Abstract
Intracellular phosphorus (P) accumulation can improve microalgal growth and lipid synthesis. However, large excess of P causes cell poisoning. This study utilized a P-fed-batch strategy to investigate its potential to improve the utilization of the excessive P, while avoiding toxic side effects. This strategy contributed to a more complete utilization of the intracellularly stored P, which enhanced the microalgae biomass by 10−15% by upregulating the brassinosteroid growth hormone gene at a P-fed-batch frequency of 2−8. Furthermore, the lipid content increased by 4−16% via upregulation of lipid synthesis-related genes. As a result, the P-fed-batch strategy significantly increased the lipid production by 13−19%. The content of saturated fatty acid increased by ~ 100%, implying improved combustibility and oxidative stability. This is the first study of this P-fed-batch strategy and provides a new concept for the complete utilization of excessive P.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microalgae are a large and heterogeneous group of microscopic algae and provide an almost untapped pool of metabolic versatility (Bumbak et al. 2011). In recent years, significant efforts focused on microalgae heterotrophic cultivation for the enhancement of valuable products, including biofuels, pigments, antioxidants, and pharmaceuticals (Perez-Garcia et al. 2011; Sureshkumar and Thomas 2019). Heterotrophic cultivation achieved 100−400% higher microalgal biomass than photoautotrophic cultivation (Basri and Maznah 2017; Kamalanathan et al. 2018). During the heterotrophic growth of microalgae, the activity of hup 1, which is related to the hexose/H+ symport system, could be stimulated to promote cell synthesis (Basri and Maznah 2017). In addition, NADPH and ATP productions showed increased capability to improve metabolic performance (Kamalanathan et al. 2018; Li et al. 2014).
However, the heterotrophic growth of microalgae usually exerts negative impacts on lipid synthesis and causes a drop of intracellular lipid content (Kamalanathan et al. 2018; Leyva and Bashan 2015; Yu et al. 2012). Nitrogen deprivation or limitation stress is commonly employed to enhance the lipid content during heterotrophic cultivation of microalgae, with an increase of as much as 15−200% (Fu et al. 2017; Shen et al. 2015). Nitrogen stress has been reported to upregulate the malic enzyme level for NADH generation, which then stimulated pyruvate and precursor-acetyl-CoA for lipid synthesis (Guarnieri et al. 2013; Li et al. 2013; Zhang et al. 2007). Unfortunately, nitrogen deprivation or limitation stress results in a scarification of microalgal biomass (Shen et al. 2015; Zhu et al. 2014), and the total lipid production still did not achieve a satisfactory improvement.
In such a situation, sufficient phosphorous (P) supply has been suggested to combat nitrogen stress. The biomass increased by 60−95% with P supplementation, instead of decreasing by 50.5% under sole nitrogen deprivation (Fu et al. 2017). Excessive P was absorbed by microalgae, which then led to the formation of polyphosphates (poly-P) in cells, enriching phosphoanhydride bonds and providing surplus energy for cell growth and lipid synthesis (Fu et al. 2017; Meza et al. 2015). P abundance (25 mg/L) also improved the cellular anabolism by upregulating the genes for growth hormones.
However, the effects of P supplementation become negative and could even become toxic for cells if P supplementation becomes excessive (Li et al. 2018; Wang et al. 2018). Consequently, the excessively accumulated P bonds to intracellular components, thus damaging organelles, which leads to a 40% decrease in cell growth and a 70% loss of cell viability (Li et al. 2018). Interestingly, these phenomena were matched by the cytotoxicity of highly concentrated organic carbon substrates during the cultivation of microalgae (Bumbak et al. 2011; Schmidt et al. 2005). High levels of organic compounds (e.g., 20−100 g/L glucose) significantly inhibited microalgal growth by 20−50% (Sansawa and Endo 2004), and higher levels even caused complete inhibition of microalgal growth (Bumbak et al. 2011; Xiong et al. 2008). To overcome such cytotoxicity, the fed-batch culture mode, where food is offered several times in batches for cultivation, was adopted to promote the electron transfer rate and increase the biomass 3−10 fold. Thus, the balance between nutrient requirement and supply could be improved (Cristobal Garcia-Canedo et al. 2016). This study proposes that P-fed-batch not only might avoid the toxicity of overly excessive P but also could further improve P utilization to benefit microalgal growth and lipid production.
P intracellular accumulation improved microalgal growth and lipid synthesis; however, a large excess of P is toxic for microalgal cells. The ideal situation is to sufficiently utilize P as an energy source, while avoiding nutrient imbalance, to achieve enhanced biomass synthesis and lipid production. Here, a fed-batch strategy was used to explore the use of excessive P for microalgal energy conversion. This study verified the advantages of this P-fed-batch strategy to enhance microalgal lipid production. Poly-P characterization and real-time polymerase chain reaction technology were employed to identify the mechanisms underlying the P-fed-batch strategy on microalgal growth and lipid synthesis.
Materials and methods
Microalgae
Chlorella regularis var. minima (C. regularis, FACHB-729) was utilized in this study. The microalgae seeds were preserved in Wuhan, China, and were purchased from the Freshwater Algae Culture Collection of the Institute of Hydrobiology.
Purification and cultivation
C. regularis was isolated and purified via the spread plate method to remove bacterial and fungal contamination; then, the BG11 medium was inoculated with purified C. regularis for photoautotrophic cultivation in an incubator (ZWYR-2102C, Zhicheng, Shanghai, China). The culture temperature was 25 ± 1 °C, at a stirring rate of 160 rpm, and a photoperiodicity of 14:10 (light for 14 h and dark for 10 h). On the 6th day, the microalgal cells grew to the logarithmic phase and were inoculated into a heterotrophic medium. The heterotrophic medium included 10-g/L glucose as source of organic carbon, and the other composition was identical to that of the BG11 medium. When C. regularis reached to logarithmic phase again, cells were collected via centrifugation (6000×g, 5 min at 4 °C) for the experiments in this study.
Experimental design
The effects of a P-fed-batch strategy on C. regularis were studied in the modified BG11 medium. The initial nitrogen (NO3−-N) level was 50 mg/L to initiate nitrogen limitation stress during cultivation (Fu et al. 2017). The final total PO43−-P was a 48-mg/L medium, and P was equally divided into 2, 4, and 8 parts and supplied to C. regularis during cultivation (see details in Fig. 1). Glucose served as organic carbon in heterotrophic cultivation, and the final concentration was 10 g/L. This experiment was carried out in 250-mL glass flasks, and the working volume was 150 mL. The flasks were sealed with sterilized air filter membrane (blowholes 0.22 μm, Solarbio, China) to avoid contamination. The microalgae in flasks were cultivated at 25 ± 1 °C and stirred at 160 rpm in the dark. Each experimental condition was conducted in triplicate and lasted for 8 days, and experiments were carried out twice.
Experimental design of phosphorus-fed-batch. Phosphorus was equally divided into 2, 4, and 8 parts and supplied to C. regularis during cultivation. The final total phosphorous amounts were identical for all protocols (48-mg/L medium). The initial cell densities of all protocols were identical (2 × 107 cells/mL).
Analytical and calculated methods
Biomass productivity and nutrient uptake
During the experiment, 20-mL suspended liquid was sampled every 2 days and then filtered using a 0.45-μm polyether sulfone membrane (Jinteng, Tianjin, China). The C. regularis cells retained by the filter membrane were dried at 105 °C to achieve constant weight for biomass evaluation. The filtrate was collected for analyses of chemical oxygen demand (COD), glucose, dissolved inorganic nitrogen (DIN), and dissolved inorganic phosphorus (DIP). The measurement procedures were identical to a previous report (Fu et al. 2017).
Growth and nutrient utilization were evaluated via biomass productivity and absorption rate, respectively, which were calculated with Eq. (1) and Eq. (2):
where m1 and m2 (g/L) represent the biomass concentrations of dry microalgae at t1 and t2, respectively, while t1 and t2 (d) represent the first and second sampling time, respectively, in the linear interval of Fig. 3a.
where C1 and C2 (mg/L) represent the concentration of DIP at T1 and T2, respectively. T1 and T2 (d) represent the start and end point of each batch in Fig. 2a, respectively.
Phosphorus (P) utilization profiles versus cultivation time marked with P uptake rates (a‚ the values were P uptake rate, mg/(L•d)); all results were obtained based on experiments in triplicate. 31P NMR spectra (b) and poly-P fluorescent images (c) of C. regularis cells under different P-fed-batch frequencies. In fluorescent images, all the DNA in microalgae was stained blue, microalgal cells are blue, and turquois represent poly-P. The scale bar is 5 μm
31P Nuclear magnetic resonance
For 31P nuclear magnetic resonance (NMR) spectroscopy analysis, samples were extracted with perchloric acid, and the extracts were dissolved using deuterium oxide to lock the signal. P spectra were measured via 31P NMR (Varian Co., USA) with a 5-mm probe at 400 MHz at room temperature (Fu et al. 2017).
Poly-P fluorescence staining
Poly-P was stained and observed with a confocal laser scanning microscope (CLSM, Olympus FV-1000, FV1000 Viewer software, Japan). Before imaging, the cells were fixed in 4% paraformaldehyde for 12 h and washed with PBS solution to remove paraformaldehyde; 5-μL cell suspension was dropped on a microslide and dried at 30 °C; then, the dried cells were stained with 4,6-diamidino-2-phenylindole (DAPI, 50-μg/mL concentration) in the dark for 10 min, followed by washing with distilled water to remove residual DAPI. The excitation wavelength of CLSM was set to 405 nm, and the emission spectral range of 530–540 nm for poly-P detection and 460–480 nm was used for DNA observation (Chu et al. 2014). In the fluorescence staining experiment, DAPI combined to both poly-P and DNA, but their combinations had different fluorescent colors, poly-P was turquois and DNA was blue (Nishikawa et al. 2006).
Lipid production and fatty acid content
The total lipids of cells were extracted via a chloroform-methanol reagent and quantified via the gravimetric method (Bligh and Dyer 1959). Lipid production was calculated with Eq. 3:
where m (mg/L) represents the biomass and p (%) represent lipid content; both were collected on day 8.
For fatty acid analysis, lyophilized microalgae were pretreated with a mixture reagent of acetyl chloride and methanol for transesterification, and then the fatty acid composition was analyzed with a gas chromatograph (Agilent 6890, USA). The pretreatment operation and chromatographic parameters were described in detail in our previous study (Fu et al. 2017).
Real-time polymerase chain reaction
The primers of genes related to C. regularis growth and lipid accumulation were designed based on prior transcriptome data (Fu et al. 2017; Zhou et al. 2017). The target growth gene ROT3 was used with 18S as reference gene; the target lipid synthesis genes were BC, BCCP, CT, and DGAT1, and NADH was used as a reference gene. The primers are shown in Table S1. The real-time polymerase chain reaction (RT-PCR) experiments were conducted using an Applied Biosystems StepOne Real-time PCR system and fluorescent dye SYBR Green. The RT-PCR program used the following steps: initial incubation at 95 °C for 3 min, and then 45 cycles were started, which contained melting at 95 °C for 7 s, annealing at 57 °C for 10 s, and extending at 72 °C for 15 s.
Statistical analysis
An independent-sample t test was used to analyze the statistical significance of differences in the data. Differences were significant at p < 0.05. All statistical analyses were performed using the SPSS software package (IBM SPSS statistic 20.0).
Results and discussion
P uptake and poly-P nuclei formation
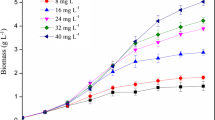
Figure 2a shows the effects of P-fed-batch on P uptake characteristics, compared with the traditional sole fed mode (control). Independent of P nutrient injection frequency, all of the supplied P was consumed by microalgal cells. However, the uptake rates (in mg/(L•d)) and the intracellular P forms differed. With low P fed frequency (control and P-fed-batch-2), P uptake rates were much higher than protocols with higher P fed frequency (P-fed-batch-4 and P-fed-batch-8); the former could be twice as high as the latter.
Excessive P was over-adsorbed by the cells and stored as intracellular poly-P, which was confirmed by the peaks at − 17.0 to − 23.3 ppm in the 31P NMR spectra (Moreno et al. 2000) and the poly-P staining images (Fig. 2 b and c) in control and P-fed-batch-2 on day 4. Interestingly, even though the totally supplied P amount was identical for all protocols, poly-P nuclei were scarcely detected for the P-fed-batch modes at higher frequencies of 4 and 8 times on day 4. The poly-P tends to accumulate in excessive P conditions; P concentration was a trigger of P uptake and poly-P formation. Acid-soluble only accumulated in 15- and 30-mg P/L experiments for Scenedesmus, while 5-mg P/L did not show this phenomenon (Brown and Shilton 2014). In addition, the abundance of poly-P depended on the P concentration (Powell et al. 2008).
This study supplied excessive P (referred to Chlorella cell compositions, C106N16P1) to all protocols at different frequencies (Falkowski 2000). Excessive P was almost completely consumed in all protocols; however, poly-P was not present in all protocols. Poly-P contained phosphoanhydride bonds that are rich in energy. Therefore, poly-P could serve as an energy source and P precursor pool for cell growth and metabolism (Chu et al. 2013; Nishikawa et al. 2006). It is worth noting that the utilization of poly-P by microalgal cells first requires degradation of poly-P to a short-chain structure via catalysis of polyphosphate kinase and adenosine triphosphatase (Harold 1966), followed by hydrolyzation to orthophosphate for the final utilization. Poly-P disappeared in both control and P-fed-batch-2 until day 8 (Fig. 2c), indicating a slow dissolution process. This implied a less efficient degradation and hydrolyzation process of the poly-P nucleus for storage beyond capacity. In the control, when P was depleted in the environment, poly-P experienced complicated degradation and hydrolyzation to generate orthophosphate, which can then be utilized by microalgae. In P-fed-batch-2, most P (24 mg/L, after each supply) was directly used by cells instead of poly-P storage. The small amount of poly-P could provide energy and serve as a P source for microalgae.
The advantage of the P-fed-batch mode was that the supplied orthophosphate may have directly served cell growth because of the low P concentration after each fed time; alternatively, the formed poly-P nuclei might have been too small to be observed and hydrolyzed quickly for direct utilization by cells. In this context, little poly-P was detected for P-fed-batch-4 and P-fed-batch-8 throughout (Fig. 2c). However, because the P supply was low (12 mg/L, P-fed-batch-4; 6 mg/L, P-fed-batch-8) for each supply compared with the high biomass after microalgae entered the logarithmic growth phase, the small amount of P was quickly exhausted and no P was available within a short period, subsequently causing negative effects on cell growth in P-fed-batch-4 and P-fed-batch-8. This deduction was supported by the microalgae growth profiles.
P-fed-batch improved the total lipid production
P-fed-batch improved cell growth
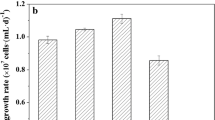
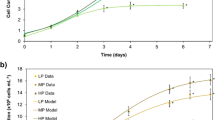
C. regularis cell growth profiles are shown in Fig. 3a. All P-fed-batch protocols achieved enhanced maximum microalgal biomass (~ 10%) on day 4. Moreover, the biomass productivities in P-fed-batch protocols were 10–15% higher than in control (Fig. 3b), and the difference between each P-fed-batch protocol and control was not significant. However, when P-fed-batch protocols serve as one P-fed-batch group and were then compared with the control, the difference was statistically significant (p < 0.05), indicating that biomass productivity was improved when using a P-fed-batch strategy. COD and DIN consumptions were consistent with biomass results (Fig. S2). These results indicated that the P-fed-batch strategy promoted microalgae growth. In particular, P-fed-batch-2 showed the maximum biomass productivity of 1.92 g/(L•d), which increased by 15% compared with control. The biomass productivity was 3−4-fold higher than that of other heterotrophic microalgae. Neochloris oleoabundans utilized glucose as carbon source and achieved a biomass productivity of 0.47 g/(L•d) (Morales-Sanchez et al. 2013), while the biomass productivity of Chlorella was cultivated in glucose-fed-batch mode of 0.35–0.45 g/(L•d) (Cheirsilp and Torpee 2012). The phenomenon of P-fed-batch-2 was higher than that of P-fed-batch-4 and P-fed-batch-8, which could be attributed to P uptake and utilization. In the control, microalgae took in excessive P and stored it in cells as poly-P, which could serve as energy and P pool. Poly-P utilization could not be easily compared with PO43−-P. In P-fed-batch-2, P was sufficient in the environment and intracellular poly-P was appropriate for the growth microalgae; while in P-fed-batch-4 and P-fed-batch-8, the P dosage in each batch was relatively lower to quickly increase biomass. The P deficiency consequently had negative effects on microalgae growth.
P-fed-batch promoted lipid accumulation
Lipid contents of C. regularis cells under different P-fed-batch frequencies are shown in Fig. 4. Clearly, the lipid content increased with P-fed-batch frequency, either on day 4 or on day 8. P-fed-batch-8 achieved the maximum lipid content of 40.5% at day 8, which was 16% more than the traditional sole fed mode. Moreover, the lipid accumulation improvement of P-fed-batch-8 was statistically significant (p < 0.05) compared with control. Combining the microalgae biomass results, the calculated total lipid production of the P-fed-batch protocols was 13–19% higher than the traditional mode of P provision (1.52 g/L), while the total supplied P amounts were identical for all protocols. These results confirmed that the P-fed-batch strategy was efficient to improve the lipid heterotrophic production of C. regularis under nitrogen limitation stress. The high lipid production is the goal of microalgae bioenergy, and P-fed-batch-2 achieved the maximum lipid production of 1.82 g/L, which was significantly (p < 0.01) higher than control; therefore, this frequency was an ideal mode for application.
For P-fed-batch protocols, the lipid content was 36.5–40.5% and the lipid production was 1.72–1.82 g/L, which far exceed the levels reported in other fed-batch reports (Cheirsilp and Torpee 2012; Morales-Sanchez et al. 2013). When marine Chlorella sp. and Nannochloropsis sp. were cultivated with glucose-fed-batch, their lipid accumulation was also improved compared with the batch mode (Cheirsilp and Torpee 2012), which supports the findings of the present study. However, their highest lipid content was 22–25% and lipid production was 0.81–0.90 g/L (Cheirsilp and Torpee 2012), indicating that the lipid accumulation was only 50–60% of that of the present study. When Neochloris oleoabundans was cultivated with nitrogen-fed-batch with glucose as heterotrophic carbon source, the lipid content was 33.7% (Morales-Sanchez et al. 2013), which is also lower than the results of the present study. Therefore, the P-fed-batch strategy is an effective method to promote microalgae lipid accumulation, which may be the result of the gene expression of lipid synthesis.
RT-PCR analysis
Figure 5 shows the effects of P-fed-batch cultivation of C. regularis on the typical growth genes based on the RT-PCR analysis. The extracted RNA was eligible from the gel electrophoresis results (Fig. S2); the gene expression was analyzed with relative quantification, and the amplification plot and melt curves of reference genes is shown in Fig. S3 and Fig. S4, respectively. The growth hormone gene ROT3 was related to brassinosteroid biosynthesis, and, according to the RT-PCR results, the ROT3 gene was more abundantly expressed for P-fed-batch-2 protocols, which was 1.5-fold higher than control (Fig. 5). This was consistent with the biomass growth profiles shown in Fig. 3.
Lipid synthesis gene expression changes are shown in Fig. 5. Acetyl-CoA was converted to malonyl-CoA via catalysis of acetyl-CoA carboxylase (ACCase) in the first step of fatty acid synthesis (Zhang and Powles 2006). For C. regularis cells, ACCase is composed of carboxyl transferase (CT), biotin carboxylase (BC), and the biotin carboxylase carrier protein (BCCP) (Konishi et al. 1996). On the whole, the expression of these key genes related to ACCase showed an increase tendency in P-fed-batch protocols, but the differences of BC and CT was not significant, when compared with the control. It is worth noting that the BCCP gene was most abundantly expressed in P-fed-batch-8, the expression of which was about 13-fold higher than in the control (Fig. 5). The lipid content was also highest in P-fed-batch-8, and the result was consistent with the BCCP expression. Diacylglycerol acyltransferase (DGAT) is the rate-limiting enzyme in triacylglycerol (TAG) synthesis. Here, diacylglycerol (DAG) is added to acyl of fatty acid and TAG is catalyzed by DGAT (Goncalves et al. 2016; Li-Beisson et al. 2015). The expression of the DGAT1 gene was slightly higher in P-fed-batch-4 and P-fed-batch-8 compared with control, but the increase trend was not statistically significant. The results of lipid synthesis gene expression suggest that the increase of batch frequency of P addition enhanced the role of ACCase by upregulating the gene expression of BCCP but not of DGAT. In this way, the microalgal lipid synthesis was improved in the P-fed-batch strategy.
Fatty acid components
P-fed-batch frequency affected FAME composition, as shown in Table 1. The percentage of C16:0 for higher P-fed-batch frequency was much higher than that of the control; C16:0 for P-fed-batch-4 and P-fed-batch-8 was even 9-fold higher than that of the control. Furthermore, in P-fed-batch-4 and P-fed-batch-8, the percentages of the unsaturated fatty acids of C16:1, C18:1, C18:2, and C18:3 generally decreased compared with the control. These results indicate that C. regularis tended to synthesize saturated fatty acids instead of unsaturated fatty acids. The contents of saturated fatty acid methyl esters for P-fed-batch-4 and P-fed-batch-8 reached 53.39% and 52.91%, respectively. These were almost 2-fold higher compared with the control. The enhanced degrees of fatty acid saturation are more preferable for biodiesel fuel (Tan and Lin 2011) and better meet the property requirements of biofuel for the USA (ASTM D6751) and Europe (EN 14214) (Hoekman et al. 2012). More importantly, P-fed-batch protocols presented a higher cetane number, lower average unsaturation, and lower iodine value (Table S2), indicating that the thus obtained biofuel had better combustibility and oxidative stability (Tan and Lin 2011).
Conclusions
This study explored a P-fed-batch strategy for cultivating microalgae in heterotrophic conditions. This P-fed-batch strategy affected phosphorus uptake and storage by microalgal cells, further leading to marked differences in microalgal growth and lipid accumulation. Employing P-fed-batch strategy, microalgae biomass production improved by 10–15%, and maximum biomass productivity was increased in P-fed-batch-2 with 5.04 g/L. Lipid accumulation also increased by 4–16%, and the highest lipid content was observed in P-fed-batch-8 with 40.5%. Considering both microalgae biomass and lipid content, the P-fed-batch strategy significantly enhanced lipid production by 13–19% based on different fed frequencies, and P-fed-batch-2 achieved the maximum lipid production of 1.82 g/L. The enhancement could be attributed to the upregulation of brassinosteroid growth hormone genes and biotin carboxylase carrier protein lipid synthesis genes. Moreover, the saturated fatty acid ratio in the FAMEs increased with P-fed-batch strategy, and the lipid achieved better combustibility and oxidative stability. This study provides a novel method for a more complete utilization of excessive P, while avoiding P toxicity for microalgal biofuel production.
References
Basri EM, Maznah WOW (2017) Differential growth and biochemical composition of photoautotrophic and heterotrophic Isochrysis maritima: evaluation for use as aquaculture feed. J Appl Phycol 29:1159–1170. https://doi.org/10.1007/s10811-017-1054-1
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. https://doi.org/10.1139/o59-099
Brown N, Shilton A (2014) Luxury uptake of phosphorus by microalgae in waste stabilisation ponds: current understanding and future direction. Rev Environ Sci Biotechnol 13:321–328. https://doi.org/10.1007/s11157-014-9337-3
Bumbak F, Cook S, Zachleder V, Hauser S, Kovar K (2011) Best practices in heterotrophic high-cell-density microalgal processes: achievements, potential and possible limitations. Appl Microbiol Biotechnol 91:31–46. https://doi.org/10.1007/s00253-011-3311-6
Cheirsilp B, Torpee S (2012) Enhanced growth and lipid production of microalgae under mixotrophic culture condition: effect of light intensity, glucose concentration and fed-batch cultivation. Bioresour Technol 110:510–516. https://doi.org/10.1016/j.biortech.2012.01.125
Chu FF, Chu PN, Cai PJ, Li WW, Lam PKS, Zeng RJ (2013) Phosphorus plays an important role in enhancing biodiesel productivity of Chlorella vulgaris under nitrogen deficiency. Bioresour Technol 134:341–346. https://doi.org/10.1016/j.biortech.2013.01.131
Chu FF, Shen XF, Lam PKS, Zeng RJ (2014) Optimization of CO2 concentration and light intensity for biodiesel production by Chlorella vulgaris FACHB-1072 under nitrogen deficiency with phosphorus luxury uptake. J Appl Phycol 26:1631–1638. https://doi.org/10.1007/s10811-013-0200-7
Cristobal Garcia-Canedo J, Cristiani-Urbina E, Flores-Ortiz CM, Ponce-Noyola T, Esparza-Garcia F, Olivia Canizares-Villanueva R (2016) Batch and fed-batch culture of Scenedesmus incrassatulus: effect over biomass, carotenoid profile and concentration, photosynthetic efficiency and non-photochemical quenching. Algal Res 13:41–52. https://doi.org/10.1016/j.algal.2015.11.013
Falkowski P (2000) Rationalizing elemental ratios in unicellular algae. J Phycol 36:3–6. https://doi.org/10.1046/j.1529-8817.2000.99161.x
Fu L, Cui X, Li Y, Xu L, Zhang C, Xiong R, Zhou D, Crittenden JC (2017) Excessive phosphorus enhances Chlorella regularis lipid production under nitrogen starvation stress during glucose heterotrophic cultivation. Chem Eng J 330:566–572. https://doi.org/10.1016/j.cej.2017.07.182
Goncalves EC, Wilkie AC, Kirst M, Rathinasabapathi B (2016) Metabolic regulation of triacylglycerol accumulation in the green algae: identification of potential targets for engineering to improve oil yield. Plant Biotechnol J 14:1649–1660. https://doi.org/10.1111/pbi.12523
Guarnieri MT, Nag A, Yang S, Pienkos PT (2013) Proteomic analysis of Chlorella vulgaris: potential targets for enhanced lipid accumulation. J Proteome 93:245–253. https://doi.org/10.1016/j.jprot.2013.05.025
Harold FM (1966) Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev 30:772–794
Hoekman SK, Broch A, Robbins C, Ceniceros E, Natarajan M (2012) Review of biodiesel composition, properties, and specifications. Renew Sust Energ Rev 16:143–169. https://doi.org/10.1016/j.rser.2011.07.143
Kamalanathan M, Chaisutyakorn P, Gleadow R, Beardall J (2018) A comparison of photoautotrophic, heterotrophic, and mixotrophic growth for biomass production by the green alga Scenedesmus sp (Chlorophyceae). Phycologia 57:309–317. https://doi.org/10.2216/17-82.1
Konishi T, Shinohara K, Yamada K, Sasaki Y (1996) Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol 37:117–122. https://doi.org/10.1093/oxfordjournals.pcp.a028920
Leyva LA, Bashan Y, de-Bashan LE (2015) Activity of acetyl-CoA carboxylase is not directly linked to accumulation of lipids when Chlorella vulgaris is co-immobilised with Azospirillum brasilense in alginate under autotrophic and heterotrophic conditions. Ann Microbiol 65:339-349. https://doi.org/10.1007/s13213-014-0866-3
Li Y, Yuan Z, Mu J, Chen D, Feng B (2013) Proteomic analysis of lipid accumulation in Chlorella protothecoides cells by heterotrophic N deprivation coupling cultivation. Energy Fuel 27:4031–4040. https://doi.org/10.1021/ef4000177
Li Y, Han F, Xu H, Mu J, Chen D, Feng B, Zeng H (2014) Potential lipid accumulation and growth characteristic of the green alga Chlorella with combination cultivation mode of nitrogen (N) and phosphorus (P). Bioresour Technol 174:24–32. https://doi.org/10.1016/j.biortech.2014.09.142
Li Q, Fu L, Wang Y, Zhou D, Rittmann BE (2018) Excessive phosphorus caused inhibition and cell damage during heterotrophic growth of Chlorella regularis. Bioresour Technol 268:266–270. https://doi.org/10.1016/j.biortech.2018.07.148
Li-Beisson Y, Beisson F, Riekhof W (2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82:504–522. https://doi.org/10.1111/tpj.12787
Meza B, de- Bashan LE, Hernandez J-P, Bashan Y (2015) Accumulation of intra-cellular polyphosphate in Chlorella vulgaris cells is related to indole-3-acetic acid produced by Azospirillum brasilense. Res Microbiol 166:399–407. https://doi.org/10.1016/j.resmic.2015.03.001
Morales-Sanchez D, Tinoco-Valencia R, Kyndt J, Martinez A (2013) Heterotrophic growth of Neochloris oleoabundans using glucose as a carbon source. Biotechnol Biofuel 6:100. https://doi.org/10.1186/1754-6834-6-100
Moreno B, Urbina JA, Oldfield E, Bailey BN, Rodrigues CO, Docampo R (2000) 31P NMR spectroscopy of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Evidence for high levels of condensed inorganic phosphates. J Biol Chem 275:28356–28362. https://doi.org/10.1074/jbc.M003893200
Nishikawa K, Machida H, Yamakoshi Y, Ohtomo R, Saito K, Saito M, Tominaga N (2006) Polyphosphate metabolism in an acidophilic alga Chlamydomonas acidophila KT-1 (Chlorophyta) under phosphate stress. Plant Sci 170:307–313. https://doi.org/10.1016/j.plantsci.2005.08.025
Perez-Garcia O, Escalante FME, de- Bashan LE, Bashan Y (2011) Heterotrophic cultures of microalgae: metabolism and potential products. Water Res 45:11–36. https://doi.org/10.1016/j.watres.2010.08.037
Powell N, Shilton AN, Pratt S, Chisti Y (2008) Factors influencing luxury uptake of phosphorus by microalgae in waste stabilization ponds. Environ Sci Technol 42:5958–5962. https://doi.org/10.1021/es703118s
Sansawa H, Endo H (2004) Production of intracellular phytochemicals in Chlorella under heterotrophic conditions. J Biosci Bioeng 98:437–444. https://doi.org/10.1263/jbb.98.437
Schmidt RA, Wiebe MG, Eriksen NT (2005) Heterotrophic high cell-density fed-batch cultures of the phycocyanin-producing red alga Galdieria sulphuraria. Biotechnol Bioeng 90:77–84. https://doi.org/10.1002/bit.20417
Shen XF, Chu FF, Lam PKS, Zeng RJ (2015) Biosynthesis of high yield fatty acids from Chlorella vulgaris NIES-227 under nitrogen starvation stress during heterotrophic cultivation. Water Res 81:294–300. https://doi.org/10.1016/j.watres.2015.06.003
Sureshkumar P, Thomas J (2019) Strategic growth of limnic green microalgae with phycoremediation potential for enhanced production of biomass and biomolecules for sustainable environment. Environ Sci Pollut Res 26:34702–34712. https://doi.org/10.1007/s11356-018-4012-9
Tan Y, Lin J (2011) Biomass production and fatty acid profile of a Scenedesmus rubescens-like microalga. Bioresour Technol 102:10131–10135. https://doi.org/10.1016/j.biortech.2011.07.091
Wang Y, Yu J, Wang P, Deng S, Chang J, Ran Z (2018) Response of energy microalgae Chlamydomonas reinhardtii to nitrogen and phosphorus stress. Environ Sci Pollut Res 25:5762–5770. https://doi.org/10.1007/s11356-017-0931-0
Xiong W, Li X, Xiang J, Wu Q (2008) High-density fermentation of microalga Chlorella protothecoides in bioreactor for microbio-diesel production. Appl Microbiol Biotechnol 78:29–36. https://doi.org/10.1007/s00253-007-1285-1
Yu X, Zhao P, He C, Li J, Tang X, Zhou J, Huang Z (2012) Isolation of a novel strain of Monoraphidium sp and characterization of its potential application as biodiesel feedstock. Bioresour Technol 121:256–262. https://doi.org/10.1016/j.biortech.2012.07.002
Zhang XQ, Powles SB (2006) The molecular bases for resistance to acetyl co-enzyme A carboxylase (ACCase) inhibiting herbicides in two target-based resistant biotypes of annual ryegrass (Lolium rigidum). Planta 223:550–557. https://doi.org/10.1007/s00425-005-0095-x
Zhang Y, Adams IP, Ratledge C (2007) Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology-Sgm 153:2013–2025. https://doi.org/10.1099/mic.0.2006/002683-0
Zhou D, Zhang C, Fu L, Xu L, Cui X, Li Q, Crittenden JC (2017) Responses of the microalga Chlorophyta sp to bacterial quorum sensing molecules (N-acylhomoserine lactones): aromatic protein induced self-aggregation. Environ Sci Technol 51:3490–3498. https://doi.org/10.1021/acs.est.7b00355
Zhu S, Huang W, Xu J, Wang Z, Xu J, Yuan Z (2014) Metabolic changes of starch and lipid triggered by nitrogen starvation in the microalga Chlorella zofingiensis. Bioresour Technol 152:292–298. https://doi.org/10.1016/j.biortech.2013.10.092
Funding
This research was financially supported by the National Natural Science Foundation of China (51708095, 51578117, 51722803), Fundamental Research Funds for the Central Universities (2412017QD027, 2412018ZD013, 2412018ZD042), the Science and Technology Project of Jilin Province (20180520168JH), and China Postdoctoral Science Foundation (2017M611302, 2018T110241).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Ta Yeong Wu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 7714 kb)
Rights and permissions
About this article
Cite this article
Fu, L., Yan, G., Li, Y. et al. Phosphorus supply via a fed-batch strategy improves lipid heterotrophic production of Chlorella regularis. Environ Sci Pollut Res 27, 31677–31685 (2020). https://doi.org/10.1007/s11356-020-09495-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-09495-5