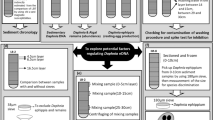
Abstract
Understandings of fish spawning are important for the effective riverine ecosystem and fisheries resources management. Application of environmental DNA (eDNA) analysis would allow fish spawning evaluation at a high temporal and spatial resolution due to its less-labor intensive property, compared to the conventional collection methods. However, the relative performance of the two eDNA-based methods used to evaluate fish spawning, pre and post comparisons of eDNA concentrations and nuclear DNA to mitochondrial DNA ratio, has not been compared in the field. We evaluated whether spawning events of Ayu Plecoglossus altivelis could be identified from day (pre-spawning) to night (post-spawning) changes in eDNA concentrations using nuclear DNA (RAG1, nuDNA) and nuDNA/mitochondrial DNA ratio in comparison to the conventional mitochondrial DNA (cyt b, mtDNA). In addition, the relative sensitivity of nuDNA concentrations in spawning evaluation was compared with mtDNA concentrations. Water samples were collected biweekly in the lower Takatsu River, western Japan, between early October and early December 2020, covering the spawning season of the fish species. Generalized linear mixed-effects models (GLMM) showed that Ayu spawning could be detected by nuDNA concentrations likewise by mtDNA concentrations, while nuDNA/mtDNA ratio could not detect spawning earlier in the spawning season. GLS models showed that the day-to-night increase in the eDNA concentrations was more profound for nuDNA than for mtDNA during the main spawning period of the fish. Our findings insist that the application of nuDNA concentrations better serves as a tool for evaluating Ayu spawning intensity in eDNA analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Conservation and rehabilitation of fish spawning habitats are important aspects of the riverine ecosystem and fisheries stock management (Maitland 1995; Wheaton et al. 2004; Cooke et al. 2012). Since many fish species aggregate around the limited sections within channels where spawning takes place, degradation, loss, or blocking of the migration routes to and from the spawning habitats would negatively affect the within river population of the fish species (Johnston 1999; Allibone et al. 2010). Widely adopted methods for detecting spawning events or evaluating spawning conditions of fishes in streams include above/under water counting of the spawning adults, observations and counting of redds, underwater observations of the egg density in the spawning grounds, and collection of drifting eggs or larvae with plankton nets (Takahashi et al. 1999; Grabowski and Isely 2007; Bylemans et al. 2017; Inui et al. 2018). Direct observations require skilled surveyors for collecting reliable data. The collection methods need to sample the eggs or larvae that may be vulnerable to physical pressure or cause disturbance to the spawning ground during underwater or wading surveys, potentially resulting in the wash down of the laid eggs. In addition, conventional surveys are labor intensive, inhibiting detailed sampling at short temporal or spatial intervals (c.f. Bylemans et al. 2017; Yoshida et al. 2019).
Environmental DNA (eDNA) analysis can provide a less intrusive and less time-consuming alternative for surveying fish spawning. eDNA analysis is a technique that can estimate the presence/absence or relative abundance of the target species from the DNA fragments present in environmental samples, including soil, water, and air (Rees et al. 2014; Barnes and Turner 2016). There are three major steps in environmental DNA analysis; environmental sample collection, DNA extraction from the environmental samples, and PCR of the extracted DNA using species-specific primers. In the case of aquatic samples, the only step done in the field for eDNA analysis is water sampling. This property of eDNA analysis enables non-intrusive surveys of fishes that do not require the physical collection of the target species. In addition, eDNA surveys could be carried out at short temporal intervals or at multiple spawning sites on the same day (c.f. Tillotson et al. 2018; Thalinger et al. 2019; Inui et al. 2021).
Two methods have been proposed for identifying fish spawning events in the field, with the relative abundance of the spawning adults taken into consideration. One relies on the marked increase in eDNA concentrations between pre- and post-spawning events within the same day. This approach has been shown effective for fishes known to spawn in a limited time within a day, Ayu Plecoglossu altivelis (Temminck and Schlegel 1846) (e.g. Inui et al. 2018; Yoshida et al, 2019) and two Medaka species Oryzias latipes and O. sakaizumii (Tsuji and Shibata 2021), based on field surveys. In Ayu, the spawning detection technique was applied to identify spawning sites or the spawning period in the surveyed rivers and to evaluate the relative intensity of spawning among dates, sites, or different time periods on the same day (Yoshida et al, 2019; Saito et al. 2020; Inui et al. 2021). These studies used mitochondrial DNA (hereafter mtDNA) as the target region for primers in PCR. The main reasons include the abundance of DNA copy numbers per cell and the abundance of sequence data available online leading to the relative ease in primer development (Foran 2006; Minamoto et al. 2017; Bylemans et al. 2018; Jo et al. 2020). Another is based on the ratio of nuclear DNA (hereafter nuDNA) and mtDNA. Bylemans et al. (2017) conducted tank experiments simulating spawning by milt supplementation using a gravel spawner, Macquarie Perch Macquaria australasica, as the target species. NuDNA/mtDNA ratio of the fish increased immediately after the simulated spawning and remained detectable for 40–60 h depending on fish density. However, when water samples were taken prior to and during the spawning season of the fish in a river, a relatively high nuDNA/mtDNA ratio was observed only in one of the two spawning period surveys.
To date, the relative sensitivity of the two eDNA-based approaches used for fish spawning detection or evaluation has not been compared in the field. In the approach based on eDNA concentration increase, the use of nuDNA has not been attempted. NuDNA may yield better sensitivity than mtDNA, as copy numbers of nuDNA are fixed among the cells within each individual (Long and Dawid 1980) unlike mtDNA. The number of mitochondria in sperms, the main source of eDNA increase during the spawning events (Tsuji and Shibata 2021), is an order of magnitude smaller compared to the numbers in somatic cells (e.g. Gwo et al. 1994; Moraes 2001; Islam and Akhter 2012). In the approach using nuDNA/mtDNA ratio, further refinements may be needed to accomplish stable detection of spawning events in the field.
Ayu can be a great target species to compare and validate the two eDNA-based methods used for fish spawning surveys in the field. Ayu has a high economic value as a fisheries resource and as a target species for recreational fishing in Japan (Takahashi and Azuma, 2016; Ministry of Agriculture, Forestry and Fisheries 2021), making this species especially important for stock management within rivers and streams. Ayu is a diadromous fish species found in coastal streams and seas in East Asia, distributed from northern Vietnam in the south to Hokkaido in Japan in the north (Nishida 2005; Tran et al. 2017). During the growing season in freshwater habitats, Ayu feeds on benthic algae during the daytime and rests in slowly flowing areas around boulders or in pools during the night (Miyadi 1960; Takahashi 2009). In the fall, the entire population of Ayu migrate downstream to the lower reaches for spawning (Iguchi et al. 1998; Lucas et al. 2001) and die there by the end of the spawning season (Nishida 2005). Spawning usually takes place in the evening to earlier in the night in gravel beds located in riffles (Ishida 1961; Takahashi and Azuma 2016; Yoshida et al. 2018), meaning that the amount of eDNA containing materials released from the fish individuals, dominantly the sperms, increase day to night near and downstream of the spawning site.
The present study aimed to compare the relative detectability of fish spawning between the two eDNA-based approaches and the relative sensitivity of nuDNA compared to that of mtDNA in spawning evaluation using Ayu as the target species. We carried out water sampling surveys covering the Ayu spawning season in the Takatsu River, western Japan. Pre- (daytime) and post-spawning (nighttime) concentrations of Ayu eDNA were quantified for mtDNA and nuDNA. Then, nuDNA/mtDNA ratio was calculated to evaluate the relative performance of mtDNA concentrations, nuDNA concentrations, and nuDNA/mtDNA ratio in the spawning detection of Ayu using generalized linear models or generalized linear mixed-effects models. The relative sensitivity of nuDNA in comparison to mtDNA for spawning evaluation was tested by constructing generalized least squares models and carrying out the model selection.
Materials and methods
Field surveys
Takatsu River flows from the western Chugoku Mountains and pours into the Sea of Japan at Masuda, Shimane Prefecture, western Japan (Fig. 1). The main channel length is 81 km, with a catchment area of 1090 km2. The Ayu population in this river is comprised of wild and stocked fish (Terakado et al. 2016). Most of the weirs and low-head dams in the lower to middle reaches of the Takatsu River are low enough or are equipped with functioning fishways so many of the Ayu could ascend the obstacles during its upstream migration from the sea or the released site, and vice versa in the fall prior to spawning (Hamano 2016; Terakado et al. 2016). In recent years, Ayu stocked in the Takatsu River do not include seedlings originating from the Lake-Biwa population (A. Okino and H. Terakado, personal communication) that has a spawning season starting about one month earlier than the wild Ayu in western Japan (Nishida 2005). The sampling sites (St.a–c) were set in the riffles of the three major spawning sites in the Takatsu River (Terakado et al. 2015; Inui et al. 2021; Figs. 1, 2).
Environmental DNA concentrations reflect fish biomass in the reaches upstream of the sampled sites at differing ranges depending on discharge (e.g. Wilcox et al. 2016). Key factors affecting the dynamics of eDNA-containing materials include settling, diffusion, advection, and resuspension (Barnes and Turner 2016). The distances between the sampling sites in this study were approximately 1 km, which is thought to be within the range of downstream transport of the eDNA-containing materials in previous studies undertaken in rivers (e.g., Pont et al. 2018; Mizumoto et al. 2022). Dynamics simulations in Kobayashi et al. (2021) showed that in the lower Takatsu River, a large proportion (ca. 95%) of eDNA-containing materials (mucus and feces) released by Ayu in the riffles and runs settled in deep pools located within ca. 1 km downstream under base-flow conditions (discharge < 32.5 m3/s). Saito et al. (2020) conducted eDNA surveys using mtDNA primer–probe set to compare daytime to nighttime increases between one hour and three hours after the sunset during the Ayu spawning season in 2019, at the same survey sites as the present study. Daytime to the nighttime increase observed in an upper reach site was not always observed similarly at the site located downstream, 1 h and 3 h after sunset. Based on these reports, we assumed that in the Takatsu River, the effects of eDNA release in the upstream sites were reasonably small, compared to the eDNA released in the riffle where water samples were taken.
Biweekly water samplings for Ayu eDNA analysis were done at the three sites in the lower Takatsu River (Figs. 1, 2). The sampling period was between October 1, 2020, and December 1, 2020 (Fig. 3), covering the spawning season of Ayu in the river (Terakado et al. 2015; Inui et al. 2021; Shimane Prefectural Fisheries Technology Center 2022). Three liters of surface water were taken with a bucket or a dipper near the lower end of riffles in each sampling site twice a day; once in the daytime (two to five hours before sunset, hereafter daytime samples), and once in the nighttime. Nighttime water sampling was conducted between one and two hours after sunset (hereafter nighttime samples) in this study, as the increase in eDNA concentration due to spawning may cease by three to four hours after sunset, especially on cloudy days (Saito et al. 2020). The three-liter surface water was dispensed into three separate one-liter plastic bags with screw caps (DP16-TN1000, Yanagi, Nagoya, Japan). Next, 1 ml of benzalkonium chloride (Takeda Pharmaceutical, Osaka, Japan; final concentration in the samples: 0.01% w/v) was added to each bag sample and mixed well for suppressing potential DNA breakdown caused by microbial activities (Yamanaka et al. 2017). To minimize the mechanical breakdown of eDNA, the packed water samples were filtered on-site using a portable water pump (DP0105-XF-0001j, Nitto Kohki, Tokyo, Japan) and filter cartridges (PP-47, ADVANTEC, Tokyo, Japan) equipped with GF/F fiberglass filters (φ: 47 mm, pore size: 0.7 μm; GE Healthcare Japan) (c.f. Yamanaka et al. 2016). One liter of water sample was filtered with each fiberglass filter. After filtration, each fiberglass filter was immediately frozen by sandwiching the wrapped filters with ice packs capable of maintaining − 16 °C degrees for over 16 h (Neo-Ice, Kawai-Giken, Ichinomiya, Japan). In the laboratory, filter samples were stored in a freezer at − 20 °C until eDNA extraction. One liter of deionized water was treated with benzalkonium chloride on-site and then filtered in the same manner as the remaining samples to check for potential Ayu DNA contaminations. The distilled water samples serve as filtration and DNA extraction negative controls but only partly as field negative controls, as the bottles were not opened at the position of water sampling. All the non-disposable water sampling and filtration equipment were bleached (final concentration: 5% v/v) and then thoroughly rinsed with DNA-free deionized water before usage. An exception was the water sampling bucket, which was covered with a new plastic bag prior to sampling at a different site.
Discharge and water temperature in the lower Takatsu River (above) and the tidal difference at Hamada, a city ca. 35 km northeast of the surveyed sites (below). Vertical lines show each survey date. Spring tides are shown with bold horizontal lines, accompanied by the full moon or new moon symbols (see Fig. 1).
Water temperature was measured on the sampling occasions and recorded at St.a (Fig. 1) every 15 min by a temperature logger (Water Temp Pro v.2, Onset, Bourne, MA, USA). Discharge data for the surveyed reaches were calculated from depth measured hourly at two nearby gauge stations (Ministry of Land, Infrastructure, Transport and Tourism 2021) and the H-Q equation for each gauge station. Tidal difference data for Hamada (a city located ca. 30 km northeast of the river mouth of the Takatsu River) were taken from an online archive (Japan Meteorological Agency, 2021).
eDNA analysis
Prior to eDNA extraction, fiberglass filters were pre-centrifuged at 5000g for 1 min to minimize the amount of benzalkonium chloride remaining in the filters that reduces fish eDNA yield (Tsuji et al. 2022). eDNA were then extracted from the filters using a DNA extraction kit (DNeasy Blood & Tissue Kit; QIAGEN, Hilden, Germany) and Salivette tubes (Sarstedt, Nümbrecht, Germany), following Tsuji et al. (2022) and the manufacturer’s instructions, as follows. Each fiberglass filter was placed in a separate Salivette tube, treated with 200 μl Buffer AL and 20 μl proteinase K solution, and then incubated at 56 °C for 30 min. Next, The Salivette tubes with filters were centrifuged at 5000g for 5 min. After adding 220 μl Tris–EDTA (TE) buffer to the filter, the samples were re-centrifuged at 5000g for 1 min. Following the addition of 200 μl ethanol, the solution was transferred to a spin column. The solution in a spin column was purified following the manufacturer’s instructions for DNeasy Kit (QIAGEN). At last, total eDNA was eluted in 100 μl buffer AE. The extracted eDNA samples were stored at -20 °C until running PCR.
The copy numbers of Ayu eDNA were quantified using two species-specific primer–probe sets targeted to mtDNA (cytochrome b, cyt b; Yamanaka and Minamoto 2016) or nuDNA (recombination activating gene 1, RAG1; Tsuji et al. in prep.). We initially attempted to develop a species-specific primer–probe set for Ayu targeting multi-copy region nuDNA. However, of the target regions tested (internal transcribed spacer 1, ITS1; ITS2; 18S ribosomal RNA gene; 5.8S), none qualified to be species-specific, due to low inter-specific variations and high GC content of the sequence, especially for the ITS regions. Thus, an already available nuDNA primer–probe set targeting a single copy region, RAG1 (López et al. 2004), was used instead (Tsuji et al. in prep.). Species specificity of the nuDNA primer–probe set was tested by running PCR on a web-based DNA database, primer BLAST (National Center for Biotechnology Information, National Institute of Health; https://www.ncbi.nlm.nih.gov/). Next, PCR was run using DNA extracted from tissues of Ayu and two Osmerifoms, the Japanese Smelt Hypomesus nipponensis McAllister, 1963 and the Japanese Ice Fish Salangichthys microdon (Bleeker 1860). Both species belong to sister families of Plecoglossidae (López et al. 2004) and are the only Osmeriform species found in rivers and streams around the Takatsu River (Hamano, 2016). In both tests, only the Ayu DNA was amplified, confirming the species specificity of the nuDNA primer–probe set developed by Tsuji et al. (in prep.).
Quantitative PCR was performed in triplicate using a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Plasmid DNA that contains the target region at pre-set concentrations (3 × 101–105 copies per reaction for mtDNA and 3 × 101–104 copies per reaction for nuDNA, respectively) and PCR negative controls (ultrapure water) were also amplified in all qPCR runs. All reactions were performed in a 15-μL total volume, comprising of 1 μl primer–probe mix (900 nM of primers and 125 nM probe), 7.5 μl TaqMan Environmental Master Mix 2.0 (Life Technologies, Carlsbad, CA, USA), 0.1 μl AmpErase Uracil N-Glycosylase (UNG; Thermo Fisher Scientific, Waltham, MA, USA), 4.4 μL sterilized water (Nacalai Tesque, Kyoto, Japan), and 2 μL eDNA solution. The qPCR conditions were as follows: UNG incubation at 50 °C for 2 min, denaturation at 95 °C for 10 min, followed by 55 cycles of 95 °C for 15 s and 60 °C for 60 s. The eDNA concentration of each sample was the average of the triplicate estimated based on the standard line constructed from the results of the standard samples. The efficiency of qPCR ranged 85.54–101.79% (Supplementary Table 1). The R2 values of the standard lines were 1.00 in all the quantitative PCR runs (Supplementary Table 1). Ayu DNA was not detected from any of the PCR negative controls. However, Ayu DNA was detected from the filtration and DNA extraction negative control samples at concentrations of 0–29 copies/μl (see Supplementary Table 2). Therefore, eDNA concentrations of the samples were calibrated by subtracting the average eDNA concentrations of the corresponding negative control sample.
Statistical analyses
Daytime and nighttime mtDNA concentrations were compared, survey date separately, using generalized linear mixed-effects models (GLMM, Bolker et al. 2009), to check if spawning events likely have taken place on that day. In GLMM, mtDNA was used as the response variable, whereas sampling hour (day/night) was used as the explanatory variable. Since three replicates of water samples were taken at the same position in each site, data from the same site are likely to be correlated, violating the independence assumption in generalized linear models (GLM) (Zuur et al. 2009). Here, both within-site correlation and site-specific trends of the response variable were incorporated into the model as site-specific random intercepts (Pinheiro and Bates 2000). The probability distribution of the response variable was taken to be following gamma distribution with the log link function. If the effect of the night is significant with positive coefficients, the corresponding survey date was designated as the “main spawning period” in the following analyses. Daytime and nighttime nuDNA concentrations were compared, sampling date separately, as in mtDNA. NuDNA/mtDNA ratio was compared day to night using GLM with quasi-binomial distribution, as the inclusion of the site-specific random intercept did not resolve overdispersion in GLMM based on binomial probability distribution with the logit link function.
Next, models were constructed with generalized least squares (GLS) to evaluate the relative sensitivity of nuDNA concentrations for Ayu spawning surveys compared to mtDNA. GLS is an extension of ordinary linear models (LM) that can account for heterogeneity in the response variable (here, variance increases in accordance with mtDNA concentration increase; Pinheiro and Bates 2000). GLM or GLMM were not used here, as an increase in mtDNA concentration is likely to be linearly reflected in nuDNA concentration, resulting in a normally distributed error structure when fitted with LM. The response variable was the nuDNA concentrations, whereas the explanatory variables were mtDNA concentration, day/night, water temperature, and whether the sampling date was in the “main spawning period” or not. The first-order interaction terms between the mtDNA concentrations and all the remaining explanatory variables were also used in the full model. The presence of heterogeneity was checked by comparing the AIC of the GLS and LM that included all the explanatory variables (Zuur et al. 2009). The absence of multicollinearity was also checked prior to the analyses based on the correlation coefficients between the explanatory variables (r < 0.7) and the variance inflation factor (VIF; VIF < 10, Supplementary Table 3). An explanatory variable was taken to be affecting nuDNA concentrations if that variable was in the best model selected after best-subset model selection based on AIC, with a significant Wald test statistic based on t-distribution for that valuable. All statistical analyses were carried out using R ver. 4.1.1 (R Core Team 2021) with the packages car, lme4, MuMIn, and nlme.
Results
Environmental conditions
The water temperature within the surveyed reaches ranged 7–23 °C from September to December (Fig. 3). By mid-October, water temperature became constantly lower than 20 °C throughout the day. Water temperature of the three sampling sites showed similar values on each survey date. Flooding events, a trigger for downstream migration of Ayu prior to spawning (Iguchi et al. 1998; Takahashi and Azuma 2016), were observed in September (Fig. 3). Since no typhoons approached or passed close by to the surveyed region in 2020, no other major floods occurred during the surveyed period.
Day to night changes in each index
In a nighttime sample collected at St.b on October 15, an Ayu larva was trapped on a fiberglass filter. Thus, the sample was omitted from the further analyses. Results of GLMM showed that Ayu mtDNA concentrations (c.f. Supplementary Table 1) significantly increased from day to night on two out of the five sampling dates (October 15 and November 15; Fig. 4, Table 1; raw data and efficacy of each PCR run shown in Supplementary Tables 1, 2). On November 2, when data for St.c were excluded from the analysis, nighttime mtDNA concentrations were shown to be significantly higher than the daytime concentrations (Table 1). Therefore, samplings between October 15 and November 15 were designated as the “main spawning period” in the following analyses. Similarly, results of GLMM showed that nuDNA concentrations rose significantly from day to night on October 15, November 2, and 15, while not on October 2 (Fig. 4; Table 2). GLMM based on gamma distribution could not be constructed for the survey on December 1, due to many observations of zeros (c.f. Supplementary Table 1). NuDNA/mtDNA ratio rose significantly from day to night on November 2 and 15, but not on October 2, 15, and December 1 (Fig. 4; Table 3).
Day-night comparisons of mitochondrial DNA concentrations (left), nuclear DNA concentrations (center), and nuclear DNA/mitochondrial DNA ratio (right) for Ayu in the lower Takatsu River. Bars and plots show the means of the three 1-L water samples at each site and values of individual water samples, respectively
Relationships between mtDNA and nuDNA concentrations
Data from December 1 were not used here because of low nuDNA concentrations, indicating that most of the adults have died after spawning by then. GLS models with constant variance increase in accordance with the mtDNA concentration increase showed lower AIC than the LM (ΔAIC = 58.5). Thus, the variance term was applied to all the models in the subsequent analyses. The best model included all the explanatory variables except for the interaction term between water temperature and mtDNA concentrations. No models were within 2 ΔAIC from the best model. The Wald test statistics were significant for all the variables selected in the best model except for an intercept term, October 2. The results of the model selection and the Wald test statistics indicated that all the selected variables are essential for explaining nuDNA-mtDNA relationship, whereas the contribution of water temperature to the relationship is marginal. Inspecting the slope terms, a positive relationship was found between mtDNA and nuDNA concentrations. The effect of the interaction term between mtDNA concentrations and night was also positive, showing that the day-to-night increase in eDNA concentrations was more profound in nuDNA than in mtDNA (Fig. 5). The effect of the interaction term between mtDNA and October 2 was negative, indicating that the relative increase in nuDNA concentrations compared to mtDNA was greater in the “main spawning period”, which was set as the baseline in the analysis.
Relationships between mitochondrial DNA concentrations and nuclear DNA concentrations. Left: day, right: night. The black and gray lines show the fitted trends of nuclear DNA concentrations based on the best generalized least squares model (Table 4) for the main spawning period (October 15–November 15) and October 2, respectively. Note that the range of the x and y-axis (upper right) differs between the two graphs due to higher mitochondrial and nuclear DNA concentrations in the nighttime
Discussion
Our results showed that day/night comparison of nuDNA concentrations is effective for detecting Ayu spawning events as in the conventional method based on mtDNA concentrations. An increase in fish eDNA concentrations from daytime to nighttime can be due to a simple increase in fish abundance or fish activity levels rather than spawning, but it is unlikely in Ayu. Ayu is a diurnal fish species that feed on benthic algae growing on the rocky substrate during the daytime and rests at the night (Miyadi 1960; Takahashi 2009), so the fish activity level is considerably lower at the night (Minh-Nyo et al. 1991). Spawning takes place mainly in riffles from evening to early night, while fish not ready for spawning stay in slowly flowing areas near the spawning sites (Nishida 2005; Takahashi 2009). Consequently, it is reasonable to assume that the increase in eDNA concentrations from daytime to nighttime is due to spawning behavior.
Next, we consult the validity of day–night comparisons of eDNA concentrations for identifying spawning events based on comparisons with collection survey results. Collection of drifting larvae near the spawning site is a widely adopted method to survey the stock status of Ayu, including the spawning period and relative intensity of spawning (e.g. Tago 1999; Tran et al. 2018). In the Takatsu River, drifting Ayu larvae were collected biweekly between October and December in 2020 at St.a, with a clear peak in early- to mid-November (Shimane Prefectural Fisheries Technology Center 2022). It takes ca. 10–14 days for Ayu larvae to hatch, with its duration dependent on water temperature (Kashiwagi et al. 1986). Consequently, the peak in Ayu spawning in the Takatsu River in 2020 is estimated to be between mid-October to early November. The survey dates with significant day/night increase in nuDNA concentrations (surveys between October 15 and November 15) covered the peak drifting period of Ayu larvae, as well as the trend shown by mtDNA (Fig. 4; Tables 1, 2). Based on the general ecology of Ayu and temporal concordance of relative abundance between drifting larvae collection and the eDNA surveys, it is reasonable to convey that day to night increase of both mtDNA and nuDNA concentrations serve as indices for Ayu spawning.
Ayu may not use all the spawning sites at the same intensity throughout the spawning season, due to changes in fish abundance levels, fish sizes, fish conditions, and the physical environment of the substrate (Ishida 1964; Aizawa 2012; Terakado et al. 2015). Inferring from mtDNA and nuDNA concentrations, fish abundance level and spawning intensity tended to be higher in St.b compared to the other two sites (Fig. 4). St.b is the most intensely used spawning site in recent decades (Terakado et al. 2015). Although we lack quantitative data, the substrate of St.b was dominated by gavels and pebbles, the suitable particle sizes for the Ayu’s spawning site (Ishida 1961; Takahashi and Azuma 2016; Yoshida et al. 2018), which likely have contributed to higher fish abundance and spawning intensity at St.b. On November 2, daytime mtDNA and nuDNA concentrations were higher than the nighttime at St.c (Fig. 4). At St.c, the largest number of the predatory birds within the survey period (more than a dozen of egrets Egretta spp. and the Great Cormorant Phalacrocorax carbo, respectively) were sighted inside the channel during the daytime. The abundance of the predatory birds, in addition to the precipitations, might have increased daytime fish DNA shedding at St.c on November 2. Although eDNA concentration rose day to night on November 15 (Fig. 4; Tables 1, 2), the number of the larvae collected two weeks later (the estimated hatching period of the eggs laid in mid-November) was 1/3 to 1/4 of the peak period for (Shimane Prefectural Fisheries Technology Center 2022). The results could be explained by the Ayu stock enhancement activities in the surveyed river. In the Takatsu River, spawning adults were released in St.b late in the spawning period (November 9–10, 2020) (Takatsu River Fisheries Cooperative Association, personal communication). During the field surveys on November 15, the school of Ayu and its spawning activities were observed in St.b. Hence, the released fish individuals, in addition to the wild Ayu individuals and their spawning activities, likely have contributed to increases in eDNA concentrations on November 15 at St.b.
NuDNA/mtDNA ratio was shown capable of detecting Ayu spawning events on November 2 and 15, but not on October 15 (Fig. 4; Table 3) within the “spawning period”. On October 15, nuDNA/mtDNA ratio did not rise day to night, but when inspected separately, both nuDNA and mtDNA concentrations rose day to night on this survey date (Tables 1, 2). Possible sources of night-specific Ayu eDNA increase other than spawning adults include hatching larvae and eggs, carcasses, and spermatozoa and eggs released during the spawning. Hatching occurs soon after sunset in Ayu (Tago 1999; Takahashi and Azuma 2016). However, both mtDNA and nuDNA concentrations were low in the survey on December 1 (Fig. 4), when eggs lain ca. two weeks earlier are hatching (c.f. Kashiwagi et al. 1986). Since mid-November recorded high eDNA concentrations (Fig. 4), it is unlikely that the eggs and hatching larvae contributed greatly to mtDNA increase during the night. Carcasses are also thought to have a small contribution in the present study, unlike in Sockeye Salmon Oncorhynchus nerka (Tillotson et al. 2018), considering that only a few dead Ayu were spotted near the riverbanks during the field surveys. Therefore, spermatozoa released during the spawning events, the major source of eDNA increase during and soon after the spawning (Bylemans et al. 2017; Tsuji and Shibata 2021) are of the primary concern.
In Ayu, larger individuals and males mature earlier and migrate downstream earlier to the spawning grounds located in the lower reaches (Iguchi et al. 1998; Takahashi and Azuma 2016). This result in larger fish sizes and a more male-biased sex ratio around the spawning ground in the early spawning season compared to the later period (Ishida 1959; Tachihara and Kimura 1991; Takahashi and Azuma 2016). Due to the biased sex ratio, only a part of the males are expected to participate successfully in spawning, insisting that the proportion of the males releasing spermatozoa is also limited earlier in the spawning season. The relative abundance of larger individuals would also yield a less profound daytime to nighttime increase in nuDNA/mtDNA ratio in the earlier spawning season (Fig. 4; Table 3). The number of mitochondria per cell decrease with aging/ increase in body sizes (Moraes 2001; Hartman et al. 2011). Hence, shedding rates of mtDNA per unit weight tend to be smaller in larger individuals within the same species (Maruyama et al. 2014; Takeuchi et al. 2019). Jo et al. (2020) demonstrated that in the Japanese Jack Mackerel Trachurus japonicus, mtDNA (cyt b)/nuDNA (ITS1) ratio is lower in the tank carrying larger-sized individuals. Since the baseline daytime nuDNA/mtDNA ratio is thought to be higher because of larger fish sizes, an increase in nuDNA/mtDNA ratio from daytime to nighttime would likely be less profound, even if spawning events were taking place between the daytime and nighttime water samplings. Limited sperm release earlier in the spawning season and/or relative abundance of larger fish could have hampered the increase in nuDNA concentrations from daytime to nighttime compared to mtDNA concentrations, making nuDNA/mtDNA ratio not a good indicator for Ayu spawning earlier in the spawning period in the present study.
NuDNA concentrations were shown to be a more sensitive indicator for Ayu spawning events compared to the conventional mtDNA. Although the concentrations of single-copy region nuDNA were lower than those of mtDNA, the magnitude of day/night increase was greater in nuDNA than in mtDNA during the “main spawning period” (Fig. 5; Table 4). Such property of nuDNA would be beneficial for detecting spawning events and evaluating spawning intensity when day/night rise in mtDNA is small. In fact, such a situation was observed at St.c on November 15 in the present study (Fig. 4). The fact that the interaction term between mtDNA concentrations and water temperature was not selected in the best GLS model indicates that the nuDNA-mtDNA relationship does not differ substantially among the dates during the “main spawning period”. Thus, nuDNA concentrations can be used to compare the relative intensity of Ayu spawning among dates.
Comparison of pre/post-spawning eDNA levels is the key concept in the spawning detection adopted in the present study (c.f. Inui et al. 2018; Tsuji and Shibata 2021). When adopting the approach, several criteria must be met to make the comparison valid. First, the timing of spawning within a day needs to be identified. If the species spawn throughout a day, it will be challenging to make pre/post-spawning comparisons of eDNA concentrations. Second, a suitable spawning environment for the target species needs to be identified at the channel unit scale. Third, fish movement among channel units within a day needs to be observed to confirm that the population density of the target fish does not increase substantially around the spawning ground for reasons other than spawning. If these prerequisites were met/ ecological information was available, the water sampling position can be set immediate downstream of the spawning sites to detect an acute increase of mtDNA and nuDNA concentrations resulting from spawning. Screening of fish spawning using mtDNA concentrations will be beneficial, in case the spawning site or timing of spawning within a day has not been identified in the waterbody to be surveyed. Time-efficient characteristics of eDNA surveys in the field allow multiple longitudinal or temporal replicates of water samples to be acquired within a day.
Ayu forms spawning aggregations and releases spermatozoa in the flowing water column (Nishida 2005). Therefore, detection of spawning was thought to have been relatively easy, as has been depicted by constant detection of single-copy nuDNA in our study except for early December when the majority of the Ayu was thought to have died following spawning (Fig. 4). Spawning surveys based on pre/post nuDNA or mtDNA concentrations are expected to be widely adaptable for other fish species that fulfill the aforementioned criteria. Other gravel spawners, including Salmonidae and part of Cypriniade, are major candidate species for future spawning studies using eDNA analysis. The applicability of the approach to nest spawners, including Bagridae catfish, Cottidae, and Gobiidae, will be a valuable topic for future studies.
Within the nuclear DNA, the multi-copy region is present in fixed numbers of multiple copies per cell, irrespective of cell types or body conditions within each individual (López et al. 2004). In contrast, the single-copy region nuDNA is present in one copy in every cell (c.f. Long and Dawid 1980). The present study used the single-copy region in nuDNA as the target region for the primer–probe set (Tsuji et al. in prep.), due to the difficulty in designing a species-specific primer targeting the multi-copy region nuDNA. Ayu inhabits an entire stretch of rivers except for headwaters until summer and by fall, a predominant proportion of the fish migrate downstream to spawn in the riffles located in the lower reaches (Iguchi et al. 1998; Nishida 2005; Kono et al. 2017). Owing to this life-history trait, Ayu eDNA concentrations in the lower reaches become considerably higher in the fall than in other seasons (Doi et al. 2017; Kono et al. 2017; Yoshida et al. 2019), likely contributing to the good detection performance of single-copy region nuDNA for Ayu spawning in the present study. In addition, the day-to-night increase in nuDNA was more profound for nuDNA compared to mtDNA. Multi-copy region nuDNA serves as a more sensitive indicator of the fish presence or abundance in terms of detectability, sensitivity, and a higher spatial and temporal resolution because of the faster degrading rate over mtDNA or single-copy region nuDNA (Bylemans et al. 2018; Dysthe et al. 2018; Jo et al. 2020, 2021). If multi-copy region nuDNA was used, an increase in nuDNA concentrations from day to night is expected to be more profound than the mtDNA or single-copy region nuDNA used in the present study due to the greater numbers of DNA copies per cell. Thus, the application of multi-copy region nuDNA for Ayu spawning evaluation is worth consideration in future studies.
The present study compared the relative performance of nuDNA concentrations and nuDNA/mtDNA ratio in spawning detection of Ayu, in comparison with the conventional method based on mtDNA concentrations. NuDNA concentrations rose day to night during the “main spawning period”, similar in a manner to mtDNA. The results indicated that day/night comparisons of nuDNA concentrations are also valid for detecting Ayu spawning events. In contrast, nuDNA/mtDNA ratio failed to detect spawning during the early spawning period but detected spawning in the mid- to late-spawning season. The relatively small increase in nuDNA concentrations from daytime to nighttime compared to mtDNA concentrations in the earlier spawning season could have been caused by the relative abundance of larger individuals and the male-biased sex ratio during this period. The relative day/night increase of nuDNA concentrations compared to mtDNA concentrations was greater during the “main spawning period” of the fish, showing higher sensitivity of nuDNA concentrations in Ayu spawning evaluation. The higher sensitivity of nuDNA would be beneficial for evaluating Ayu spawning intensity in rivers and streams with low Ayu density, especially in the rivers with their Ayu spawning habitat degraded by anthropogenic alterations, including alterations to bed transport and flow regime, and blocking of migration routes to the spawning grounds. Future examination using multi-copy region nuDNA instead of the single-copy region nuDNA used in the present study will likely enhance the applicability of nuDNA-based evaluation of Ayu spawning using eDNA analysis.
References
Aizawa Y (2012) Distribution of the spawning ground and spawning season of Ayu Plecoglossusαltivelis in the Tamagawa River. Bull Kanagawa Prefect Fish Tech Cent 5:15–20 (in Japanese with English abstract)
Allibone R, David B, Hitchmough R et al (2010) Conservation status of New Zealand freshwater fish, 2009. New Zeal J Mar Freshw Res 44:271–287
Barnes MA, Turner CR (2016) The ecology of environmental DNA and implications for conservation genetics. Conserv Genet 17:1–17
Bolker BM, Brooks ME, Clark CJ et al (2009) Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol 24:127–135
Bylemans J, Furlan EM, Hardy CM et al (2017) An environmental DNA-based method for monitoring spawning activity: a case study, using the endangered Macquarie Perch (Macquaria australasica). Methods Ecol Evol 8:646–655
Bylemans J, Furlan EM, Gleeson DM et al (2018) Does size matter? An experimental evaluation of the relative abundance and decay rates of aquatic environmental DNA. Environ Sci Technol 52:6408–6416
Shimane Prefectural Fisheries Technology Center (2022) The Reiwa 2nd annual report of Shimane Prefectural Fisheries Technology Center (in Japanese)
Cooke SJ, Paukert C, Hogan Z (2012) Endangered river fish: factors hindering conservation and restoration. Endanger Species Res 17:179–191
Doi H, Inui R, Akamatsu Y et al (2017) Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw Biol 62:30–39
Dysthe JC, Franklin TW, McKelvey KS et al (2018) An improved environmental DNA assay for Bull Trout (Salvelinus confluentus) based on the ribosomal internal transcribed spacer. PLoS One 13:1–11
Foran DR (2006) Relative degradation of nuclear and mitochondrial DNA: an experimental approach. J Forensic Sci 51:766–770
Grabowski TB, Isely JJ (2007) Spatial and temporal segregation of spawning habitat by Catostomids in the Savannah River, Georgia and South Carolina, U.S.A. J Fish Biol 70:782–798
Gwo JC, Lin XW, Kao YS et al (1994) The ultrastructure of Ayu Plecoglossus altivelis spermatozoon (Teleostei, Salmonifomes, Plecoglossidae). J Submicrosc Cytol Pathol 26:467–472
Hamano (ed) (2016) Mizube-no-kowaza, 2nd edn. River Section, Department of Civil Engineering & Architecture, Yamaguchi Prefectural Office, Yamaguchi (in Japanese)
Hartmann N, Reichwald K, Wittig I et al (2011) Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell 10:824–831
Iguchi K, Ito F, Yamaguchi M et al (1998) Spawning downstream migration of Ayu in the Chikuma River. Bull Natl Res Inst Fish Sci 11:75–84 (in Japanese with English abstract)
Inui R, Takahashi I, Goto M et al (2018) Monitoring the use of artificial spawning grounds for Plecoglossus altivelis altivelis at the Nahari River—focusing on the comparison between visual survey and environmental DNA analysis. Adv Riv Eng 24:333–338 (in Japanese with English abstract)
Inui R, Akamatsu Y, Kono T et al (2021) Spatiotemporal changes of the environmental DNA concentrations of amphidromous fish Plecoglossus altivelis altivelis in the spawning grounds in the Takatsu River, western Japan. Front Ecol Evol 9:1–12
Ishida R (1959) On the spawning of the Ayu, Plecoglossus altivelis T. & S.—I. Structure of the spawning shoal and spawning behaviour. Bull Jpn Soc Sci Fish 25:259–268 (in Japanese with English abstract)
Ishida R (1961) On the spawning of the Ayu, Plecoglossus altivelis T. & S.—II. Relationships between the size of spawners and the size of gravels of the spawning bed. Bull Jpn Soc Sci Fish 27:1052–1056 (in Japanese with English abstract)
Ishida R (1964) On the spawning of the Ayu, Plecoglossus altivelis T. & S.—IV. Distribution and some geographical features of the spawning ground. Bull Jpn Soc Sci Fish 30:478–485 (in Japanese with English abstract)
Islam MS, Akhter T (2012) Tale of fish sperm and factors affecting sperm motility: a review. Adv Life Sci 1:11–19
Japan Meteorological Agency (2021) Miscellaneous data and information. https://www.jma.go.jp/jma/menu/menureport.html/. Accessed 21 Mar. 2021
Jo T, Arimoto M, Murakami H et al (2020) Estimating shedding and decay rates of environmental nuclear DNA with relation to water temperature and biomass. Environ DNA 2:140–151
Jo T, Takao K, Minamoto T (2022) Linking the state of environmental DNA to its application for biomonitoring and stock assessment: targeting mitochondrial/nuclear genes, and different DNA fragment lengths and particle sizes. Environ DNA 4:1–13
Johnston CE (1999) The relationship of spawning mode to conservation of North American minnows (Cyprinidae). Environ Biol Fishes 55:21–30
Kashiwagi M, Iwai T, Yamamoto H et al (1986) Effects of temperature and salinity on egg hatch of the Ayu Plecoglossus altivelis. Bull Fac Fish Mie Univ 13:17–24
Kobayashi K, Akamatsu Y, Nakao R (2021) Examinations on spatial range where fish biomass can be grasped by environmental DNA concentrations based on fluvial material dynamics simulations. J Jpn Soc Civil Eng Ser B1 (hydraul Eng) 77:I_553-I_558 (in Japanese with English abstract)
Kono T, Akamatsu Y, Goto M et al (2017) Quantification of Plecoglossus altivelis using environmental DNA and trial of monitoring of downstream migration. Adv Riv Eng 23:669–674 (in Japanese with English abstract)
Long EO, Dawid IB (1980) Repeated genes in eukaryotes. Annu Rev Biochem 49:727–764
López JA, Chen WJ, Ortí G (2004) Esociform phylogeny. Copeia 2004:449–464
Lucas MC, Baras E, Thom TJ et al (2001) Migration of freshwater fishes. Blackwell Science, Oxford
Maitland PS (1995) The conservation of freshwater fish: past and present experience. Biol Conserv 72:259–270
Maruyama A, Nakamura K, Yamanaka H et al (2014) The release rate of environmental DNA from juvenile and adult fish. PLoS One 9:1–13
Minamoto T, Uchii K, Takahara T et al (2017) Nuclear internal transcribed spacer-1 as a sensitive genetic marker for environmental DNA studies in common carp Cyprinus carpio. Mol Ecol Resour 17:324–333
Minh-Nyo M, Tabata M, Oguri M (1991) Circadian locomotor activity in Ayu Plecoglossus altivelis. Nippon Suisan Gakkaishi 57:979–979
Ministry of Agriculture, Forestry and Fisheries (2021) The Reiwa 2nd census of fisheries. https://www.maff.go.jp/j/tokei/kouhyou/kensaku/bunya6.html/. Accessed 27 Jan. 2022
Ministry of Land, Infrastructure, Transport, and Tourism (2021) Water Information System. http://www1.river.go.jp/. Accessed 21 Mar. 2021
Miyadi D (1960) Stories of Ayu. Iwanami Shoten Publishers, Tokyo (in Japanese)
Mizumoto H, Kishida O, Takai K et al (2022) Utilizing environmental DNA for wide-range distributions of reproductive area of an invasive terrestrial toad in Ishikari river basin in Japan. Biol Inv 24:1199–1211
Moraes CT (2001) What regulates mitochondrial DNA copy number in animal cells? Trends Genet 17:199–205
Nishida M (2005) Ayu. In: Kawanabe H, Mizuno H, Hosoya K (eds) Freshwater fishes of Japan, 2nd edn. Yama-Kei Publishers, Tokyo, pp 66–79 (in Japanese)
Pinheiro JC, Bates DM (2000) Mixed-effects models in S and S-plus. Springer-Verlag, New York
Pont D, Rocle M, Valentini A et al (2018) Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci Rep 8:1–13
R Core Team (2021) R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna. https://www.R-project.org
Rees HC, Maddison BC, Middleditch DJ et al (2014) The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. J Appl Ecol 51:1450–1459
Saito M, Inui R, Kono T et al (2020) Examinations on appropriate time of a day for evaluating spawning intensity of Plecoglossus altivelis altivelis using environmental DNA in the Takatsu River, western Japan. J Jpn Soc Civil Eng Ser B1 (hydraul Eng) 76:I_1315-I_1320 (in Japanese with English abstract)
Tachihara K, Kimura S (1991) Maturity and spawning of land locked Ayu Plecoglossus altivelis altivelis in Lake Ikeda, Kagoshima Prefecture. Nippon Suisan Gakkaishi 57:2187–2192 (in Japanese with English abstract)
Tago Y (1999) Diurnal fluctuation, body length and area of hatching ground of Ayu larvae during seaward migration in the Shou River. Aquacult Sci 47:201–207 (in Japanese with English abstract)
Takahashi I (2009) Rivers where Natural Ayu Grow. Tsukiji Shokan Publishers, Tokyo, Tokyo (in Japanese)
Takahashi I, Azuma K (2016) The book on wild Ayu. Tsukiji Shokan Publishers, Tokyo (in Japanese)
Takahashi I, Azuma K, Hiraga H et al (1999) Different mortality in larval stage of Ayu Plecoglossus altivelis by birth dates in the Shimanto estuary and adjacent coastal waters. Fish Sci 65:206–210
Takeuchi A, Iijima T, Kakuzen W et al (2019) Release of eDNA by different life history stages and during spawning activities of laboratory-reared Japanese Eels for interpretation of oceanic survey data. Sci Rep 9:1–9
Terakado H, Murayama T, Sota K et al (2015) Transition of distribution and the riverbed condition of spawning grounds of Ayu, Plecoglossus altivelis altivelis, in the Takatsu River, Shimane Prefecture. Rep Shimane Prefect Fish Technol Cent 8:1–28 (in Japanese)
Terakado H, Murayama T, Kanaiwa M (2016) Estimation of the mixed ratio of native and released Ayu Plecoglossus altivelis altivelis caught in the Takatsu River, Shimane Prefecture, Japan. Nippon Suisan Gakkaishi 82:911–916 (in Japanese with English abstract)
Thalinger B, Wolf E, Traugott M et al (2019) Monitoring spawning migrations of potamodromous fish species via eDNA. Sci Rep 9:1–11
Tillotson MD, Kelly RP, Duda JJ et al (2018) Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol Conserv 220:1–11
Tran HD, Iida M, Maeda K (2018) Downstream migration of newly hatched Ayu (Plecoglossus altivelis) in the Tien Yen River of northern Vietnam. Environ Biol Fishes 100:1329–1341
Tsuji S, Shibata N (2021) Identifying spawning events in fish by observing a spike in environmental DNA concentration after spawning. Environ DNA 3:190–199
Tsuji S, Nakao R, Saito M et al (2022) Pre-centrifugation before DNA extraction mitigates extraction efficiency reduction of environmental DNA caused by the preservative solution (benzalkonium chloride) remaining in the filters. Limnol 23:9–16
Wheaton JM, Pasternack GB, Merz JE (2004) Spawning habitat rehabilitation—II. Using hypothesis development and testing in design, Mokelumne River, California, U.S.A. Int J River Basin Manag 2:21–37
Wilcox TM, McKelvey KS, Young MK et al (2016) Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol Conserv 194:209–216
Yamanaka H, Minamoto T (2016) The use of environmental DNA of fishes as an efficient method of determining habitat connectivity. Ecol Indic 62:147–153
Yamanaka H, Motozawa H, Tsuji S et al (2016) On-site filtration of water samples for environmental DNA analysis to avoid DNA degradation during transportation. Ecol Res 31:963–967
Yamanaka H, Minamoto T, Matsuura J et al (2017) A simple method for preserving environmental DNA in water samples at ambient temperature by addition of cationic surfactant. Limnol 18:233–241
Yoshida K, Maeno S, Takahashi S et al (2018) Physical environmental evaluation of the spawning for Ayu based on estimation of flow field and sediment behavior using ALB data in the lower Asahi River. J Jpn Soc Civil Eng Ser B1 (hydraul Eng) 74:I_421-I_426 (in Japanese with English abstract)
Yoshida K, Inui R, Udagawa R et al (2019) Spawning habitat potential evaluation for Ayu using an environmental DNA analysis in the lower Asahi River with degraded riverbed. J Jpn Soc Civil Eng Ser B1 (hydraul Eng) 75:I_529-I_534 (in Japanese with English abstract)
Zuur AF, Ieno EN, Walker NJ et al (2009) Mixed effects models and extensions in ecology with R. Springer, New York
Acknowledgements
We thank M. Amimoto, T. Kodama, and K. Kobayashi, Yamaguchi University, for field or analytical assistance. We also thank A. Okino, H. Terakado, and Y. Taniguchi, Shimane Prefectural Fisheries Technology Center, and T. Hatama, Yamaguchi Prefectural Fisheries Research Center, for valuable information on the Ayu in the Takatsu River. Takatsu River Fisheries Cooperative Association provided us with permission to conduct this study and valuable information on the Ayu stock management in the Takatsu River. Thorough and constructive comments from two anonymous reviewers helped improve the manuscript. This work was partly supported and funded by the Hamada Office of River and National Highway, Ministry of Land, Infrastructure, Transport, and Tourism, as a part of the research project, “A study on the effects of the sediment replenishment on the environment in the Gonokawa River”.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saito, M., Tsuji, S., Nakao, R. et al. Comparative study on nuclear and mitochondrial DNA of Ayu Plecoglossus altivelis for environmental DNA-based spawning evaluation. Landscape Ecol Eng 19, 55–67 (2023). https://doi.org/10.1007/s11355-022-00519-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11355-022-00519-5