbiol 21:1–17. https://doi.org/10.1111/1462-2920.14370
Hoffman K, Bienhold C, Buttigieg PL et al (2020) Diversity and metabolism of Woeseiales bacteria, global members of marine sediment communities. ISME J 14:1042–1056. https://doi.org/10.1038/s41396-020-0588-4
Hori T, Aoyagi T, Itoh H, Narihiro T, Oikawa A, Suzuki K et al (2015) Isolation of microorganisms involved in reduction of crystalline iron(III) oxides in natural environments. Front Microbiol 6:386. https://doi.org/10.3389/fmicb.2015.00386
Hu YF, Yang X, Li J, Lv N, Liu F, Wu J et al (2016) The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl Environ Microbiol 82:6672–6681. https://doi.org/10.1128/AEM.01802-16
Hu Z, Lee JW, Chandran K, Kim S, Sharma K, Khanal SK (2014) Influence of carbohydrate addition on nitrogen transformations and greenhouse gas emissions of intensive aquaculture system. Sci Total Environ 470–471(1):193–200. https://doi.org/10.1016/j.scitotenv.2013.09.050
Huang D, Xue W, Zeng G, Wan J, Chen G, Huang C et al (2016) Immobilization of cd in river sediments by sodium alginate modified nanoscale zero-valent iron: impact on enzyme activities and microbial community diversity. Water Res 106(1):15–25. https://doi.org/10.1016/j.watres.2016.09.050
Jin J, Wang Y, Wu Z, Hergazy A, Lan J, Zhao L et al (2017) Transcriptomic analysis of liver from grass carp (Ctenopharyngodon idellus) exposed to high environmental ammonia reveals the activation of antioxidant and apoptosis pathways. Fish Shellfish Immun 63:444–451. https://doi.org/10.1016/j.fsi.2017.02.037
Jordan TE, Valiela I (1982) A nitrogen budget of the ribbed mussel, Geukensia demissa, and its significance in nitrogen flow in a New England Salt Marsh. Limnol Oceanogr 27(1):75–90. https://doi.org/10.4319/lo.1982.27.1.0075
Khan S, Cao Q, El-Latif Hesham A, Xia Y, He JZ (2007) Soil enzymatic activities and microbial community structure with different application rates of Cd and Pb. J Environ Sci 19(7):834–840. https://doi.org/10.1016/S1001-0742(07)60139-9
Kümmerer K (2009) Antibiotics in the aquatic environment–a review–part I. Chemosphere 75:417–434. https://doi.org/10.1016/j.chemosphere.2008.11.086
Larouche JR, Bowden WB, Giordano R, Flinn MB, Crump BC (2012) Microbial biogeography of arctic streams: exploring influences of lithology and habitat. Front Microbiol 3:309. https://doi.org/10.3389/fmicb.2012.00309
Li CC, Quan Q, Gan YD, Dong JY, Fang JH, Wang LF, Liu J (2020) Effects of heavy metals on microbial communities in sediments and establishment of bioindicators based on microbial taxa and function for environmental monitoring and management. Sci Total Environ 749:141555. https://doi.org/10.1016/j.scitotenv.2020.141555
Li H, Li X, Qiang L, Ying L, Song J, Zhang Y (2017) Environmental response to long-term mariculture activities in the Weihai coastal area. China Sci Total Environ 601(1):22–31. https://doi.org/10.1016/j.scitotenv.2017.05.167
Li W, Wang MM, Bian XM, Guo JJ, Cai L (2016) A high-level fungal diversity in the intertidal sediment of Chinese seas presents the spatial variation of community composition. Front Microbiol 7(5995):2098. https://doi.org/10.3389/fmicb.2016.02098
Li YX, Huang RQ, Hu LL, Zhang CF, Xu XR, Song L, Wang ZY, Pan XL, Christakos G, Wu JP (2022) Microplastics distribution in different habitats of Ximen Island and the trapping effect of blue carbon habitats on microplastics. Marine Pollut Bull 181:113912. https://doi.org/10.1016/j.marpolbul.2022.113912
Liaw A, Wiener M (2002) Classification and regression by random forest. R News 2:18–22
Liu X, Yu S (2022) Anthropogenic metal loads in nearshore sediment along the coast of China mainland interacting with provincial socioeconomics in the period 1980–2020. Sci Total Environ 839:156286. https://doi.org/10.1016/j.scitotenv.2022.156286
Lu SD, Sun YJ, Lu BY, Zheng DY, Xu SW (2020) Change of abundance and correlation of Nitrospira inopinata-like comammox and populations in nitrogen cycle during different seasons. Chemosphere 241:125098. https://doi.org/10.1016/j.chemosphere.2019.125098
Martínez JL (2008) Antibiotics and antibiotic resistance genes in natural environments. Science 321(5887):365–367. https://doi.org/10.1126/science.1159483
Mußmann M, Pjevac P, Krüger K, Dyksma S (2017) Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J 11:1276–1281. https://doi.org/10.1038/ismej.2016.185
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59(3):695–700. https://doi.org/10.0000/PMID7683183
Nogales B, Lanfranconi MP, Piña-Villalonga JM, Bosch R (2011) Anthropogenic perturbations in marine microbial communities. FEMS Microbiol Rev 35:275–298. https://doi.org/10.1111/j.1574-6976.2010.00248.x
Noguchi H, Park J, Takagi T (2006) MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34(19):5623–5630. https://doi.org/10.1093/nar/gkl723
Nordlund LM, de la Torre-Castro M, Erlandsson J, Conand C, Muthiga N, Jiddawi N et al (2014) Intertidal zone management in the Western Indian Ocean: assessing current status and future possibilities using expert opinions. Ambio 43:1006–1019. https://doi.org/10.1007/s13280-013-0465-8
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al (2019) Vegan: community ecology package. R package version 2:5–6. https://cran.r-project.org/web/packages/vegan
Omont A, Elizondo-González R, Quiroz-Guzmán E, Escobedo-Fregoso C, Hernández-Herrera R, Peña-Rodríguez A (2020) Digestive microbiota of shrimp Penaeusvannamei and oyster Crassostreagigas co-cultured in integrated multi-trophic aquaculture system. Aquaculture 521:735059 https://doi.org/10.1016/j.aquaculture.2020.735059
Otwell AE, Callister SJ, Zink EM, Smith RD, Richardson RE (2016) Comparative proteomic analysis of Desulfotomaculum reducens MI-1: insights into the metabolic versatility of a gram-positive sulfate-and metal-reducing bacterium. Front Microbiol 7:191. https://doi.org/10.3389/fmicb.2016.00191
Pan X, Lin L, Zhang W, Dong L, Yang Y (2020) Metagenome sequencing to unveil the resistome in a deep subtropical lake on the Yunnan-Guizhou Plateau, China. Environ Pollut 263:114470. https://doi.org/10.1016/j.envpol.2020.114470
Quero GM, Cassin D, Botter M, Perini L, Luna GM (2015) Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Front Microbiol 6:1–15. https://doi.org/10.3389/fmicb.2015.01053
Shen J, Luo YL, White PJ, Sun G, Li M et al (2022) The exacerbation of soil acidification correlates with structural and functional succession of the soil microbiome upon agricultural intensification. Sci Total Environ 828:154524. https://doi.org/10.1016/j.scitotenv.2022.154524
Shi J, Li X, He T, Wang J, Wang Z, Li P, Lai Y, Sanganyado E, Liu W (2018) Integrated assessment of heavy metal pollution using transplanted mussels in eastern Guangdong, China. Environ Pollut 243:601–609. https://doi.org/10.1016/j.envpol.2018.09.006
Somenahally AC, Mosher JJ, Yuan T, Podar M, Phelps TJ, Brown SD, Yang ZK, Hazen TC, Arkin AP, Palumbo AV (2013) Hexavalent chromium reduction under fermentative conditions with lactate stimulated native microbial communities. PloS One 8(12):e83909. https://doi.org/10.1371/journal.pone.0083909
Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G (2011) Environmental and gut Bacteroidetes: the food connection. Front Microbio 2:93. https://doi.org/10.3389/fmicb.2011.00093
Tipayno SC, Truu J, Samaddar S, Truu M, Preem JK, Oopkaup K, Espenberg M, Chatterjee P, Kang Y, Kim K, Sa T (2018) The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol Evol 8:6157–6168. https://doi.org/10.1002/ece3.4170
Tomczyk-Żak K, Zielenkiewicz U (2015) Microbial diversity in caves. Geomicrobiol J 33:20–38. https://doi.org/10.1080/01490451.2014.1003341
Wang F, Yao J, Si Y, Chen H, Russel M, Chen K, Qian Y, Zaray G, Bramanti E (2010) Short-time effect of heavy metals upon microbial community activity. J Hazard Mater 173:510–516. https://doi.org/10.1016/j.jhazmat.2009.08.114
Wang J, Yuan S, Tang L, Pan X, Shen C (2019) Contribution of heavy metal in driving microbial distribution in a eutrophic river. Sci Total Environ 712:136295. https://doi.org/10.1016/j.scitotenv.2019.136295
Williams KP, Sobral BW, Dickerman AW (2007) A robust species tree for the alphaproteobacteria. J Bacteriol 189(13):4578–4586. https://doi.org/10.1128/JB.00269-07
Xia LZ, Yang LZ, Yan MC (2004) Nitrogen and phosphorus cycling in shrimp ponds and the measures for sustainable management. Environ Geochem Health 26:245–251. https://doi.org/10.1023/B:EGAH.0000039587.64830.43
Xu N, Tan G, Wang H, Gai X (2016) Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur J Soil Biol 74:1–8. https://doi.org/10.1016/j.ejsobi.2016.02.004
Yan C, Wang F, H Geng, Liu H, Pu S, Tian Z, Chen H, B Zhou, Yuan R, Yao J (2020) Integrating high-throughput sequencing and metagenome analysis to reveal the characteristic and resistance mechanism of microbial community in metal contaminated sediments. Sci Total Environ 707:136116, https://doi.org/10.1016/j.scitotenv.2019.136116
Yang P, He Q, Huang J, Tong C (2015) Fluxes of greenhouse gases at two different aquaculture ponds in the coastal zone of southeastern China. Atmos Environ 115:269–277. https://doi.org/10.1016/j.atmosenv.2015.05.067
Yang P, Lai DYF, Jin BS, Bastviken D, Tan LS, Tong C (2017) Dynamics of dissolved nutrients in the aquaculture shrimp ponds of the min river estuary, China: concentrations, fluxes and environmental loads. Sci Total Environ 603–604:256–267. https://doi.org/10.1016/j.scitotenv.2017.06.074
Yang YF, Li CH, Nie XP, Tang DL, Chung IK (2004) Development of mariculture and its impacts in Chinese coastal waters. Rev Fish Biol Fisher 14:1–10. https://doi.org/10.1007/s11160-004-3539-7
Yang YY, Song WJ, Lin H, Wang WB, Du LN, Xing W (2018) Antibiotics and antibiotic resistance genes in global lakes: a review and meta-analysis. Environ Int 116:60–73. https://doi.org/10.1016/j.envint.2018.04.011
Ye L, Zhang T, Wang T, Fang Z (2012) Microbial structures, functions, and metabolic pathways in wastewater treatment bioreactors revealed using high-throughput sequencing. Environ Sci Technol 46:13244–13252. https://doi.org/10.1021/es303454k
Yu Z, He Z, Tao X et al (2016) The shifts of sediment microbial community phylogenetic and functional structures during chromium (VI) reduction. Ecotoxicology 25:1759–1770. https://doi.org/10.1007/s10646-016-1719-6
Yuan SY, Liu C, Liao CS, Chang BV (2003) Occurrence and microbial degradation of phthalate esters in Taiwan river sediment. Chemosphere 49(10):1295–1299. https://doi.org/10.1016/S0045-6535(02)00495-2
Zhang B, Wu X, Tai X, Sun L, Wu M, Zhang W, Chen X, Zhang G, Chen T, Liu G, Dyson P (2019a) Variation in actinobacterial community composition and potential function in different soil ecosystems belonging to the Arid Heihe River Basin of Northwest China. Front Microbiol 10:2209. https://doi.org/10.3389/fmicb.2019.02209
Zhang H, Han X, Tian Y, Xu Y, Li Y, Chai Y, Xu X, Sanganyado E (2019b) Relationship analysis of anaerobic fermentation parameters exposed to elevated chromium (VI). Environ Prog Sustain Energy 13212. https://doi.org/10.1002/ep.13212
Zhang JY, Buhe CL, Yu DW, Zhong H, Wei YS (2020) Ammonia stress reduces antibiotic efflux but enriches horizontal gene transfer of antibiotic resistance genes in anaerobic digestion. Bioresour Technol 295:122191. https://doi.org/10.1016/j.biortech.2019.122191
Zhao DY, Shen F, Zeng J, Huang R, Yu ZB, Wu QLL (2016) Network analysis reveals seasonal variation of co-occurrence correlations between Cyanobacteria and other bacterioplankton. Sci Total Environ 573:817–825. https://doi.org/10.1016/j.scitotenv.2016.08.150
Zhao Z, Wang J, Chen J, Liu G, Lu H et al (2017) Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ Pollut 220:909–918. https://doi.org/10.1016/j.envpol.2016.10.075
Zhou ZC, Zheng J, Wei YY, Chen T, Dahlgren RA, Shang X, Chen H (2017) Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters. Environ Sci Pollut Res 24:23753–23762. https://doi.org/10.1007/s11356-017-0032-0
Zhuang M, Sanganyado E, Li P, Liu W (2019) Distribution of microbial communities in metal-contaminated nearshore sediment from eastern Guangdong, China. Environ Pollut 250:482–492. https://doi.org/10.1016/j.envpol.2019.04.041
Zhuang WQ, Maszenan AM, Tay S (2002) Bacillus naphthovorans sp. nov. from oil-contaminated tropical marine sediments and its role in naphthalene biodegradation. Appl Microbiol Biot 58(4):547–553. https://doi.org/10.1007/s00253-001-0909-0
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 31971523) and National Key Research and Development Project by MOST of China (grant numbers 2016YFA0601001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible editor: Elena Romano
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yue, Y., Rong, H., Yang, Z. et al. Microbial diversity and functional profiling in coastal tidal flat sediment with pollution of nutrients and potentially toxic elements. J Soils Sediments 23, 2935–2950 (2023). https://doi.org/10.1007/s11368-023-03511-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-023-03511-0