Abstract
The assessment of the direct impact of breakdown products of pesticide components on aquatic wildlife is ecotoxicologically relevant, but frequently disregarded. In this context, the evaluation of the genotoxic hazard posed by aminomethylphosphonic acid (AMPA—the major natural degradation product of glyphosate) to fish emerges as a critical but unexplored issue. Hence, the main goal of the present research was to assess the AMPA genotoxic potential to fish following short-term exposures (1 and 3 days) to environmentally realistic concentrations (11.8 and 23.6 μg L−1), using the comet and erythrocytic nuclear abnormalities (ENA) assays, as reflecting different levels of damage, i.e. DNA and chromosomal damage, respectively. Overall, the present findings pointed out the genotoxic hazard of AMPA to fish and, subsequently, the importance of including it in future studies concerning the risk assessment of glyphosate-based herbicides in the water systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Most studies concerning the effects of pesticides on (non-target) aquatic organisms have been focused either on the active ingredients or the commercial products as a whole. However, in the environment, the parental compounds present in those formulations may suffer modifications of their chemical structure, originating products with different toxic properties. The previous perspective triggered studies with the transformation products of endosulfan (a broad-spectrum insecticide) (Hoang et al. 2011) and dichlobenil (a broad-spectrum herbicide) (Björklund et al. 2011), pointing out their potential risk to aquatic biota. Therefore, the assessment of the direct impact of chemicals that may occur in the water systems as breakdown products of the former ingredients should be considered ecotoxicologically unavoidable and thereby included in the priorities of researchers and public authorities.
Glyphosate [N-(phosphono-methyl-glycine)] is the active ingredient of the most widely used non-selective post-emergence herbicides in the world. Formulations containing glyphosate are heavily used in agriculture, urban landscaping and forestry (Kolpin et al. 2006; Landry et al. 2005). Though it can be intentionally applied to control emergent and floating aquatic vegetation, surface runoff following terrestrial uses is known to be the primary way of glyphosate transfer to surface waters. Studies on environmental fate of glyphosate indicated that it tends to strongly bind to organic matrices, like sediment of aquatic systems, and rapidly degrading (Feng et al. 1990). The soil sorption and the degradation of glyphosate exhibit a great variation depending on soil composition and properties (Gimsing et al. 2004), as well as the factor leachability (Borggaard and Gimsing 2008). Once in the aquatic environment, glyphosate can be naturally converted into sarcosine and aminomethylphosphonic acid (AMPA) (Al-Rajab et al. 2008; Landry et al. 2005). Of these two, AMPA has the highest occurrence in water, showing an environmental mobility and persistence greater than glyphosate (Kolpin et al. 2006), being thus assumed as the major breakdown product (Williams et al. 2000). Studies concerning its occurrence in water reported the importance of microorganism in the metabolization of this compound (Rueppel et al. 1977), while chemical degradation and photodecomposition seemed to be minor routes (Mallat and Barceló 1998).
The relative rapid degradation of glyphosate (half-life from 7 to 14 days) in the aquatic environment (Giesy et al. 2000) can, apparently, limit the environmental risk associated. However, this is highly questionable due to the appearance of its metabolites, namely AMPA, which has been found in levels ranging 3.6–60 μg L−1 (Battaglin et al. 2005; Struger et al. 2008).
Considering the abundance of studies reporting the occurrence of AMPA as a pollutant in the aquatic environment, it would be expected that its effects on organisms have already been extensively explored. Bearing this in mind, only a study performed with the Pacific oyster (Crassostrea gigas) demonstrates the toxicity of AMPA in aquatic organisms (Mottier et al. 2013). Thus, this is a matter almost completely uncovered and relatively little is known about the biological activity of this compound (Mañas et al. 2009), and thus, it is quite surprising its categorization by some regulatory agencies as “not of toxicological concern” (E.U. 2002; USEPA 1993).
The analysis of DNA alterations in aquatic organisms have been shown as a highly suitable method for evaluating the environmental genotoxic contamination, allowing the detection of exposure to low concentrations of contaminants, including pesticides, in a wide range of species (Scalon et al. 2010). Hence, and despite the considerable amount of studies addressing glyphosate and Roundup® (a glyphosate-based herbicide) genotoxic potential to fish (Cavalcante et al. 2008; Çavas and Könen 2007; Guilherme et al. 2010; 2012a; 2012b), the risk posed to genome integrity by AMPA remains unknown. The only study carried out in this direction was applied to mammalian models (human cell lines and mice), clearly demonstrating its genotoxic action (Mañas et al. 2009). To the authors’ knowledge, no studies were performed concerning the genotoxicity of AMPA in aquatic organisms.
Thus, the main goal of the present research was to assess the genotoxic potential of AMPA, as the major breakdown product of glyphosate, in blood cells of Anguilla anguilla L, following short-term exposures (1 and 3 days) to environmentally realistic concentrations (11.8 and 23.6 μg L−1). Genotoxic end points such as comet and erythrocytic nuclear abnormalities (ENAs) assays were adopted, since they reflect different levels of genetic damage, i.e. DNA and chromosomal damage, respectively. The comet assay detects DNA strand breaks and alkali-labile sites (Andrade et al. 2004; Lee and Steinert 2003), representing an early sign of damage that might be subject to a repair process. In order to achieve a better understanding of DNA-damaging mechanisms, an extra step was added to the standard procedure of comet assay where nucleoids were incubated with DNA lesion-specific repair enzymes, highlighting specifically oxidised DNA bases. Complementary, the ENA assay, based on the detection of micronuclei and other nuclear anomalies (Pacheco and Santos 1997), signals chromosome breakage (clastogenicity) or loss and mitotic spindle apparatus dysfunction (aneugenicity) (Fenech 2000; Stoiber et al. 2004), which are hardly reparable lesions. Hence, ENA assay displays later and less transient alterations when compared with those detected by the comet assay.
Material and methods
Chemicals
AMPA and all chemicals needed to perform the comet assay and the ENAs test were obtained from the Sigma-Aldrich Chemical Company (Madrid, Spain). DNA lesion-specific repair enzymes, namely formamidopyrimidine DNA glycosylase (FPG) and endonuclease III (Endo III), were purchased from Prof. Andrew Collins (University of Oslo; Norway).
Test animals and experimental design
European eel (A. anguilla L.) juvenile specimens with an average weight 0.25 ± 0.02 g (glass eel stage) were captured at the Minho river mouth, Caminha, Portugal. Eels were acclimated to laboratory for 20 days and kept in 20-L aquaria under a natural photoperiod (10L: 14D), in aerated, filtered, dechlorinated and recirculating tap water, with the following physico-chemical conditions: salinity 0, temperature 20 ± 1 °C, pH 7.1 ± 0.3, nitrate 23 ± 0.1 mg L−1, nitrite 0.03 ± 0.02 mg L−1, ammonia 0.2 ± 0.04 mg L−1, dissolved oxygen 8.1 ± 0.2 mg L−1. During this period, fish were daily fed with fish roe.
The experiment was carried out in 1-L aquaria, in a semi-static mode, under the conditions described for the acclimation period. After acclimation, 72 eels were divided into 6 groups, corresponding to three test conditions and two exposures times. Thus, fish were exposed to 11.8 and 23.6 μg L−1 AMPA (groups A1 and A2, respectively). Another group was kept with clean water—control (group C). For each test condition, 1- and 3-day exposures were carried out. Water medium in 3-day aquaria was daily renewed (100 %). The concentrations of glyphosate adopted previously by Guilherme and co-workers (Guilherme et al. 2012b) served as a basis to determine the AMPA concentrations currently tested. Taking this as a starting point, the concentration of AMPA was calculated assuming that it results from a glyphosate conversion on a basis of 1:1 mol. Thus, for instance, it was assumed that 17.9 μg (1.065 × 10−7 mol) of glyphosate corresponds to 11.8 μg (1.065 × 10−7 mol) of AMPA. A stock solution of AMPA was prepared using deionized water just before its addition to the exposure water. The experiment was carried out using triplicate (n = 3) groups of 4 fish for each condition/time.
Fish were not fed during experimental period. Fish were sacrificed by cervical transaction at the post-opercular region and blood collected from the heart using heparinised capillary tubes. Blood smears were immediately prepared for ENA assay. For comet assay, 2 μL of blood was immediately diluted in 1 mL of ice-cold phosphate-buffered saline (PBS) to prepare a cell suspension, which was kept on ice up to further procedure.
Evaluation of genetic damage
Comet assay
The conventional alkaline version of the comet assay was performed according to the methodology of Collins (2004) and adapted by Guilherme et al. (2010), with the proper adjustments to assay procedure with one extra step of digesting the nucleoids with endonucleases. In order to significantly increase the throughput of the assay, a system of eight gels per slide was adopted, based on a model created by Shaposhnikov et al. (2010) and adapted by Guilherme et al. (2012b). Briefly, 20 μL of cell suspension (previously prepared in PBS) was mixed with 70 μL of 1 % low melting point agarose (in PBS). Eight drops with 6 μL of cell suspension were placed onto the precoated slide (with 1 % normal melting point agarose) as two rows of 4 (4 groups of 2 replicates), without coverslips, containing each gel approximately 1,500 cells. The gels were left for ±5 min at 4 °C in order to solidify agarose and then immersed in a lysis solution (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris, 1 % Triton X-100, pH 10) at 4 °C, for 1 h. After lysis of agarose-embedded cells, slides were washed three times with enzyme buffer (0.1 M KCl, 0.5 mM EDTA, 40 mM HEPES, 0.2 mg.mL−1 bovine serum albumin, pH 8) at 4 °C.
Three sets of slides were prepared: two sets were incubated with endonucleases FPG and EndoIII that convert oxidised purines and pyrimidines into DNA single-strand breaks, respectively (Azqueta et al. 2009), and a third set was incubated only with buffer. Hence, 30 μL of each enzyme diluted in buffer was applied in each gel, along with a coverslip, prior to incubation at 37 °C for 30 min in a humidified atmosphere. Then, the coverslips were removed and slides gently placed in the electrophoresis tank (Sub-Cell® GT, Bio-Rad), immersed in electrophoresis solution (±20 min) for alkaline treatment. DNA migration was performed at a fixed voltage of 25 V, a current of 300 mA (power supply PowerPac™, Bio-Rad) that results in 0.7 V cm−1 (achieved by adjusting the buffer volume in the electrophoresis tank). The slides were stained with ethidium bromide (20 μg L−1).
Slides with eight gels each, and 50 nucleoids per gel, were observed, using a Leica DMLB fluorescence microscope (×400 magnification). The DNA damage was quantified by visual classification of nucleoids into five comet classes, according to the tail intensity and length, from 0 (no tail) to 4 (almost all DNA in tail) (Collins 2004). The total score expressed as a genetic damage indicator (GDI) was calculated multiplying the percentage of nucleoids in each class by the corresponding factor, according to this formula:
GDI values were expressed as arbitrary units in a scale of 0 to 400 per 100 scored nucleoids (as average value for the 2 gels observed per fish). When the comet assay was performed with additional FPG and EndoIII steps, GDI values were obtained in the same way but the parameter designated GDIFPG and GDIEndoIII, respectively. Additional DNA breaks corresponding to net enzyme-sensitive sites (NSSFPG or NSSEndoIII) were also expressed. These parameters were calculated based on the difference between GDIFPG and GDI or GDIEndoIII and GDI.
Besides GDI, the frequency of nucleoids observed in each comet class was also expressed, as recommended by Azqueta et al. (2009). In order to improve the expression of the DNA damage extent (Çavas and Könen 2007; Palus et al. 1999), the subtotal frequency of nucleoids with medium (class 2), high (class 3) and complete (class 4) damaged DNA was also calculated (2 + 3 + 4).
ENA assay
This assay was carried out in mature peripheral erythrocytes according to the procedure of Pacheco and Santos (1996). Briefly, one blood smear per animal was fixed with methanol for 10 min and stained with Giemsa (5 %) for 30 min. From each smear, 1,000 erythrocytes were scored under ×1,000 magnification (microscope Olympus BX-50) to determine the frequency of the following nuclear lesion categories: kidney-shaped nuclei (K), lobed nuclei (L), binucleate or segmented nuclei (S) and micronuclei (MN). In addition, notched nuclei (N) were also scored as suggested by Fenech (2000) and Ayllon and Garcia-Vazquez (2001). Final results were expressed as the mean value (‰) of the sum for all the lesions observed (K + L + S + N + MN).
Statistical analysis
Statistica 7.0 software (StatSoft, Inc., OK, USA) was used for statistical analysis. All data were first tested for normality (Shapiro-Wilk test) and homogeneity of variance (Levene’s test) to meet statistical demands. One-way analyses of variance (ANOVA), followed by Dunnett test as post hoc comparison, was applied to compare treated with control groups, within the same exposure duration. Two-way ANOVA was applied to test the effect of the factors concentration and exposure time on the levels of DNA damage, as well as on the frequency of nuclear abnormalities. The Tukey test was applied as post hoc comparison. In all the analyses, differences between means were considered significant when p < 0.05 (Zar 1996). Statistical treatments were carried out using the means of replicate groups. The relationship between the assessed parameters was explored using linear regression analyses. The correlation coefficient (r) was calculated, and its statistical significance (p) was determined from the table of critical values for the correlation coefficient (Zar 1996).
Results
Non-specific DNA damage
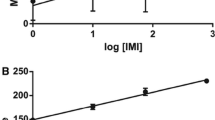
Analysing GDI values after 1 day of exposure (Fig. 1), both AMPA groups showed significant increases, relative to the control. The 3 days of exposure (Fig. 1) revealed that only the higher concentration of AMPA (A2) induced significant DNA damage, in comparison with the control. Neither concentration nor time related significant differences were observed; however, a decrease tendency was displayed by all treatments in relation to time.
Mean values of genetic damage indicator (GDI) measured by the standard (alkaline) comet assay in blood cells of A. anguilla exposed to 11.8 and 23.6 μg L−1 aminophosphoric acid (AMPA; A1, A2), during 1 and 3 days (replicate aquaria, n = 3). Bars represent the standard error. Statistically significant differences (p < 0.05) are: (*) in relation to control (C), within the same exposure time
Considering the results in terms of individual DNA damage classes (Table 1), after 1 day of exposure, it was perceptible that only control group (C) displayed a prevalence of classes 1 and 2. Differently, both concentrations of AMPA (A1, A2) presented higher frequencies in classes 2 and 3. Moreover, the subtotal of damaged nucleoids (2 + 3 + 4) revealed significantly higher values in both treated groups, in comparison with control.
After 3 days of exposure (Table 1), the group A1, like the control (C), displayed classes 1 and 2 as the most frequent. In a different way, classes 2 and 3 presented higher frequencies for A2 group. The subtotal of damaged nucleoids (2 + 3 + 4) displayed significantly higher values only for the group A2, compared to control.
Comparing 1- and 3-day results (Table 1), a general time-related increase tendency was observed in the frequency of class 1, despite the absence of significant differences, while class 3 showed the opposite temporal variation, with a significant decrease in A2 group.
Oxidative DNA damage
The detection of oxidised bases was achieved by the analysis of the results of the comet assay with an extra step where nucleoids were incubated with DNA lesion-specific repair enzymes—FPG and EndoIII (Figs. 2 and 3).
Mean values of DNA damage, measured by comet assay in blood cells of A. anguilla exposed to 11.8 and 23.6 μg L−1 aminophosphoric acid (AMPA; A1, A2), during 1 and 3 days (replicate aquaria, n = 3). Values resulted from the assay with an extra step of digestion with formamidopyrimidine DNA glycosylase (FPG) to detect oxidised purine bases. a Overall damage (GDIFPG) with the contribution of DNA breaks corresponding to net FPG-sensitive sites (NSSFPG; black). b NSSFPG alone. Bars represent the standard error. Statistically significant differences (p < 0.05) are: (*) in relation to control (C), within the same exposure time
Mean values of DNA damage, measured by comet assay in blood cells of A. anguilla exposed to 11.8 and 23.6 μg L−1 aminophosphoric acid (AMPA; A1, A2), during 1 and 3 days (replicate aquaria, n = 3). Values resulted from the assay with an extra step of digestion with endonuclease III (EndoIII) to detect oxidised pyrimidine bases. a Overall damage (GDIEndoIII) with the contribution of additional DNA breaks corresponding to net EndoIII-sensitive sites (NSSEndoIII; dark grey). b NSSEndoIII alone. Bars represent the standard error. Statistically significant differences (p < 0.05) are: (*) in relation to control (C), within the same exposure time; (♦) between exposure times, within the same treatment
Mean frequency (‰) of erythrocytic nuclear abnormalities (ENA) in peripheral erythrocytes of A. anguilla exposed to 11.8 and 23.6 μg L−1 aminophosphoric acid (AMPA; A1, A2), during 1 and 3 days (replicate aquaria, n = 3). Bars represent the standard error. Statistically significant differences (p < 0.05) are: (*) in relation to control (C), within the same exposure time; (▲) between treatments, within the same exposure time; (♦) between exposure times, within the same treatment
FPG-associated damage
Regarding GDIFPG results, both treatments, and both exposure times, showed significantly higher damage, in comparison with the control (Fig. 2a). In a different way, NSSFPG parameter was not capable to distinguish any treatment, in relation to the control (Fig. 2b).
Neither concentration- nor time-related differences were observed for GDIFPG and NSSFPG (Fig. 2) data.
EndoIII-associated damage
After the 1-day exposure, the digestion with EndoIII (GDIEndoIII; Fig. 3a) revealed an overall damage significantly higher than the control, in both treated groups. Concerning the NSSEndoIII parameter (Fig. 3b), no significant differences were found.
Regarding the 3-day exposure, only the group corresponding to the higher concentration of AMPA (A2) showed to be significantly higher than the control (GDIEndoIII; Fig. 3a). On the other hand, the NSSEndoIII parameter (Fig. 3b) followed the pattern of 1 day of exposure, being unable to identify any AMPA effective concentration, when compared with the control.
A significant general time-related decrease was detected in GDIEndoIII values, considering both AMPA groups. Differently, and considering the NSSEndoIII parameter, no differences were found, comparing both exposure times.
Chromosomal damage
No significant alterations were found in ENA frequency following the first day of exposure (Fig. 4). On the other hand, considering the 3-day exposure, a significant increase for the higher concentration of AMPA (A2) was observed, in relation to the control. This exposure condition was the only one able to indicate a concentration dependence, showing a significantly higher chromosomal damage for the group A2 when compared to the group A1 (Fig. 4). Moreover, it was perceptible a time-related increase for the higher concentration of AMPA (A2).
The results in terms of individual analysis of each nuclear lesion category (Table 2) showed no differences in what concerns to 1 day of exposure. Contrarily, the 3-day exposure revealed that L and S categories, as well as the subtotal (K + L + S + N), were significantly higher than the control, when the higher concentration of AMPA (A2) was considered. Similar to what was noticed for ENA frequency (Fig. 4), L category and the subtotal displayed significant concentration- and time-related increases. Kidney-shaped nuclei (K) was the most commonly detected nuclear abnormality in all experiment groups, except in the higher AMPA concentration, after the 3-day exposure (A2) where L category exhibited the highest frequency (Table 2).
Discussion
Despite being considered as practically nontoxic to fish (USEPA 1993), glyphosate’s genotoxic potential to this group of aquatic organisms was recently and unequivocally demonstrated in A. anguilla (Guilherme et al. 2012b). The fast conversion of glyphosate into its breakdown product AMPA seems to be a silent problem to the environment, since this metabolite has not been taken into account when the impact of this pesticide was under evaluation. Though its persistence is higher than glyphosate, until now, AMPA occurrence in the environment has been neglected and its toxicity largely ignored. Consequently, concerns regarding its possible health and environmental hazard have emerged, justifying further research in this direction. Hence, the present study appears as the first study assessing the genotoxic risk of AMPA to fish.
The concentrations of AMPA currently tested were calculated on the basis of environmentally realistic concentrations of glyphosate (Guilherme et al. 2012b), considering a total degradation into its metabolite. Keeping this in mind and the scarcity of data published so far, the following discussion will be mainly focused on the interpretation of the current findings, having as background the available data concerning its precursor—glyphosate.
The genotoxicity of AMPA was assessed by two genotoxic end points (comet and ENA), in order to reflect genetic damage at different levels as stated in the introduction. In line with Mañas et al. (2009), the comet assay could be considered a biomarker of genotoxic exposure (measuring damage which may be repaired), while the ENA assay is a biomarker of genotoxic effect (signalising irreparable lesions).
In terms of non-specific DNA damage, depicted in GDI values, AMPA showed its genotoxic potential in both concentrations. Despite being statistically non-significant, a time-related decrease tendency in GDI values can be noticed. Adding to this tendency, the lack of a significant GDI increase for the lower concentration (A1) after 3 days, it can be suggested that fish had the capacity to adapt to the genotoxic stimulus, allowing blood cells to avoid the damage expression as GDI. This idea is confirmed by the analysis of the DNA damage classes individually, as most of the AMPA treatments displayed classes 2 and 3 as the most prevalent, contrarily to A1 group after 3 days where classes 1 and 2 prevailed. Since it is known that DNA strand breaks and alkali-labile sites detected by the comet assay represent an early sign of damage (Lee and Steinert 2003), which might be subject to a repair process (Collins 2004), this DNA damage reduction may be explained by the intervention of DNA-repair system and/or by the catabolism of heavily damaged cells. An increased splenic erythrophagia was previously associated to intense genetic damage in A. anguilla (Pacheco and Santos 2002). Despite not being tested, this hypothesis cannot be excluded, considering the specificities of blood in terms of the modulation of the cell population’s renewal. These processes have been previously presented by Saleha Banu et al. (2001) to explain reductions in comet tail-length after 2 and 3 days and a return to control levels, after 4 days, in blood cells of Tilapia mosambica exposed to an organophosphate pesticide.
The comparison of DNA-damaging effects of AMPA presently detected (as GDI) with those described for its precursor glyphosate (Guilherme et al. 2012b) demonstrated a similar pattern for both compounds. The unique noticeable difference is related to its recovery capacity from the damage caused. Thus, after 3 days of exposure, fish showed to be able to recover from the damage induced by the exposure to 35.7 μg L−1 of glyphosate (the equivalent concentration to the highest concentration of AMPA) after 1 day, while considering the metabolite, fish were only able to recover from the exposure to the lowest concentration (corresponding to 17.9 μg L−1 of glyphosate). Thus, the idea that the metabolite (AMPA) is less toxic than the parental compound (glyphosate) as previously mentioned in a report of the European Commission (E.U. 2002) cannot be corroborated, at least concerning genotoxicity evaluation.
In order to understand a particular damaging action, namely DNA oxidation, the comet assay was improved with an extra step with two DNA lesion-specific repair enzymes. Thus, data on the DNA breaks scored after the incubation with endonucleases also pointed out the genotoxicity of AMPA (in all treatments and exposure times, except for GDIEndoIII in A1 group after 3 days of exposures). Surprisingly, when only the additional breaks corresponding to net enzyme-sensitive sites were considered, none of the conditions revealed significant levels of oxidative damage. However, the use of this methodology allows the detection of a genotoxic risk resulting from unspecific (alkali-labile sites and single-strand breaks associated with incomplete excision repair sites) and specific (bases oxidation) damage jointly, as well as the isolation of the oxidative DNA damage. The additional step of the assay also improves the possibility to identify a damaging action that could have been masked by the breaks score as GDI only.
Looking specifically to GDIFPG parameter after 1 day of exposure, the results demonstrated to be in accordance with those obtained for GDI. However, when 3-day results as GDIFPG were considered, the group A1 kept its genotoxic action, pointing out an inability of adaptation to the genotoxic stimulus, contrarily to what had been supposedly demonstrated in GDI results. Accordingly, it can be suggested that the oxidative damage seems to be more difficult to repair when compared to the non-specific damage, as found by Guilherme and co-workers during a post-exposure period after an exposure to the glyphosate-based herbicide Roundup® (Guilherme et al. submitted). Anyway, it can be inferred that the DNA-repair system played the principal role on the temporal recovery displayed by the GDI parameter, rather than the catabolism of heavily damaged cells. The involvement of the latter process would have affected both GDI and GDIFPG parameters (which was not the case).
As described for GDI and GDIFPG, GDIEndoIII showed significantly higher DNA damage for both AMPA treatments, considering the 1-day exposure. The GDIEndoIII results obtained after the exposure of 3 days, as observed for the GDI parameter and contrarily of GDIFPG, did not point out the lowest AMPA concentration as genotoxic.
The NSSEndoIII parameter, similarly to NSSFPG, was not able by itself to indicate AMPA as a notable inducer of oxidative damage. In this way, the potential of AMPA to exert oxidative damage, though it cannot be overlooked, seems to be limited, preventing the detection of damage when only the additional breaks corresponding to the incubation with DNA lesion-specific repair enzymes (FPG and EndoIII) are assessed.
Thus, the present results point out a limitation of the standard comet assay (GDI data), as already stated by Guilherme et al. (2012b). Likewise, previous results of non-specific DNA damage, depicted by GDI, pointed out the higher concentration of glyphosate (corresponding to the higher concentration of AMPA currently used) as non genotoxic (Guilherme et al. 2012b). This fact would be disclaimed by the results obtained as overall oxidative damage, as well as considering the enzyme-associated DNA breaks (Guilherme et al. 2012b). Contrarily to AMPA, the previous study demonstrated that glyphosate was able to induce oxidative damage measured as NSSEndoIII (in a concentration corresponding to the higher AMPA concentration tested). Thus, AMPA showed no evidences to have higher potential to oxidatively damage DNA when compared to its precursor—glyphosate. In spite of different biological models used, the present findings on DNA-damaging potential of AMPA agree with those reported by Mañas et al. (2009).
Considering the ENA assay, chromosomal damage was only found in fish exposed to the highest concentration of AMPA (A2), after 3 days. In addition, and considering this exposure length, it was possible to distinguish both AMPA concentrations. In what concerns to the individual abnormality categories, the observed differences were mostly due to the significant increase of the lobed (L) category, despite the slight contribution of S category. In addition, and following the total ENAs pattern, L category also displayed a significant increase between both treated groups, after 3 days of exposure, appearing as the only category able to distinguish between concentrations. A time-related increase was also observed for L category (A2 groups), supporting once again its contribution to the total ENAs frequency. Despite the kidney-shaped nuclei frequency assumed similar levels considering the control and treated groups, it is important to clarify that in control groups, the values were around 10‰ (10 cells in a total score of 1,000) and thus considered as low (Pacheco and Santos 1996, 1998, 2002; Guilherme et al. 2010).
The total absence of MN in the present study reinforced the usefulness of the other nuclear abnormalities scoring, as previously stated (Guilherme et al. 2010; Guilherme et al. 2008). The single score of MN may lead to a possible lack of sensitivity related to its low frequency in wild fish. As mentioned for comet assay, the present ENAs results agree with the findings of Mañas et al. (2009) who described an increase frequency of micronucleated erythrocytes (as an indication of chromosomal damage) in mice, 2 days after an i.p. injection of AMPA.
The comparative analysis of comet and MN (or ENA) assays in terms of their sensitivity is a controversial matter. It is well known that comet assay detects primary DNA lesions resulting from the balance of DNA damage (strand breaks and alkali-labile sites) and repair mechanisms, while the MN (or ENA) test reveals fixed DNA lesions or irreparable aneugenic effects (Bolognesi et al. 2004). Thus, data resulting from both assays were considered in parallel, as reflecting different types of genetic damage expression. In this perspective, current ENA data reflected a late appearance of damage when compared to comet assay, as suggested already by Wirzinger et al. (2007). This fact seems linked to the need of the exposed cell population to undergo at least one cell cycle (Udroiu 2006), which is not a requisite for comet assay. Subsequently, only comet assay demonstrated to be able of genetic damage detection after 1 day of exposure, confirming the early nature of the damaging events involved. On the other hand, ENAs, unlike comet assay, demonstrated the ability to distinguish between the two tested concentrations. Moreover, it was possible to observe a different pattern related to the temporal evolution of the induced damage. Hence, longer exposures tended to increase the magnitude of chromosomal damage, while DNA damage (comet assay) decreased pointing out a recovery phenomenon.
In order to clarify the relation between the two end points, their correlation was tested (ENAs vs. GDI, r = 0.3363). Accordingly, the absence of significant correlation reinforced the idea that the detected genetic damage could be caused by different events. Even though ENAs can be originated by DNA single-strand breaks (measured by comet assay), a diversity of processes (e.g. DNA repair) may prevent the manifestation of this causal association and, subsequently, the existence of correlation. Moreover, it cannot be ignored that ENAs could have an aneugenic origin (not detectable by comet assay) that can also justify the absence of correlations.
Briefly, it can be inferred that these two genotoxic end points provide complementary information, allowing a more effective assessment of AMPA genotoxic effects, when jointly applied. In this direction, only comet assay detected effects after 1 day of exposure, while only ENA assay reflected a concentration-effect relationship as well as the aptitude to reflect temporal variations. Accordingly, Wirzinger et al. (2007) stated previously that both are non-specific biomarkers which reflect different forms of environmental stress, recommending the application of both tests.
Overall, and bearing in mind the persistence of AMPA in water bodies, as well as presented results, the adoption of longer exposures in future studies would be interesting. Moreover, as an attempt to clarify the organisms’ ability to cope with the damage previously inflicted, follow-up studies should be extended to post-exposure periods.
Conclusions
The present findings demonstrated, for the first time in fish, the genotoxicity of AMPA, expressed both as DNA (comet assay) and chromosomal (ENA assay) damage. Overall, AMPA displayed a genotoxic potential comparable to its precursor (glyphosate), bringing to the fore a recent publication of our research group (Guilherme et al. 2012b).
In an attempt to clarify the mechanisms involved in the detected damaging action, the results indicated that AMPA did not induce a marked DNA oxidation. Nevertheless, the use of DNA lesion-specific repair enzymes as an extra step to the standard methodology of comet assay appears as a value added towards an effective assessment of genotoxic hazard.
Finally, it is strongly recommended to include AMPA in futures studies concerning the risk assessment of glyphosate-based herbicides due to its rapid appearance in the water systems and the potential risk to aquatic organisms, namely fish.
References
Al-Rajab A, Amellal S, Schiavon M (2008) Sorption and leaching of 14C-glyphosate in agricultural soils. Agron Sustain Dev 28:419–428
Andrade V, Silva J, Silva F, Heuser V, Dias J, Lu M (2004) Fish as bioindicators to assess the effects of pollution in two southern Brazilian rivers using the comet assay and micronucleus test. Environ Mol Mutagen 468:459–468
Ayllon F, Garcia-Vazquez E (2001) Micronuclei and other nuclear lesions as genotoxicity indicators in rainbow trout Oncorhynchus mykiss. Ecotoxicol Environ Saf 225:221–225
Azqueta A, Shaposhnikov S, Collins A (2009) DNA oxidation: investigating its key role in environmental mutagenesis with the comet assay. Mutat Res 674:101–108
Battaglin WA, Kolpin DW, Scribner EA, Kuivila KM, Sandstrom MW (2005) Glyphosate, other herbicides, and transformation products in midwestern streams, 2002: agricultural hydrology and water quality. American Water Resources Association, Middleburg, VA, ETATS-UNIS
Björklund E, Styrishave B, Anskjær GG, Hansen M, Halling-Sørensen B (2011) Dichlobenil and 2,6-dichlorobenzamide (BAM) in the environment: What are the risks to humans and biota? Sci Total Environ 409:3732–3739
Bolognesi C, Buschini A, Branchi E, Carboni P, Furlini M, Martino A, Monteverde M, Poli P, Rossi C (2004) Comet and micronucleus assays in zebra mussel cells for genotoxicity assessment of surface drinking water treated with three different disinfectants. Sci Total Environ 333:127–136
Borggaard O, Gimsing A (2008) Fate of glyphosate in soil and the possibility of leaching to ground and surface waters: a review. Pest Manag Sci 64:441–456
Cavalcante M, Martinez R, Sofia S (2008) Genotoxic effects of Roundup on the fish Prochilodus lineatus. Mutat Res 655(1–2):41–46
Çavas T, Könen S (2007) Detection of cytogenetic and DNA damage in peripheral erythrocytes of goldfish (Carassius auratus) exposed to a glyphosate formulation using the micronucleus test and the comet assay. Mutagenesis 22:263–268
Collins A (2004) The comet assay for DNA damage and repair. Mol Biotechnol 26:249–261
E.U. (2002) Glyphosate, Endpoints and related information 1.Toxicology and metabolism. European commission health and consumer protection directorate general. Appendix I
Fenech M (2000) The in vitro micronucleus technique. Mutat Res 455:81–95
Feng JC, Thompson DG, Reynolds PE (1990) Fate of glyphosate in a Canadian forest watershed. 2. Persistence in foliage and soils. J Agric Food Chem 38:1118–1125
Giesy J, Dobson S, Solomon K (2000) Ecotoxicological risk assessment for Roundup herbicide. Rev Environ Contam Toxicol 167:35–120
Gimsing AL, Borggaard OK, Bang M (2004) Influence of soil composition on adsorption of glyphosate and phosphate by contrasting Danish surface soils. Eur J Soil Sci 55:183–191
Guilherme S, Válega M, Pereira ME, Santos MA, Pacheco M (2008) Erythrocytic nuclear abnormalities in wild and caged fish (Liza aurata) along an environmental mercury contamination gradient. Ecotoxivol Environ Saf 70:411–421
Guilherme S, Gaivão I, Santos MA, Pacheco M (2010) European eel (Anguilla anguilla) genotoxic and pro-oxidant responses following short-term exposure to Roundup®—a glyphosate-based herbicide. Mutagenesis 25:523–530
Guilherme S, Gaivão I, Santos MA, Pacheco M (2012a) DNA damage in fish (Anguilla anguilla) exposed to a glyphosate-based herbicide—elucidation of organ-specificity and the role of oxidative stress. Mutat Res Genet Toxicol Environ 743:1–9
Guilherme S, Santos M, Barroso C, Gaivão I, Pacheco M (2012b) Differential genotoxicity of Roundup® formulation and its constituents in blood cells of fish (Anguilla anguilla): considerations on chemical interactions and DNA damaging mechanisms. Ecotoxicology 21:1381–1390
Hoang TC, Rand GM, Gardinali PR, Castro J (2011) Bioconcentration and depuration of endosulfan sulfate in mosquito fish (Gambusia affinis). Chemosphere 84:538–543
Kolpin DW, Thurman EM, Lee EA, Meyer MT, Furlong ET, Glassmeyer ST (2006) Urban contributions of glyphosate and its degradate AMPA to streams in the United States. Sci Total Environ 354:191–197
Landry D, Dousset S, Fournier J-C, Andreux F (2005) Leaching of glyphosate and AMPA under two soil management practices in Burgundy vineyards (Vosne-Romanée, 21-France). Environ Pollut 138:191–200
Lee R, Steinert S (2003) Use of the single cell gel electrophoresis/comet assay for detecting DNA damage in aquatic (marine and freshwater) animals. Mutat Res 544:43–64
Mallat E, Barceló D (1998) Analysis and degradation study of glyphosate and of aminomethylphosphonic acid in natural waters by means of polymeric and ion-exchange solid-phase extraction columns followed by ion chromatography–post-column derivatization with fluorescence detection. J Chromatogr A 823:129–136
Mañas F, Peralta L, Raviolo J, García Ovando H, Weyers A, Ugnia L, Gonzalez Cid M, Larripa I, Gorla N (2009) Genotoxicity of AMPA, the environmental metabolite of glyphosate, assessed by the comet assay and cytogenetic tests. Ecotoxicol Environ Saf 72:834–837
Mottier A, Kientz-Bouchart V, Serpentini A, Lebel JM, Jha AN, Costil K (2013) Effects of glyphosate-based herbicides on embryo-larval development and metamorphosis in the Pacific oyster, Crassostrea gigas. Aquat Toxicol 128:67–78
Pacheco M, Santos MA (1996) Induction of micronuclei and nuclear abnormalities in the erythrocytes of Anguilla anguilla L. exposed either to cyclophosphamide or to bleached kraft pulp mill effluent. Fresenius Environ Bull 5:746–751
Pacheco M, Santos MA (1997) Induction of EROD activity and genotoxic effects by polycyclic aromatic hydrocarbons and resin acids on juvenile eel Anguilla anguilla L. Ecotoxicol Environ Saf 252–259
Pacheco M, Santos M (1998) Induction of liver EROD and erythrocytic nuclear abnormalities by cyclophosphamide and PAHs in Anguilla anguilla L. Ecotoxicol Environ Saf 40:71–76
Pacheco M, Santos MA (2002) Biotransformation, genotoxic, and histopathological effects of environmental contaminants in European eel (Anguilla anguilla L.). Ecotoxicol Environ Saf 53:331–347
Palus J, Dziubaltowska E, Rydzynski K (1999) DNA damage detected by the comet assay in the white blood cells of workers in a wooden furniture plant. Mutat Res Genet Toxicol Environ 444:61–74
Rueppel ML, Brightwell BB, Schaefer J, Marvel JT (1977) Metabolism and degradation of glyphosate in soil and water. J Agric Food Chem 25:517–528
Saleha Banu B, Danadevi K, Rahman MF, Ahuja YR, Kaiser J (2001) Genotoxic effect of monocrotophos to sentinel species using comet assay. Food Chem Toxicol 39:361–366
Scalon MCS, Rechenmacher C, Siebel AM, Kayser ML, Rodrigues MT, Maluf SW, Rodrigues MAS, Silva LB (2010) Evaluation of Sinos River water genotoxicity using the comet assay in fish. Braz J Biol 70:1217–1222
Shaposhnikov S, Azqueta A, Henriksson S, Meier S, Gaivão I, Huskisson NH, Smart A, Brunborg G, Nilsson M, Collins AR (2010) Twelve-gel slide format optimised for comet assay and fluorescent in situ hybridisation. Toxicol Lett 195:31–34
Stoiber T, Bonacker D, Bohm K, Bolt H, Thier R, Degen G, Unger E (2004) Disturbed microtubule function and induction of micronuclei by chelate complexes of mercury (II). Mutat Res 563:97–106
Struger J, Thompson D, Staznik B, Martin P, McDaniel T, Marvin C (2008) Occurrence of glyphosate in surface waters of southern Ontario. Bull Environ Contam Toxicol 80:378–384
Udroiu I (2006) The micronucleus test in piscine erythrocytes. Aquat Toxicol 79:201–204
USEPA (1993) Environmental Protection Agency. Re-registration Eligibility Decision (RED): Glyphosate. Washington, DC
Williams GM, Kroes R, Munro IC (2000) Safety evaluation and risk assessment of the herbicide Roundup and its active ingredient, glyphosate, for humans. Regul Toxicol Pharmacol 31:117–165
Wirzinger G, Weltje L, Gercken J, Sordyl H (2007) Genotoxic damage in field-collected three-spined sticklebacks (Gasterosteus aculeatus L.): a suitable biomonitoring tool? Mutat Res 628:19–30
Zar J (1996) Biostatistical analysis. Prentice Hall International Inc, USA
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia (FCT; Government of Portugal) through the Research project PTDC/AAC-AMB/114123/2009 [co-financed by FCT/MCTES in its national budget component (PIDDAC) and by the European Regional Development Fund (ERDF) through COMPETE—Thematic Factors of Competitiveness Operational Programme (POFC)] and the Post-doctoral fellowship SFRH/BPD/88947/2012, as well as by Centre for Environmental and Marine Studies (CESAM).
Ethical statement
This study was conducted in accordance with the EU Directive 2010/63/EU on the protection of animals used for scientific purposes, under the supervision of a team member (Mário Pacheco) authorised by the competent authorities.
Conflict of interest
The authors declare that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Henner Hollert
Rights and permissions
About this article
Cite this article
Guilherme, S., Santos, M.A., Gaivão, I. et al. DNA and chromosomal damage induced in fish (Anguilla anguilla L.) by aminomethylphosphonic acid (AMPA)—the major environmental breakdown product of glyphosate. Environ Sci Pollut Res 21, 8730–8739 (2014). https://doi.org/10.1007/s11356-014-2803-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-014-2803-1