Abstract
The present study aimed to examine whether the physical reworking of sediments by harrowing would be suitable for favouring the hydrocarbon degradation in coastal marine sediments. Mudflat sediments were maintained in mesocosms under conditions as closer as possible to those prevailing in natural environments with tidal cycles. Sediments were contaminated with Ural blend crude oil, and in half of them, harrowing treatment was applied in order to mimic physical reworking of surface sediments. Hydrocarbon distribution within the sediment and its removal was followed during 286 days. The harrowing treatment allowed hydrocarbon compounds to penetrate the first 6 cm of the sediments, and biodegradation indexes (such as n-C18/phytane) indicated that biodegradation started 90 days before that observed in untreated control mesocosms. However, the harrowing treatment had a severe impact on benthic organisms reducing drastically the macrofaunal abundance and diversity. In the harrowing-treated mesocosms, the bacterial abundance, determined by 16S rRNA gene Q-PCR, was slightly increased; and terminal restriction fragment length polymorphism (T-RFLP) analyses of 16S rRNA genes showed distinct and specific bacterial community structure. Co-occurrence network and canonical correspondence analyses (CCA) based on T-RFLP data indicated the main correlations between bacterial operational taxonomic units (OTUs) as well as the associations between OTUs and hydrocarbon compound contents further supported by clustered correlation (ClusCor) analysis. The analyses highlighted the OTUs constituting the network structural bases involved in hydrocarbon degradation. Negative correlations indicated the possible shifts in bacterial communities that occurred during the ecological succession.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Oil spills are spectacular accidents threatening marine ecosystems as evidenced by the recent Deepwater Horizon (Gulf Mexico, 2010) disaster with approximately 700 Ktons of crude oil released. But, since the Deepwater Horizon catastrophe, the Cedre (Centre de documentation, de recherche et d'expérimentations sur les pollutions accidentelles des eaux; http://www.cedre.fr/), French institution in charge of the response to accidental water pollution, has recorded 22 less publicised marine accidents over the world. Oil spills are estimated to release more than 1.3 million tons of hydrocarbons per year in the sea, revealing the extent of the threat. Appropriate responses, which effectiveness is depending on the pollutant type (crude oil, heavy fuel oil, diesel…), must be implemented according to the environment (sea, beaches, mudflats…) in order to mitigate the ecological impact. Most cleaning methods are well adapted to sandy and rocky beaches, but they are very difficult to transpose for cleaning complex ecosystems characterised by fine, sandy-muddy sediments such as mudflats and estuarine areas, which are highly productive areas. In such areas, physical removal is impossible and oil spills are thus difficult to control (Duran and Goňi-Urriza 2010). Biological treatments attempting to manage microorganisms, main actors in hydrocarbon biodegradation (McGenity 2014; Timmis et al. 2010), have been developed and appears to be friendly adapted methods for such areas. Bio-stimulation and bio-augmentation strategies aim to enhance the microbial activities by adding nutrients and hydrocarbon-degrading microorganism, respectively (Goni-Urriza et al. 2013). Alternative bioremediation approaches should take into account the characteristic conditions constraining the ecosystem functioning and thus use them as advantages, particularly the oxic/anoxic oscillations due to tide and sediment reworking by burrowing organisms (Cravo-Laureau and Duran 2014). The bioturbation affects the sediment matrix and the inhabiting microbial communities both via burrow construction modifying the physical structure of sediments and via ventilation modifying the chemical parameters, oxygen supply and nutrients availability (Adamek and Marsalek 2013; Kristensen et al. 2012). We recently demonstrated that applying anoxic/oxic oscillations to oiled sediments resulted in the acceleration of hydrocarbon degradation when restoring oxic conditions after an anoxic period (Vitte et al. 2013; Vitte et al. 2011). Similarly, increasing the bioturbation activity by the addition of burrowing organisms such as Hediste diversicolor did not affect the hydrocarbon degradation (Stauffert et al. 2013) modifying the bacteria and archaea communities’ structures (Cravo-Laureau and Duran 2014; Stauffert et al. 2014), suggesting functional redundancy in hydrocarbon degradation. We hypothesised that physical sediments reworking such as harrowing will affect the microbial community structure in oil-contaminated sediments, which would be a strategy to manage microbial communities’ structures and activities for, at the applied point of view, increasing hydrocarbon degradation in contaminated areas. At the academic point of view, the controlled modification of microbial communities’ structures is an opportunity to gain information to decipher the rules of microbial assemblages in oil-contaminated sediments (Cravo-Laureau and Duran 2014; Morales and Holben 2011).
In the present study, mudflat sediments maintained under tidal cycles in order to ensure conditions close to those prevailing in the natural environment, were contaminated by crude oil and subjected or not to a harrowing treatment. The effects of this physical treatment on hydrocarbon degradation and on microbial communities’ structure were compared to those observed in oiled sediments without harrowing treatment, used as control.
Materials and methods
Sampling and experimental setup
Field site and experimental devices have been previously described (Stauffert et al. 2013). Muddy-sand sediments were collected at the tidal basin Aber-Benoît (Treglonou, Brittany (France), 48° 33′ 12.40″ N; 4° 32′ 8.69″ W). In order to maintain the sediment structure, sediments were collected carefully with a core collector and then transferred over geotextile membrane on mesocosm-boxes (65 cm × 50 cm × 41 cm; approximately 30 L of sediment). Six mesocosm-boxes were set up at room temperature and connected with a device supplying natural sand-filtered and UV-treated seawater from the Oceanopolis aquarium (Brest, France). Tidal cycles (rise/fall 12 h—6 h high tide and 6 h low tide) were simulated using a water level control consisting of an up and down drainage system. Seawater was renewed at each tidal cycle (20 L of water per box). At high tide, the water level was maintained at about 5 cm above the sediment surface using a faucet ballcock. In order to determine the effect of harrowing treatment on oiled sediments, two conditions were applied in triplicate (N = 6) as follows: (i) control condition (C) corresponding to oil-contaminated sediments and (ii) harrowing treatment (H) corresponding to oil-contaminated sediments in which harrowing by a rake was performed twice a week. In order to ensure a uniform contamination of sediments, the upper 2-cm layer of each mesocosm was sampled, pooled, mixed with crude oil (final concentration 24 ± 4 mg/g of wet sediment) and homogenised. This mixture was spread over the sediment surface (2-cm layer). The oil used for the experimentation was the Russian export blend crude oil (REBCO), an Ural type crude oil, distilled at 110 °C to eliminate the more volatile hydrocarbon compounds. This oil contained approximately 60 % of saturated hydrocarbons (including 6.2 % alkanes and 53.7 % unresolved complex mixture), 25 % of aromatic compounds (including 0.8 % polyaromatic hydrocarbons (PAHs) and 24 % unresolved complex mixture), 10 % of resins and 5 % of asphaltenes. The major metals are nickel (50 mg/kg) and vanadium (100 mg/kg). The tidal cycle was initiated 2 h after reloading sediments.
Sediment subsamples were collected after 63, 151 and 286 days of incubation for chemical and molecular analyses. Four core samples (0–10 cm; falcon tube, 10 mL) were collected from each mesocosm: one for chemical analyses (triplicate conditions), and three for molecular analyses (sampling replicates × triplicate conditions: nine replicates per condition at each sampling time). The subsamples were collected 10 min after reaching the low tide. The sampling hole was loaded with a plastic tube in order to maintain the sediment’s structure. Samples were immediately frozen in liquid nitrogen after sampling and stored at −80 °C until analyses. The effect of the harrowing on sediment reworking was estimated by using fluorescent particulate tracers (luminophores) as previously described (Stauffert et al. 2013). A suspension of 100 g of orange luminophores (63–125 mm) was homogeneously spread over the sediment surface of each mesocosm 1 h after reloading the sediments surface. At the end of the experiment, two cores samples were collected (PVC corers; diameter 10 cm, length 20 cm) and sliced into 1-cm-thick sediment layers for the analysis of the macrobenthic community densities and reworking activity (bioturbation).
Macrobenthic communities’ densities and bioturbation
Each 1-cm-thick layer obtained from each corer sampled at the end of the experiment was fixed with 4 % formaldehyde. The sediments were first sieved with a 500-μm mesh in order to retain macroorganisms and then with a 44-μm mesh to collect the sediment fraction containing the luminophores particles. Each sediment layer retained on the 500-μm mesh was preserved in 70 % alcohol. The macroorganisms were identified to the major taxonomic level possible and counted using a stereomicroscope at magnification ×40 (Duport et al. 2007). Each sediment layer retained on the 44-μm mesh was freeze-dried, gently crushed to powder, homogenised and luminophores were counted using a microplate reader (Biotek Synergy Mx; Majdi et al. 2014) at λex/λem 450/585 nm. This resulted in a particulate tracer distribution profile over depth, for each sediment core. The sediment reworking (i.e. production of a biodiffusion-like coefficient Db describing particle transport in the whole sedimentary column) was quantified as previously described (Stauffert et al. 2013).
Chemical analyses
Prior to analyses, each sediment core was divided into two layers (0–6 and 6–10 cm). Each layer was then split for the determination of moisture content (50 °C for 24 h), and for chemical analyses (total petroleum hydrocarbon, TPH, concentrations and alkanes/aromatics distributions).
Sediment samples were extracted with 30 mL methylene chloride at 100 °C and 2000 psi for 14 min in a Dionex ASE 350 accelerated solvent extractor. Extractions of sediment were performed twice for each sample. The organic extracts were dried over Na2SO4 (activated at 400 °C for 4 h) and concentrated to approximately 2 mL using a Syncore (Büchi, Germany). Extracts were fractionated using a solid phase extraction (SPE) cartridge (silica/cyanopropyl (SiO2/C3-CN; 1.0/0.5 g, 6 mL; Interchim, France) (Alzaga et al. 2004). Saturate and aromatic fractions were eluted simultaneously with 8 mL of methylene chloride/pentane (80:20, v/v) and concentrated to approximately 2 mL.
Extracts were injected in a gas chromatography Agilent 6890N equipped with a split/splitless injector (splitless time 1 min, flow 50 mL/min) coupled to an Agilent 5975 mass selective detector (MSD) (electronic impact 70 eV, voltage 1200 V). The injector temperature was maintained at 300 °C. The interface temperature was 300 °C. The GC/MS temperature gradient was from 50 °C (1 min) to 300 °C (20 min) at 5 °C/min. The carrier gas was helium at a constant flow of 1 mL/min. The capillary column used was an HP-5 MS 30 m × 0.25 mm ID × 0.25 μm film thickness.
TPH contents were analysed according to the standard NF EN ISO 9377-2 protocol using the GCMS in Scan mode. Different concentrations of the oil were diluted with methylene chloride and used to calibrate the Scan mode response and then to quantify the TPH extracted from the sediment samples.
n-alkanes and PAHs semi-quantifications were performed using Single Ion Monitoring (SIM) mode with the most representative fragment (saturates) or the molecular ion (PAHs) of each compound at a minimum of 1.4 cycles/s. n-alkane and PAH degradation was followed with respect to 17α(H),21β(H)-hopane (m/z = 191) used as a conserved internal biomarker during the analysis (Prince et al. 1994).
Bacterial community analyses
The sediment cores were separated in two horizons (0–6 and 6–10 cm). The PowerSoil DNA isolation kit (MoBio Laboratories) was used for metagenomic DNA extraction following the manufacturer’s recommendations. Briefly, sediments were centrifuged at 5000g for 15 min, to remove the water phase and the initial step of horizontal vortexing was performed for 30 min. DNA was eluted in 100 μL water and stored at −20 °C until use.
The primer couple 63F (5′-CAGGCCTAACACATGCAAGTC-3′), 1387R (5′-GGGCGGWGTGTAACAAGGC-3′), specific for bacteria (Marchesi et al. 1998), was used to amplify the 16S rRNA gene. The forward primer was labelled at the 5′ end with phosphoramidite fluorochrome carboxyfluorescein (FAM). The PCR conditions were as previously described (Duran et al. 2008). Briefly: initial denaturation (95 °C for 10 min) followed by 35 cycles of denaturation (95 °C for 45 s), annealing (58 °C for 45 s), and extension (72 °C for 60 s) and a terminal extension at 72 °C for 10 min. The reaction mixture (25 μL final volume) contained 12.5 μL of Amplitaq Gold 360 Master Mix 2x (Life Technologies), 0.2 μM of each primer and 1 μL of DNA template. PCR products were visualised by agarose gel electrophoresis and purified with the PCR purification kit (GE Healthcare).
The purified 16S rRNA amplified fragments were digested by 3.5 U of AluI (Takara) at 37 °C in a final volume of 10 μL for 3 h. Then, digested products (1 μL) were mixed with deionised formamide (10 μL) and 0.25 μL of internal size standard (GS-500 Rox, Applied Biosystems). Fluorescently labelled fragments were separated and detected with an ABI Prism 3130XL capillary sequencer (Applied Biosystems) run in Genetic Analyser version 3.1. Injection was performed electrokinetically at 1.2 kV for 23 s, and the runs at 15 kV were completed within 20 min. Terminal restriction fragment length polymorphism (T-RFLP) profiles were analysed using GENEMAPPER version 4.1 software (Applied Biosystems). Only terminal fragments whose size ranged from 35 to 500 bp and whose height was greater than 20 fluorescence units were considered for analysis. T-RFLP profiles were aligned by identifying and grouping homologous fragments and normalised by calculating relative abundances of each terminal-restriction fragment (T-RFs) from height fluorescence intensity, using T-REX software (Culman et al. 2009). Shannon and Evenness indexes were calculated from T-RFLP patterns using the MVSP software (Multi-Variate Statistical Package 3.12d, Kovach Computing Services, 1985–2001, UK).
Bacterial abundance
The bacterial abundance was assessed by the determination of 16S rRNA gene copy numbers. It was performed by real time PCR with Sso Advanced SYBR Green Supermix using a CFX96 Real Time System (C1000 Thermal Cycler, Bio-Rad Laboratories, 170 CA, USA). The primer set GML5F-Uni516R (Michotey et al. 2012) was used. The real time PCR cycles consisted of a denaturation step of 5 s, a hybridization step of 20 s at 56 °C with an elongation step of 40 s. Gene abundance in mesocosm DNA samples was determined in relation with a calibration standard corresponding to dilution of plasmids harbouring Gammaproteobacterial SSU rRNA gene fragment (from 9.106 to 9.103 copies). The resulting conditions lead to a Q-PCR efficiency of 97.1 % (R 2 = 0.994). At the end of the PCR reaction, the specificity of the amplification was checked from the first derivative of their melting curves.
Statistics and multivariate analyses
Data statistical analyses were performed with STATISTICA software, version 5.5 (StatSoft, USA). The analysis of variance (ANOVA) was performed in order to investigate the effects of treatments on bacterial abundances and TPH content. The comparisons of the biodiffusion coefficients (Queiros, #23) and of the macroorganisms’ densities were performed to assess differences between the experimental conditions by means of the non-parametric Kruskal-Wallis one-way analysis of variance by ranks (Kruskal and Wallis 1952).
Bacterial communities’ comparisons
In order to compare bacterial communities (T-RFLP patterns data) according to treatment and time, two dimension non-metric multidimensional scaling ordination (NMDS) based on Bray–Curtis distances was performed using the function metaMDS from the “Vegan” package in R (Oksanen et al. 2011). The significant differences between groups were tested with an analysis of similarities (ANOSIM) performed with Primer 6 software (Primer E, Plymouth, UK).
In order to determine the relationships between bacterial communities (T-RFLP patterns data) and environmental parameters (hydrocarbon concentrations and bacterial abundance), a canonical correspondence analysis (CCA) was carried out using MVSP software (Multi-Variate Statistical Package 3.12d, Kovach Computing Services, 1985–2001, UK). Clustered correlation (ClusCor) analysis was described previously (Weinstein et al. 1997). A programme for making clustered correlation is available (http://discover.nci.nih.gov.gate1.inist.fr).
Co-occurrence network for detecting microbial interactions
In order to determine the bacterial co-occurrence and mutual-exclusion relationships, a bacterial network was built by a Cytoscape plugin CoNet 1.0b6 (Co-occurrence Network inference; http://psbweb05.psb.ugent.be/conet/; Faust et al. 2012). The input matrix consisted of T-RFLP data from all the samples (triplicates of both treatments and horizons). The analysis was performed with two correlation methods (Spearman and Pearson), edge scores for the randomization with 1000 bootstraps iterations. The resulting networks were merged applying Fichers’Z P value threshold 0.05 and Bonferroni multiple-test corrections. The co-occurrence bacterial network was visualised with Cytoscape (Smoot et al. 2011).
Results and discussion
Harrowing effect on macrobenthic communities’ densities and bioturbation
Because the volume of sediments required for the analyses is rather large (about 1.2 L per core), the structure and the densities of macroorganisms as well as the luminophores distribution with depth were only determined at the end of the experiment (286 days). Although there is no significant statistical difference (chi-square = 2.4; p = 0.1213), the harrowing treatment had a negative effect on the macrofauna community, by decreasing both the number of individuals and the species richness in comparison to controls (Table 1). This could be explained by the direct killing of individuals and by the destruction of the macrofaunal habitats due to the mechanical effect of harrowing. This is further supported by the absence in the harrowed sediments of Hydrobia ulvae (Gastropoda) and Cirratulus cirratus (Polychaeta) individuals, which sediment reworking and mobility traits correspond to surficial modifiers with limited movements’ capacities (Queiros et al. 2013). These species are therefore most likely sensitive to perturbations of the sediment surface. By contrast, no significant difference (chi-square = 0.02; p = 0.8845) in the sediment mixing intensity between control (4.53 ± 1.4 cm2 y−1; mean ± SD, n = 3; Table 1) and harrowed treatments (4.13 ± 0.93 cm2 y−1; mean ± SD, n = 3; Table 1) was observable. The luminophore distribution, however, indicated that the harrowing treatment resulted in the greater homogenization of the first 4 cm of the surface sediments in comparison to controls (Fig. 1).
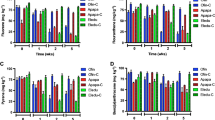
Effect of the harrowing treatment (a) compared to the control (b) on sediment reworking (luminophore distribution) and total petroleum distribution (TPH). Luminophore distribution was determined at the end of the experiment (286 days). TPH distribution was determined at 63 days (black), 151 days (white) and 286 days (grey). Error bars indicate the standard error (n = 3). Asterisk show significant differences between treatments (ANOVA p = 0.05)
Harrowing effect on hydrocarbon degradation
The mechanical mixing of the surface sediments induced the hydrocarbon compounds to be introduced deeper in the sediments in comparison to the untreated control (ANOVA p = 0.05) as shown by the distribution of total petroleum hydrocarbon (TPH) (Fig. 1). But more than 70 % of TPH were retained within the first 6 cm depth all along the experiment although the TPH proportion increased with time on the deeper horizon (6–10 cm, Fig. 1a). We thus considered the horizons 0–6 and 6–10 cm for further study.
The hopane units, normalisation using the 17α(H),21β(H)-hopane as internal standard (Prince et al. 1994), which serve as proxy to estimate petroleum biodegradation in the environment was used to follow the hydrocarbon degradation (Fig. 2a). At the end of the experimentation (286 days), the hydrocarbon (n-alkanes and PAHs) biodegradation reached 80 % in both harrowing and control conditions (Fig. 2a). Obviously, the degradation started faster in the harrowing condition with 50 % hydrocarbon degradation at 63 days whilst at the same incubation time the degradation was 23 % in the control without harrowing (Fig. 2a; ANOVA p = 0.05). Similarly, the decrease of the n-C18/phytane ratio (Fig. 2b) indicated a faster biodegradation in the harrowing condition than in the control but the highest difference was observed at day 151 (ANOVA p = 0.05). Interestingly, at 63 days, the biodegradation of PAHs was similar to that of n-alkanes (65 %) in the harrowing condition whilst it was estimated to 49 % for n-alkanes and 37 % for PAHs in the control condition (data not shown). However, at day 286, the biodegradation of n-alkanes reaching 91 % was higher than that of PAHs estimated at 68 %. This observation indicated that harrowing, by homogenising the surface of sediments, facilitated the hydrocarbon biodegradation probably enabling the sediments’ oxygenation and thus the oxygen attack of molecules, the first step of the aerobic hydrocarbon catabolism. We previously demonstrated in a bioreactor study that oxygenation was a critical step in hydrocarbon biodegradation (Vitte et al. 2013; Vitte et al. 2011), which is consistent with the aerobic biodegradation pathways so far described (Cerniglia 1992; McGenity 2014; Timmis et al. 2010). However, the harrowing treatment not only had a negative effect on the macrofauna, by decreasing the number of individuals from 77.3 ± 28.0 to 43.5 ± 13.4 ind. core−1 in control and harrowed sediments, respectively, but also by reducing the species richness from 10 (control) to 4 (harrowing) at the end of experiment. This was probably due to the mechanical effect of harrowing by direct killing of individuals and by the destruction of the macrofaunal habitats.
Biodegradation of hydrocarbon compounds indicated by hopane (a) and phytane (b) biomarkers during the incubation under harrowing (dashed line) and control (full line) conditions on the 0–6 cm horizon. The hydrocarbon content was determined at 63, 151 and 286 days. The Error bars indicate standard deviation (n = 3). Asterisk show significant differences between treatments (ANOVA p = 0.05)
Harrowing effect on bacterial community structure in oiled sediments
In order to determine the effect of the harrowing treatment on oil-degrading bacterial communities, microbiological analyses were performed at the latest stages of incubation (151 and 286 days) when the biodegradation of n-alkanes + PAHs was over 55 % in all conditions (Fig. 2a) but at distinct levels according to n-C18/phytane ratio as for the 151 days (Fig. 2b; ANOVA p = 0.05) allowing the comparison of contrasted situations. First, the bacterial abundances were determined by estimating the 16S rRNA gene copy numbers by Q-PCR. At the beginning of the experiment, the bacterial abundance was estimated to 6.5.1010 ± 5.109 16S rRNA gene copies per gram of sediment for the 0–6 cm horizon. At 151 days, the bacterial abundance was found below 4.1010 16S rRNA gene copies per gram of sediment for each condition (0–6 cm horizon; Fig. 3) showing the effect of crude oil on bacterial growth. The bacterial abundance remained stable for the control condition whilst it reached the initial level for the harrowing condition at the end of the experiment (286 days; Fig. 3a). This observation suggested that the harrowing favoured bacterial growth probably by increasing the oxygenation in the 0–6 cm horizon of the sediment. The effect of physical perturbation in marine sediments has been previously discussed (Widdicombe and Austen 2001), and the increase of diffusive oxygen uptake rate after harrowing demonstrated (Soltwedel et al. 2008). It is important to notice that in the 6–10-cm horizon, the bacterial abundance remained stable around 4.1010 16S rRNA gene copies per gram of sediment until the end of experiment in the control condition (Fig. 3b) whereas it was found lower in the harrowing condition increasing slightly at 286 days (Fig. 3a). These observations indicated that harrowing influenced the bacterial communities not only at the surface (0–6 cm) of sediments but also at lower depth (6–10 cm horizon), disturbing probably bacterial populations living in anaerobic zones by introducing both hydrocarbon compounds and oxygen in deeper horizons whilst modifying the sediment structure as previously shown (Widdicombe and Austen 2001).
The bacterial community structures were then determined by T-RFLP based on 16S rRNA genes analysis. The T-RFLP patterns showed an average of 26.5 ± 1.7 T-RFs (terminal restriction fragments; or operational taxonomic units (OTUs)). The Shannon and Evenness were almost similar (no significant differences) in all conditions with average 2.94 ± 0.2 and 0.7 ± 0.1 respectively.
The bacterial communities’ structures from the different treatments were compared by NMDS. At 151 days, the bacterial communities’ structures were similar except those from the first 0–6-cm horizon in the harrowing treatment that grouped apart (Fig. 4a; R = 0.1, p = 0.12) showing the effect of the harrowing treatment whereas the bacterial communities’ structures were found different at 286 days (Fig. 4b; R = 0.65, p = 0.002) indicating that each treatment and each horizon was characterised by a specific bacterial community. Thus, considering the 0–6 cm horizon where biodegradation occurred, this observation suggested that the bacterial communities of the harrowing treatment at 286 days were distinct to those from the control treatment although the biodegradation rates were found similar. We have reported comparable observation with sediments in which bioturbation reworking was enhanced by the addition of H. diversicolor (Cravo-Laureau and Duran 2014; Stauffert et al. 2013). Such observation also indicated that the mechanical reworking by harrowing imposed a selection above the presence of oil, resulting in a distinct bacterial community structure. It is thus likely that the harrowing was a stronger driver for the bacterial community than oil, because the effect of oil was hidden by the harrowing effect. In response to environmental perturbations, the bacterial communities may be resistant, resilient or functionally redundant (Allison and Martiny 2008; Yachi and Loreau 1999). The results reported here could be explained either by the fact that the specific degraders group was not affected or by functional redundancy in hydrocarbon degradation. The manipulation of bacterial community structure and composition, for instance by harrowing as reported here, will provide the opportunity to decipher the mechanisms underlying functional redundancy by combining in depth characterisation of bacterial composition, by high throughput sequencing, with the analysis of appropriate genes involved in hydrocarbon degradation.
Comparison of bacterial community structures by non-metric multidimensional scaling (NMDS) analyses based on T-RFLP 16S rRNA gene patterns. a and b effect of treatment: comparisons of bacterial community structures at 151 (a) and 286 (b) days from harrowing treatment (H, red) and control (C, blue) for the horizons 0–6 cm (dashed lines) and 6–10 cm (full lines). c and d temporal shifts: bacterial community structure shifts between 151 (blue) and 286 (orange) days in sediments from control (c) and harrowing (d) treatments. Analyses were performed in triplicates, which are linked by lines
The observed changes in the bacterial T-RFLP fingerprints in the harrowed mesocosms may be also explained in part by an indirect effect as harrowing modified the structure of the macrobenthic community and the density of macroorganisms (Table 1). Indeed, previous studies demonstrated that the absence/presence of specific macroorganisms like polychaetes may influence the bacterial communities’ structure not only in uncontaminated sediments (Pischedda et al. 2011) but also in oil-contaminated sediments (Cuny et al. 2007; Stauffert et al. 2013). Differences in mechanical and biological mixing in the sediments may have also induced different environmental conditions for bacterial communities.
Additionally, NMDS analyses showed a clear temporal shift of bacterial community structure between 151 and 286 days in control treatment (Fig. 4c; R = 0.44, p = 0.003) and harrowing treatment (Fig. 4d; R = 0.47, p = 0.004), which resulted to the separation of the bacterial communities of the surface (0–6 cm) from those of the deeper horizon (6–10 cm) without affecting the bacterial abundance (Fig. 3b). This observation is in accordance with ecological succession theory that hypothesises a succession of microorganisms following the succession of metabolites during the degradation (Head et al. 2006). Such modifications have been observed in sediments from different habitats including highly saline (Bordenave et al. 2004a; Bordenave et al. 2004b) and marine (Chronopoulou et al. 2013; Coulon et al. 2012) ecosystems contaminated by crude oil. However, it is important to notice that the bacterial community structures of the sediments surface (0–6 cm) at 286 days were still slightly related to those at 151 days in the harrowing-treated mesocosms (Fig. 4d), endorsing that the harrowing was strongly affecting the bacterial communities.
Bacterial assemblage in oiled sediments
In order to determine the bacterial populations (OTUs) associated with the degradation capacities, we performed canonical correspondence analyses (CCA) correlating T-RFLP patterns with the hydrocarbon content, degradation indexes and bacterial abundance. The objective being to observe the correlations between OTUs and hydrocarbons, the CCA was performed with the samples in which biodegradation occurred, i.e. the 0–6 cm horizon of both treatments (Fig. 5). The CCA, explaining 94 % of the data distribution, showed that hydrocarbon content and bacterial abundances were in opposite directions along the axis 1 (70 % of the distribution), indicating that the presence of hydrocarbons probably limited the bacterial growth. Another explanation could be that more degradation occurred where bacteria was more abundant. Three OTUs (124, 218 and 231) were associated with high hydrocarbon content; they determine the specificity of the control bacterial community (C 151d 0–6) at 151 days when the biodegradation was low in the control mesocosms (Fig. 2), as indicated by the correlation with the n-C17/pristane ratio in the CCA. These three OTUs may be tolerant to the presence of crude oil but their role in the biodegradation must be limited. In contrast, two OTUs (201 and 242) were associated with high bacterial abundance, explaining the difference of the harrowing-treated communities at 286 days (H 286d 0–6). It is likely that these two OTUs became predominant once the negative effect of hydrocarbon was attenuated; they probably play a minor role in hydrocarbon degradation. These observations are in accordance with previous studies demonstrating that some populations in hydrocarbon-degrading consortia play an auxiliary role in the bacterial community such as utilising metabolites (Dastgheib et al. 2012; Hii et al. 2009), increasing the bioavailability of the molecules (Kanaly et al. 2002), surfactant production (Ranchou-Peyruse et al. 2004) and syntrophic interactions (Hirschler-Réa et al. 2012; Kleinsteuber et al. 2008).
Canonical correspondence analysis (CCA) correlating bacterial populations (triangles, T-RFLP OTUs) and communities (blue circles, T-RFLP patterns) with the hydrocarbon content (Alk and Aro, see below), biodegradation indexes (C17/prist and C18/phyt) and bacterial abundance (Bac Abun). The analysis is based on mean of triplicate samples for T-RFLP profiles, bacterial abundance and hydrocarbon contents. The specific OTUs (T-RFs from T-RFLP profiles) are indicated in green and the common OTUs in red. Blue circles show the bacterial communities in harrowed (H) and control (C) sediments at 151 (151d) and 286 (286d) days for the horizon 0–6 cm (0–6). Hydrocarbon content: AlkT total alkane compounds, AlkL light molecular weight alkanes (<20 C atoms), AlkH high molecular weight alkanes (>20 C atoms), AroT total polyaromatic hydrocarbons (PAHs), AroL light molecular weight PAHs (<3 benzene rings), AroH high molecular weight PAHs (>3 benzene rings), Alk + Aro total of alkanes and PAHs compounds. Biodegradation indexes correspond to the ratio of n-C17 alkane/pristane (C17/prist) and n-C18 alkane/phytane (C18/phyt). The bacterial abundance (Bac Abun) corresponds to 16S rRNA gene copy numbers assessed by Q-PCR
In the CCA, most OTUs were grouped in the centre (red in Fig. 5), suggesting that they play a pivotal role in the bacterial community organisation. In order to test the hypothesis that the central OTUs in the CCA constitute the framework of the bacterial assemblage in oiled sediments, we conducted a co-occurrence network analysis. Because the objective was to determine the correlations between OTUs in oiled sediments, all the data set (both treatments and horizons) was included in the analysis in order to exclude the relationships that may occur in non-degrading conditions. Interestingly, the co-occurrence network analysis confirmed that most of these OTUs had a pivotal role in the microbial assemblage (Fig. 6), revealing three sub-networks.
Co-occurrence bacterial network showing the relationships between the OTUs. The network was inferred from T-RFLP patterns applying Pearson (straight lines) and Spearman (curved lines) correlations with 1000 bootstrap iterations, Fichers’Z P value threshold 0.05 and Bonferroni multiple-test corrections. The positive correlations are indicated by green lines and mutual exclusions by red dashed lines. Node circles correspond to OTUs, the size is proportional to the relative abundances considering all T-RFLP profiles. Numbers at the edge correspond to correlation weight
The first sub-network formed by the most abundant OTUs (194, 166) shows mutual exclusion with no positive relation with other OTUs. It is likely that these OTUs, although dominant, did not play a crucial role in the bacterial community as indicated by the CCA (Fig. 5). The second sub-network was formed by four couples (183/256, 193/199, 218/202 and 201/242) with strong positive correlations (Pearson >0.7; p value < 0.05). These couples excluded each other, suggesting ecological succession since negative associations are considered as structural shifts (Faust et al. 2012). Among them, it is important to notice that the couple formed by OTUs 201 and 242 were found in the harrowing treatment at 286 days as highlighted in the CCA (Fig. 5). Furthermore, these couples showed different correlation with hydrocarbon compounds as revealed by the clustered correlation (ClusCor) analysis (Fig. 7), suggesting different degradation capacities and thus supporting the ecological succession. The couples 183/256 and 201/242 showed no or weak correlation (−0.17 < Pearson > 0.2) and even exclusion (Pearson < −0.3) with hydrocarbon compounds, respectively. In contrast, the couple 193/199 showed correlation with all hydrocarbons (Pearson > 0.47) whereas the couple 218/202 was strongly correlated to high molecular weight (MW) alkanes (Pearson > 0.7) and low MW aromatic compounds (0.47 < Pearson > 0.58; Fig. 7).
Clustered correlation (ClusCor) between T-RFLP OTUs and chemical data. Colour legend shows Pearson correlation, red indicating high positive correlation and blue high negative correlation (exclusion). Clustering was performed applying Pearson correlation and complete linkage as cluster method. Abbreviations of the environmental parameters are those indicated in Fig. 5. Diamonds indicate OTUs of the sub-network 1, coloured circles the different OTUs couples of the sub-network 2 and triangles the OTUs of the sub-network 3
The co-occurrence network analysis did not found correlations for OTUs 124 and 231, which were associated to the hydrocarbon content in the CCA and the ClusCor analysis (Figs. 5 and 7). This observation confirms their limited role in the bacterial assemblage. It is likely that these OTUs were present transiently in the mesocosms. The third sub-network showed the central role of four OTUs (134, 164, 190 and 212) ensuring most associations, supported by positive Pearson correlations (>0.7; p value < 0.05), constituting thus the network structural bases. The ClusCor analysis showed that the OTUs 212 and 134 were correlated with all types of hydrocarbon compounds whilst OTUs 164 and 190 were correlated with high MW alkanes and excluded low MW aromatics (Fig. 7). Noteworthy, the OTU 36 was associated with the OTU 205 but showed mutual exclusion with OTUs 134 and 212, major OTUs in the frame of the network, the OTUs 36 and 205 showing mutual exclusion with hydrocarbon compounds (Fig. 7). This observation is in accordance with the NMDS and CCA analyses (Figs. 4 and 5), suggesting that this negative association may correspond to a structural shift due to the effect of petroleum. The bacterial community modification has been demonstrated in many cases after petroleum contamination and resulting in ecological succession (Bordenave et al. 2004a; Bordenave et al. 2007; Head et al. 2006; Païssé et al. 2008). Similar modifications have been also reported for other perturbations such as biotic reworking activity by polychaetes (Christensen et al. 2002; Gilbert et al. 1996; Grossi et al. 2002; Stauffert et al. 2013), crustaceans (Bertics and Ziebis 2009), meiofauna (Louati et al. 2013) and natural perturbations (Paerl et al. 2002). Although the network was constructed with low-resolution fingerprinting method comparing to high throughput sequencing methods, it demonstrates the complex interactions between bacteria that constitute bacterial communities. Further studies are required to decipher the bacterial interactions and their role in hydrocarbon degradation. The in-depth characterisation of microbial communities by high throughput sequencing technology is a promising approach (Cravo-Laureau and Duran 2014) to describe microbial assemblage that may provide useful information to manage microbial degradation capacities.
Conclusion
The control of bacterial assemblages and activities is the major challenge for the implementation of biological processes for mitigating the effect of pollutants. Several approaches including bio-stimulation and bio-augmentation have been developed to favour and enhance bacterial activities involved in biodegradation (Goni-Urriza et al. 2013). In this study, we demonstrated that the mechanical sediment reworking by harrowing favoured the initiation of bacterial hydrocarbon degradation activities. However, the intensive harrowing treatment performed in this study, applied twice a week, had a damaging effect on the macrofauna that may be deleterious for the whole ecosystem functioning. The usefulness of such harrowing treatment should thus be carefully examined before to envisage applying it for bioremediation processes. But overall, the harrowing treatment resulted in a distinct bacterial assemblage than that observed in the control. At the academic point of view, studying the modification of bacterial communities is a useful way to understand the assemblage rules driving bacterial communities’ organisation; harrowing is an easy way to manipulate bacterial communities.
References
Adamek Z, Marsalek B (2013) Bioturbation of sediments by benthic macroinvertebrates and fish and its implication for pond ecosystems: a review. Aquacult Int 21:1–17
Allison SD, Martiny JBH (2008) Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105:11512–11519
Alzaga R, Montuori P, Ortiz L, Bayona JM, Albaigés J (2004) Fast solid-phase extraction-gas chromatography-mass spectrometry procedure for oil fingerprinting: application to the prestige oil spill. J Chromatogr 1025:133–138
Bertics VJ, Ziebis W (2009) Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME J 3:1269–1285
Bordenave S, Fourçans A, Blanchard S, Goni-Urriza MS, Caumette P, Duran R (2004a) Structure and functional analyses of bacterial communities changes in microbial mats following petroleum exposure. Ophelia 58:195–203
Bordenave S, Jézéquel R, Fourçans A, Budzinski H, Merlin FX, Fourel T, Goñi-Urriza M, Guyoneaud R, Grimaud R, Caumette P, Duran R (2004b) Degradation of the “Erika” oil. Aquat Living Resour 17:261–267
Bordenave S, Goñi-Urriza MS, Caumette P, Duran R (2007) Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat. Appl Environ Microbiol 73:6089–6097
Cerniglia CE (1992) Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351–368
Christensen M, Banta GT, Andersen O (2002) Effects of the polychaetes Nereis diversicolor and Arenicola marina on the fate and distribution of pyrene in sediments. Mar Ecol Prog Ser 237:159–172
Chronopoulou PM, Fahy A, Coulon F, Païssé S, Goñi Urriza M, Acuña Alvarez L, McKew BA, Lawson T, Timmis KN, Duran R, Underwood GJC, McGenity TJ (2013) Impact of a simulated oil spill on benthic phototrophs and nitrogen-fixing bacteria in mudflat mesocosms. Environ Microbiol 15:242–252
Coulon F, Chronopoulou P-M, Fahy A, Sandrine P, Goñi-Urriza M, Peperzak L, Acuña Alvarez L, McKew BA, Brussaard CPD, Underwood GJC, Timmis KN, Duran R, McGenity TJ (2012) Central role of dynamic tidal biofilms dominated by aerobic hydrocarbonoclastic bacteria and diatoms in the biodegradation of hydrocarbons in coastal mudflats. Appl Environ Microbiol 78:3638–3648
Cravo-Laureau C, Duran R (2014) Marine coastal sediments microbial hydrocarbon degradation processes: contribution of experimental ecology in the omics’era. Front Microbiol 5:39
Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH (2009) T-REX: Software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10
Cuny P, Miralles G, Cornet-Barthaux V, Acquaviva M, Stora G, Grossi V, Gilbert F (2007) Influence of bioturbation by the polychaete Nereis diversicolor on the structure of bacterial communities in oil contaminated coastal sediments. Mar Pollut Bull 54:452–459
Dastgheib SMM, Amoozegar MA, Khajeh K, Shavandi M, Ventosa A (2012) Biodegradation of polycyclic aromatic hydrocarbons by a halophilic microbial consortium. Appl Microbiol Biotechnol 95:789–798
Duport E, Gilbert F, Poggiale JC, Dedieu K, Rabouille C, Stora G (2007) Benthic macrofauna and sediment reworking quantification in contrasted environments in the Thau Lagoon. Estuar Coast Shelf Sci 72:522–533
Duran R, Goňi-Urriza MS (2010) Impact of pollution on microbial mats. In: Timmis K (ed) Handbook of Hydrocarbon and Lipid Microbiology. Springer Berlin Heidelberg, pp 2339-2348. doi:10.1007/978-3-540-77587-4_170
Duran R, Ranchou-Peyruse M, Menuet V, Monperrus M, Bareille G, Goñi MS, Salvado JC, Amouroux D, Guyoneaud R, Donard OFX, Caumette P (2008) Mercury methylation by a microbial community from sediments of the Adour estuary (Bay of Biscay, France). Environ Pollut 156:951–958
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C (2012) Microbial co-occurrence relationships in the human microbiome. PLoS Comp Biol 8
Gilbert F, Stora G, Bertrand JC (1996) In situ bioturbation and hydrocarbon fate in an experimental contaminated Mediterranean coastal ecosystem. Chemosphere 33:1449–1458
Goni-Urriza M, Cravo-Laureau C, Duran R (2013) Microbial bioremediation of aquatic environments. In: Férard J, Blaise C (eds) Encyclopedia of aquatic ecotoxicology, vol 2. Springer, Berlin, pp 709–720
Grossi V, Massias D, Stora G, Bertrand JC (2002) Burial, exportation and degradation of acyclic petroleum hydrocarbons following a simulated oil spill in bioturbated Mediterranean coastal sediments. Chemosphere 48:947–954
Head IM, Jones DM, Röling WF (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182
Hii YS, Law AT, Shazili NAM, Abdul-Rashid MK, Lee CW (2009) Biodegradation of Tapis blended crude oil in marine sediment by a consortium of symbiotic bacteria. Int Biodeterior Biodegrad 63:142–150
Hirschler-Réa A, Cravo-Laureau C, Casalot L, Matheron R (2012) Methanogenic octadecene degradation by syntrophic enrichment culture from brackish sediments. Curr Microbiol 65:561–567
Kanaly RA, Harayama S, Watanabe K (2002) Rhodanobacter sp strain BPC1 in a benzo a pyrene-mineralizing bacterial consortium. Appl Environ Microbiol 68:5826–5833
Kleinsteuber S, Schleinitz KM, Breitfeld J, Harms H, Richnow HH, Vogt C (2008) Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol Ecol 66:143–157
Kristensen E, Penha-Lopes G, Delefosse M, Valdemarsen T, Quintana CO, Banta GT (2012) What is bioturbation? The need for a precise definition for fauna in aquatic sciences. Mar Ecol Prog Ser 446:285–302
Kruskal WH, Wallis WA (1952) Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621
Louati H, Ben Said O, Soltani A, Got P, Mahmoudi E, Cravo-Laureau C, Duran R, Aissa P, Pringault O (2013) The roles of biological interactions and pollutant contamination in shaping microbial benthic community structure. Chemosphere 93:2535–2546
Majdi N, Bardon L, Gilbert F (2014) Quantification of sediment reworking by the Asiatic clam Corbicula fluminea Muller, 1774. Hydrobiologia 732:85–92
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
McGenity TJ (2014) Hydrocarbon biodegradation in intertidal wetland sediments. Curr Opin Biotechnol 27:46–54
Michotey V, Guasco S, Boeuf D, Morezzi N, Durieux B, Charpy L, Bonin P (2012) Spatio-temporal diversity of free-living and particle-attached prokaryotes in the tropical lagoon of Ahe atoll (Tuamotu Archipelago) and its surrounding oceanic waters. Mar Pollut Bull 65:525–537
Morales SE, Holben WE (2011) Linking bacterial identities and ecosystem processes: can ‘omic’ analyses be more than the sum of their parts? FEMS Microbiol Ecol 75:2–16
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H (2011) vegan: Community ecology package. R package version 2.0-2. http://CRAN.R-project.org/package=vegan
Paerl HW, Dyble J, Twomey L, Pinckney JL, Nelson J, Kerkhof L (2002) Characterizing man-made and natural modifications of microbial diversity and activity in coastal ecosystems. Antonie Van Leeuwenhoek 81:487–507
Païssé S, Coulon F, Goñi Urriza M, Peperzak L, McGenity TJ, Duran R (2008) Structure of bacterial communities along a hydrocarbon contamination gradient in a coastal sediment. FEMS Microbiol Ecol 66:295–305
Pischedda L, Militon C, Gilbert F, Cuny P (2011) Characterization of specificity of bacterial community structure within the burrow environment of the marine polychaete Hediste (Nereis) diversicolor. Res Microbiol 162:1033–1042
Prince RC, Elmendorf DL, Lute JR, Hsu CS, Haith CE, Senius JD, Dechert GJ, Douglas GS, Butler EL (1994) 17-Alpha(H),21-beta(H)-hopane as a conserved internal marker for estimating the biodegradation of crude-oil. Environ Sci Technol 28:142–145
Queiros AM, Birchenough SNR, Bremner J, Godbold JA, Parker RE, Romero-Ramirez A, Reiss H, Solan M, Somerfield PJ, Van Colen C, Van Hoey G, Widdicombe S (2013) A bioturbation classification of European marine infaunal invertebrates. Ecol Evol 3:3958–3985
Ranchou-Peyruse A, Moppert X, Hourcade E, Hernandez-Raquet G, Caumette P, Guyoneaud R (2004) Characterization of brackish anaerobic bacteria involved in hydrocarbon degradation: a combination of molecular and culture-based approaches. Ophelia 58:255–262
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Soltwedel T, Lansard B, Gilbert F, Hasemann C, Bell E, Sablotny B, Eagle M, Kershaw P, Rabouille C (2008) An “Integrated Sediment Disturber” (ISD) to study the impact of repeated physical perturbations on sediment geochemistry and the small benthic biota. Limnol Oceanogr Methods 6:307–318
Stauffert M, Cravo-Laureau C, Jézéquel R, Barantal S, Cuny P, Gilbert F, Cagnon C, Militon C, Amouroux D, Mahdaoui F, Bouyssiere B, Stora G, Merlin FX, Duran R (2013) Impact of oil on bacterial community structure in bioturbated sediments. PLoS ONE 8
Stauffert M, Duran R, Gassie C, Cravo-Laureau C (2014) Response of archaeal communities to oil spill in bioturbated mudflat sediments. Microb Ecol 67:108–119
Timmis KN, McGenity TJ, Meer JR, Lorenzo V (2010) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin
Vitte I, Duran R, Jezequel R, Caumette P, Cravo-Laureau C (2011) Effect of oxic/anoxic switches on bacterial communities and PAH biodegradation in an oil-contaminated sludge. Environ Sci Pollut Res 18:1022–1032
Vitte I, Duran R, Hernandez-Raquet G, Mounier J, Jézéquel R, Bellet V, Balaguer P, Caumette P, Cravo-Laureau C (2013) Dynamics of metabolically active bacterial communities involved in PAH and toxicity elimination from oil-contaminated sludge during anoxic/oxic oscillations. Appl Microbiol Biotechnol 97:4199–4211
Weinstein JN, Myers TG, Oconnor PM, Friend SH, Fornace AJ, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, Buolamwini JK, vanOsdol WW, Monks AP, Scudiero DA, Sausville EA, Zaharevitz DW, Bunow B, Viswanadhan VN, Johnson GS, Wittes RE, Paull KD (1997) An information-intensive approach to the molecular pharmacology of cancer. Science 275:343–349
Widdicombe S, Austen MC (2001) The interaction between physical disturbance and organic enrichment: an important element in structuring benthic communities. Limnol Oceanogr 46:1720–1733
Yachi S, Loreau M (1999) Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci U S A 96:1463–1468
Acknowledgments
We acknowledge the support of the French programme ANR DECAPAGE (project ANR-CESA-2011-006 01). We would like to thank all partners of the DECAPAGE project and MELODY group for their useful discussions. We acknowledge the Regional Platform for Environmental Microbiology PREMICE supported by the Aquitaine Regional Government Council (France) and the urban community of Pau-Pyrénées (France). We thank the Direction Générale de l’Armement (Edgar) and the Aquitaine Regional Government Council (France) for financial support to JA. We acknowledge B. Bouyssiere, S. Guasco, M.S. Goñi-Urriza and J. Gury for technical assistance during sampling campaign.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Duran, R., Bonin, P., Jezequel, R. et al. Effect of physical sediments reworking on hydrocarbon degradation and bacterial community structure in marine coastal sediments. Environ Sci Pollut Res 22, 15248–15259 (2015). https://doi.org/10.1007/s11356-015-4373-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-4373-2