Abstract
Purpose
Whole-body (WB) dynamic positron emission tomography (PET) enables imaging of highly quantitative physiological uptake parameters beyond the standardized uptake value (SUV). We present a novel dynamic WB anthropomorphic PET simulation framework to assess the potential of 2-deoxy-2-[18F]fluoro-D-glucose ([18F]FDG) net uptake rate constant (Ki) imaging in characterizing tumor heterogeneity.
Procedures
Validated heterogeneous [18F]FDG tumor kinetics were modeled within the XCAT phantom (ground truth). Thereafter, static (SUV) and dynamic PET data were simulated and reconstructed, followed by indirect WB Patlak Ki imaging. Subsequently, we compared the methods of affinity propagation (AP) and automatic segmentation with active contour (MASAC) to evaluate the impact of tumor delineation. Finally, we extracted the metabolically active tumor volume (MATV), Dice similarity coefficient (DSC), and the intratumoral heterogeneity metrics of the area under the cumulative intensity histogram curve (CIHAUC), homogeneity, entropy, dissimilarity, high-intensity emphasis (HIE), and zone percentage (ZP), along with the target-to-background (TBR) and contrast-to-noise ratios (CNR).
Results
Ki images presented higher TBR but lower CNR compared to SUV. In contrast to MASAC, AP segmentation resulted in smaller bias for MATV and DSC scores in Ki compared to SUV images. All metrics, except for ZP, were significantly different in AP segmentation between SUV and Ki images, with significant correlation observed for MATV, homogeneity, dissimilarity, and entropy. With MASAC segmentation, CIHAUC, homogeneity, and dissimilarity were significantly different between SUV and Ki images, with all metrics, except for HIE and ZP, being significantly correlated. In ground truth images, increased heterogeneity was observed with Ki compared to SUV, with a high correlation for all metrics.
Conclusions
A novel simulation framework was developed for the assessment of the quantitative benefits of WB Patlak PET on realistic heterogeneous tumor models. Quantitative analysis showed that WB Ki imaging may provide enhanced TBR and facilitate lesion segmentation and quantification beyond the SUV capabilities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Positron emission tomography (PET) imaging employing 2-deoxy-2-[18F]fluoro-D-glucose ([18F]FDG) is commonly used in clinical oncology for diagnosis, staging, restaging, radiation therapy treatment planning, and the assessment of treatment outcome [1,2,3]. Beyond the currently established static PET imaging protocols, the analysis of dynamic PET data might also enable the extraction of highly reproducible kinetic features of the [18F]FDG uptake for in-depth and quantitative characterization of tumor glucose metabolism over the course of treatment. Several investigators have demonstrated the feasibility of utilizing dynamic PET imaging in the clinic for the objective characterization of a spectrum of kinetic metabolic parameters in oncologic malignancies [4, 5]. However, these techniques were restricted to single bed positions, thus preventing their application to whole-body (WB) imaging to quantify primary and metastatic malignancies across multiple tissues in the same exam [6]. Recently, WB dynamic [18F]FDG PET imaging protocols have started gaining interest in clinical oncology owing to their ability to deliver highly quantitative parametric WB PET images, beyond the conventional standardized uptake value (SUV) metric, by tracking both the spatial and temporal distributions of the [18F]FDG uptake over multiple bed positions [7].
In the meantime, a number of intratumoral uptake heterogeneity metrics in [18F]FDG PET have recently been shown to correlate with treatment response [8, 9]. The use of these metrics can be extended to parametric imaging, where they may exhibit different effects compared to SUV images that can be clinically useful. Previous studies reported significant differences in the metabolically active tumor volume (MATV) scores between SUV and parametric images [10, 11]. Tixier et al. [9] assessed a number of metrics linked to tumor uptake heterogeneity in non-small cell lung cancer and reported high correlations for all metrics between SUV and parametric images. However, the lack of ground truth (GT) in clinical studies made it difficult to draw further conclusions. Therefore, the systematic comparison of the performance of a range of oncologic image-derived PET metrics between SUV and parametric images, where the GT is known a priori, is highly desirable.
The purpose of the present work is to develop a novel WB dynamic anthropomorphic [18F]FDG PET simulation framework supporting realistic tumor heterogeneity models to assess the performance of a wide spectrum of advanced PET image metrics in WB Patlak-derived uptake rate constant (Ki) imaging, with respect to SUV. In addition, two automated segmentation algorithms were employed to assess the impact of tumor delineation on the extracted features.
Materials and Methods
Dynamic Anthropomorphic Phantom Simulation
To perform realistic simulations, we constructed dynamic WB PET anthropomorphic emission and attenuation maps from the extended cardio-torso (XCAT) phantom along with a respiratory motion model [12]. In addition, we modeled a heterogeneous set of [18F]FDG time-activity curves (TACs) for a range of tissues (Fig. 1) using a validated two-tissue compartment model and a set of [18F]FDG kinetic parameter values reported in the literature (Table 1). Subsequently, each of the generated TACs was assigned to its corresponding tissue region in the XCAT phantom.
Noise-free TACs for different tissues derived from the kinetic parameters shown in Table 1 for a 2-compartment kinetic model.
According to a previously proposed clinical WB dynamic PET acquisition protocol [18], we first considered a dynamic PET acquisition (1st phase) centered over the heart corresponding to the first 6 min post-injection (p.i.) of [18F]FDG, including 12 × 10 s and 12 × 20 s frames. This is followed by simulation of six unidirectional (head-to-thighs) WB passes (2nd phase) consisting of seven bed positions, each scanned for 45 s. An input function model reported by Feng et al. [19] was adopted in our simulations, sampled at the mid-time points of the first 24 cardiac frames of the first phase and the six subsequent cardiac frames of the second phase.
Subsequently, realistic tumor shapes with three different levels of [18F]FDG uptake were modeled and incorporated within the extended cardiac-torso (XCAT) phantom according to a technique proposed by Le Maitre et al. to reflect the intratumoral uptake heterogeneity typically observed in clinical oncologic PET studies [20]. Specifically, different irregularly shaped heterogeneities were segmented using an adaptive thresholding technique within a set of modeled tumors (7 lung tumors and 6 cases with laryngeal squamous cell carcinoma from Louvain database) [21, 22]. The low-activity region of the tumor, which we assume contains both high and middle activity concentration levels, was segmented using a thresholding value of 0.5 (TLow) on normalized images on [0–1] scale. Then, the value at the center of the tumor (THigh) was used as a threshold to segment the high-activity region within the tumor and the middle value ((TLow + THigh) ∕ 2) was used as a threshold to segment the middle-activity region for the tumor. Finally, the 3D mesh of these segmented contours was reconstructed using Amide software [23] and converted into three non-uniform rational B-spline (NURBS) surfaces using Rhinoceros software (CADLINK, France), where each surface represented a specific activity level.
We adopted a normal respiratory breathing cycle of 5 s in our simulation and divided it into 10 bins, each representing one of the phases of the periodic respiratory cycle. Then, we utilized the respiratory motion modeling tools of the XCAT phantom to produce for each dynamic PET frame (a given bed position and given pass) 10 respective pairs of emission and attenuation maps, with each pair reflecting the unique anatomy of the tissues at a specific respiration phase. A common respiratory periodic motion pattern was used with a maximum diaphragm motion of 1.5 cm, a maximum anterior-posterior expansion of 0.5 cm, and a normal respiratory cycle of 5 s, as typically observed in our clinical studies. Each emission map was built by assigning the average [18F]FDG activity concentration value for the respective time window of that bed frame, assuming negligible changes in the TACs within the scanning period (45 s) of a given bed frame during each WB pass (Fig. 1). Subsequently, the above 10 emission maps at each dynamic frame were averaged to produce a single emission map for that frame to model the effects of respiratory motion in our simulated dynamic PET data. Ultimately, a noise-free realistic dynamic emission XCAT phantom was created by repeating the above steps across all bed positions and passes included in our simulated dynamic WB PET scan protocol. In addition, the 10 attenuation maps at each bed position were also averaged to align the effects of respiratory motion between the simulated emission and attenuation maps.
Thereafter, the PET sinograms for each frame were generated using an analytical fully 3D forward projector reflecting the Siemens Biograph™ mCT PET system’s detection geometry. Scatter and random count effects were not included in our simulation as these were deemed to have no or limited impact on the outcome. The attenuation coefficient maps were then applied to the emission sinograms. Subsequently, the attenuated PET data were scaled with a global factor to match the nominal sensitivity of the mCT PET scanner and quantitative levels of Poisson noise were added, equivalent to a 45 s acquisition time per bed.
The noisy projection data were later reconstructed using an ordered subsets expectation maximization (OSEM) algorithm employing 10 subsets and 40 sub-iterations, using the open-source Software for Tomographic Image Reconstruction (STIR) platform [24]. The matrix size of the image data was 200 × 200 with a pixel size of 0.50 × 0.41 × 0.41 cm3. The Patlak ordinary least squares (OLS) regression was then applied on the dynamic PET images to estimate Ki images representing the tracer influx rate constant Ki macro-parameter [25]. The flowchart illustrating the above four steps of our simulation framework is shown in Fig. 2. Furthermore, conventional static SUV PET images were generated from a simulated PET dataset involving a single WB pass at 70 min p.i. and lasting 180 s at each bed position.
In total, 13 different lesions (volume range 6.64–69.34 cm3) were simulated in the lungs followed by application of a Gaussian smoothing filter of 2 mm on an image matrix of 200 × 200 voxels for both SUV and Ki images. The GT in SUV images was defined as the noise-free static XCAT images at 70 min p.i. The respective GT in Ki images was obtained after conducting Patlak OLS regression across six noise-free dynamic XCAT images at different time points.
PET Image Segmentation Algorithms
As reported in earlier studies [26] and more recently in the American Association of Physicists in Medicine (AAPM) Report No. 211, a high variability is observed between different segmentation methods [27]. To assess the impact of tumor delineation, two different segmentation methods, namely a method for automatic segmentation using an active contour model (MASAC) [28] and an affinity propagation algorithm (AP) [29], were employed. These algorithms underwent extensive testing in our lab and were chosen owing to their accuracy and consistency as reported in previous studies using phantom and clinical studies. In particular, the parameter lambda in the implementation of MASAC was set to 3 [28] whereas the default parameters were used for AP with the largest regional intensity grouping as the segmentation result.
Evaluation Metrics
The MATV, Dice similarity coefficient (DSC), and several intratumoral heterogeneity features were included for the comparison of SUV and Ki images. The DSC was used to assess the accuracy of the geographical match/mismatch between the segmented volumes and the GT [30, 31], whereas the heterogeneity features were selected because of their reproducibility and robustness as reported in previous studies [8, 9]. Specifically, the area under the cumulative intensity histogram curve (CIHAUC) was considered as a global heterogeneity indicator (with low values indicating a higher degree of heterogeneity), whereas the homogeneity, entropy, dissimilarity, high-intensity emphasis (HIE), and zone percentage (ZP) were chosen as the local heterogeneity-related features. The local heterogeneity features defined below were derived using Pyradiomics software package [32].
where Ng is the number of gray level intensities and p(i, j) is the (i, j) element in the normalized gray level co-occurrence matrix, representing the number of times for the combination of levels i and j to occur in two pixels in the image, separated by a distance of δ pixels in direction α. In our study, Ng is set to 64 gray levels and δ is 1 over all 13 spatial directions (26-connectivity in 3D).
where Ng is the number of gray level intensities in the gray level size zone matrix, Ns is the number of discrete zone sizes, Np is the number of voxels, and P(i, j) represents the number of one or more connected gray level zones that share the gray level i and size j in the image.
Additionally, the target-to-background ratio (TBR) and contrast-to-noise ratio (CNR) were employed to assess the contrast and noise in SUV and Ki images, as defined below:
where MeanT and MeanB are the mean values for the tumor (target) and background regions, respectively, and StdB is the standard deviation of the background region.
Statistical Analysis
Statistical analysis was conducted using the SPSS 24.0 commercial software package (IBM, Chicago, USA). A non-parametric Wilcoxon test was used to assess if the differences between each pair of methods compared in this study were significant or not, with a P value of 0.05 denoting significance. The correlation of various evaluated metrics between SUV and Ki images was assessed using the non-parametric Spearman analysis, with a P value of 0.05 determining statistical significance. Statistical analysis results were presented using box-and-whisker plots, providing lower to upper quartiles (25th to 75th percentile, central box), the median (middle line of the box), and the outliers, as identified in Tukey’s method (1.5 × inter-quartile range) [33]. In addition, scatter plots were also used to explore the relationships between different groups of data.
Results
SUV vs. Ki Image Segmentation
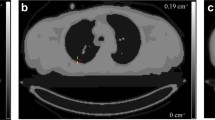
The contours extracted by both segmentation methods of a representative realistic anthropomorphic phantom study are presented in Fig. 3. It can be seen that both segmentation techniques could delineate the tumors properly on either SUV or Ki images. Similar results were also observed in other cases included in this study. The average TBR in Ki images for all lesions (6.25) was significantly enhanced compared to that (4.77) in SUV images (P < 0.002), whereas SUV images achieved significantly better average CNR (34.36) over Ki images (13.70, P < 0.002).
Compared to SUV images, Ki images yielded smaller average bias in MATV (SUV − 52.77 %, Ki − 31.62 %) and DSC (SUV − 36.00 %, Ki − 25.00 %) with AP, whereas no significant differences were observed in MATV and DSC for MASAC algorithm (Table 2 and Fig. 4). Except for ZP, most heterogeneity metrics were significantly different between SUV and Ki images for AP segmentation, and a similar trend was also observed for CIHAUC, homogeneity, and dissimilarity with MASAC segmentation (Table 2 and Fig. 5). Besides, a general trend of correlation between SUV and Ki images was observed for MATV, homogeneity, dissimilarity, and entropy with AP segmentation, whereas most metrics were found to be significantly correlated between SUV and Ki images, except for HIE and ZP, when using MASAC segmentation algorithm (Table 3, Fig. 6 and Supplemental Material S1–S2).
SUV vs. Ki Images in Noise-Free GT
Most heterogeneity features, except dissimilarity, exhibited statistically significant differences between SUV and Ki noise-free GT images (Table 2). More specifically, slightly decreased CIHAUC (− 6.28 %), homogeneity (− 3.99 %), and HIE (− 3.95 %) along with increased entropy (2.21 %) and ZP (4.52 %) were observed in Ki noise-free GT images compared with SUV GT images (Fig. 5). Besides, it could be observed from Table 3 that all heterogeneity features were highly correlated between SUV and Ki noise-free GT images (P < 0.001).
AP vs. MASAC Segmentation
There is no significant difference in the MATV metric between the two segmentation algorithms for Ki images (Table 4). MASAC yielded smaller bias in DSC (− 20.00 %) for Ki images and smaller bias in MATV (− 33.75 %) and DSC (− 19.00 %) for SUV images, compared to AP.
Segmentation Results vs. Noise-Free GT
Compared to noise-free GT images, most metrics derived from segmentation results, except CIHAUC and entropy in some cases, were significantly different in either SUV or Ki images (Table 5). In addition, both MASAC and AP presented lower MATV, homogeneity, and HIE, and higher ZP scores for SUV and Ki images compared to GT (Figs. 4 and 5).
Discussion
A clinically feasible WB dynamic PET acquisition protocol enabling highly quantitative multi-parametric PET imaging across multiple bed positions was presented in previous studies [7, 34]. In this work, we developed a multi-bed dynamic 4D XCAT-based realistic simulation framework supporting tumor heterogeneity modeling, to be utilized for (i) evaluation of WB 4D PET image reconstruction and segmentation algorithms, (ii) optimization of dynamic WB PET acquisition and image analysis methods, (iii) modeling of the WB pharmacokinetic properties of novel drugs under development, and (iv) assessment of the usefulness of a wide range of quantitative metrics in Ki vs. SUV images. Moreover, our framework allowed the extraction of useful conclusions by enabling the assessment of a wide range of metrics under noise-free conditions. In particular, the Ki noise-free GT images showed increased heterogeneity, in terms of lower CIHAUC, homogeneity, and HIE, and higher entropy and ZP, compared to SUV GT images, thereby indicating that heterogeneity features can be different between SUV and Ki images. Independent of the PET segmentation algorithm and the type of images analyzed (SUV vs. Ki), a lower homogeneity, a lower HIE, and a higher ZP were observed on simulated noisy against noise-free GT images. This is attributed to the relatively higher degree of heterogeneity expected for noisy PET images.
Furthermore, our findings indicate that WB Ki imaging can provide enhanced TBR as well as an additional set of highly quantitative tumor features, beyond the static features currently supported with the respective SUV image metrics (Fig. 3). Our results are consistent with observations made in previous studies. In particular, Chen et al. [35] reported that gradient-based tumor delineation method may be more accurate on Patlak Ki parametric maps compared to conventional static SUV images using magnetic resonance imaging as the GT. Furthermore, Llan et al. [36] found that Ki images present better tumor-to-liver contrast compared with SUV images. Finally, Wangerin et al. [37] assessed the variations during the PET imaging process using a series of linked simulations and found that Ki images were associated with superior receiver operating characteristic performance compared to SUV images.
In our study, significant correlations were also found between SUV and Ki images, regardless of the segmentation method. It should be noted that parametric Ki images may still be complementary to SUV even when a high correlation is observed between the two images, as they are essentially different quantities, each providing information that cannot be deduced from the other. This is because Ki imaging measures the tracer net uptake rate during a relatively long scan time window post-injection, whereas SUV imaging measures the average absolute uptake of the tracer within a relatively short scan time window post-injection. However, a systematic evaluation of the clinical usefulness of the added information derived from WB Ki imaging is beyond the scope of the current work. We are planning to utilize the findings of this study to evaluate the same metrics using a large clinical database acquired with a recently proposed combined SUV/Patlak imaging framework [38, 39].
We have also observed that the MATV was systematically underestimated on both SUV and Ki images regardless of the segmentation algorithm. It should be noted that the presence of respiratory motion is expected to have amplified the actual MATV due to motion blurring. Therefore, for routine clinical operation, respiratory motion and its varying effect on SUV and Ki metrics should be carefully considered.
A number of practical limitations are associated with this study. Firstly, the potential efficiency variance across all PET detector pairs was not modeled. We also assumed no scatter and random effects in our analytic simulations. Furthermore, a regular periodic respiratory motion pattern was adopted. Patient’s irregular free breathing would have caused asymmetric blurring of focal lesions, thereby resulting in less predictable artifacts. Moreover, the number of evaluated cases in this study may not be sufficient. Nevertheless, we aim at a systematic follow-up study using a larger sample to investigate the effect of different acquisition/reconstruction protocols on various clinical PET scanners, across a typical range of lesion sizes and contrasts observed in clinical studies. In addition, the time-averaging of attenuation maps in our study is adopted to simplify the simulation, which may not reflect the actual process of attenuation as the measurement model in the attenuation map is non-linear. Finally, in the absence of guidelines on optimal reconstruction parameters for parametric Ki images, both Ki and SUV images have been reconstructed using the same iteration numbers. Further investigations of optimal reconstruction protocols for parametric imaging are warranted.
Conclusion
A dynamic multi-bed PET simulation framework was developed based on the 4D XCAT anthropomorphic model, respiratory motion and tumor heterogeneity models, and validated 18F-FDG kinetic parameters to enable the systematic evaluation of the clinical usefulness of WB parametric PET imaging for various types of oncologic malignancies beyond the currently established SUV metric. Our results showed that Ki images may provide enhanced TBR and further facilitate lesion segmentation and quantification beyond the SUV capabilities, thereby demonstrating the potential of hybrid SUV/Ki imaging, in terms of lesion quantification.
References
Chung MK, Jeong HS, Park SG, Jang JY, Son YI, Choi JY, Hyun SH, Park K, Ahn MJ, Ahn YC, Kim HJ, Ko YH, Baek CH (2009) Metabolic tumor volume of [18F]-fluorodeoxyglucose positron emission tomography/computed tomography predicts short-term outcome to radiotherapy with or without chemotherapy in pharyngeal cancer. Clin Cancer Res 15:5861–5868
La TH, Filion EJ, Turnbull BB et al (2009) Metabolic tumor volume predicts for recurrence and death in head-and-neck cancer. Int J Radiat Oncol Biol Phys 74:1335–1341
Obara P, Pu YL (2013) Prognostic value of metabolic tumor burden in lung cancer. Chin J Cancer Res 25:615–622
Kristian A, Revheim ME, Qu H, Mælandsmo GM, Engebråten O, Seierstad T, Malinen E (2013) Dynamic 18F-FDG-PET for monitoring treatment effect following anti-angiogenic therapy in triple-negative breast cancer xenografts. Acta Oncol 52:1566–1572
Dimitrakopoulou-Strauss A, Pan L, Strauss LG (2012) Quantitative approaches of dynamic FDG-PET and PET/CT studies (dPET/CT) for the evaluation of oncological patients. Cancer Imaging 12:283–289
Zaidi H, Karakatsanis N (2018) Towards enhanced PET quantification in clinical oncology. Br J Radiol 91:20170508
Karakatsanis NA, Casey ME, Lodge MA, Rahmim A, Zaidi H (2016) Whole-body direct 4D parametric PET imaging employing nested generalized Patlak expectation-maximization reconstruction. Phys Med Biol 61:5456–5485
Hatt M, Majdoub M, Vallieres M, Tixier F, le Rest CC, Groheux D, Hindie E, Martineau A, Pradier O, Hustinx R, Perdrisot R, Guillevin R, el Naqa I, Visvikis D (2015) 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med 56:38–44
Tixier F, Vriens D, Cheze-Le Rest C et al (2016) Comparison of tumor uptake heterogeneity characterization between static and parametric 18F-FDG PET images in non-small cell lung cancer. J Nucl Med 57:1033–1039
Visser EP, Philippens ME, Kienhorst L et al (2008) Comparison of tumor volumes derived from glucose metabolic rate maps and SUV maps in dynamic 18F-FDG PET. J Nucl Med 49:892–898
Cheebsumon P, van Velden FH, Yaqub M et al (2011) Measurement of metabolic tumor volume: static versus dynamic FDG scans. EJNMMI Res 1:35
Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW (2010) 4D XCAT phantom for multimodality imaging research. Med Phys 37:4902–4915
Vriens D, Disselhorst JA, Oyen WJG, de Geus-Oei LF, Visser EP (2012) Quantitative assessment of heterogeneity in tumor metabolism using FDG-PET. Int J Radiat Oncol Biol Phys 82:E725–E731
Dimitrakopoulou-Strauss A, Georgoulias V, Eisenhut M, Herth F, Koukouraki S, Mäcke HR, Haberkorn U, Strauss LG (2006) Quantitative assessment of SSTR2 expression in patients with non-small cell lung cancer using 68Ga-DOTATOC PET and comparison with 18F-FDG PET. Eur J Nucl Med Mol Imaging 33:823–830
Choi Y, Hawkins RA, Huang SC, Brunken RC, Hoh CK, Messa C, Nitzsche EU, Phelps ME, Schelbert HR (1994) Evaluation of the effect of glucose-ingestion and kinetic-model configurations of FDG in the normal liver. J Nucl Med 35:818–823
Lin KP, Huang SC, Choi Y, Brunken RC, Schelbert HR, Phelps ME (1995) Correction of spillover radioactivities for estimation of the blood time-activity curve from the imaged lv chamber in cardiac dynamic FDG PET studies. Phys Med Biol 40:629–642
Sachpekidis C, Mai EK, Goldschmidt H, Hillengass J, Hose D, Pan L, Haberkorn U, Dimitrakopoulou-Strauss A (2015) 18F-FDG dynamic PET/CT in patients with multiple myeloma patterns of tracer uptake and correlation with bone marrow plasma cell infiltration rate. Clin Nucl Med 40:E300–E307
Karakatsanis NA, Lodge MA, Tahari AK, Zhou Y, Wahl RL, Rahmim A (2013) Dynamic whole-body PET parametric imaging: I. Concept, acquisition protocol optimization and clinical application. Phys Med Biol 58:7391–7418
Feng D, Huang SC, Wang X (1993) Models for computer simulation studies of input functions for tracer kinetic modeling with positron emission tomography. Int J Biomed Comput 32:95–110
Le Maitre A, Segars WP, Marache S et al (2009) Incorporating patient-specific variability in the simulation of realistic whole-body F-18-FDG distributions for oncology applications. Proc IEEE 97:2026–2038
Wanet M, Lee JA, Weynand B, de Bast M, Poncelet A, Lacroix V, Coche E, Grégoire V, Geets X (2011) Gradient-based delineation of the primary GTV on FDG-PET in non-small cell lung cancer: a comparison with threshold-based approaches, CT and surgical specimens. Radiother Oncol 98:117–125
Daisne JF, Duprez T, Weynand B, Lonneux M, Hamoir M, Reychler H, Grégoire V (2004) Tumor volume in pharyngolaryngeal squamous cell carcinoma: comparison at CT, MR imaging, and FDG PET and validation with surgical specimen. Radiology 233:93–100
Loening AM, Gambhir SS (2003) AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging 2:131–137
Thielemans K, Tsoumpas C, Mustafovic S, Beisel T, Aguiar P, Dikaios N, Jacobson MW (2012) STIR: software for tomographic image reconstruction release 2. Phys Med Biol 57:867–883
Patlak CS, Blasberg RG, Fenstermacher JD (1983) Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab 3:1–7
Zaidi H, El Naqa I (2010) PET-guided delineation of radiation therapy treatment volumes: a survey of image segmentation techniques. Eur J Nucl Med Mol Imaging 37:2165–2187
Hatt M, Lee J, Schmidtlein CR et al (2017) Classification and evaluation strategies of auto-segmentation approaches for PET: report of AAPM Task Group No. 211. Med Phys 44:e1–e42
Zhuang M, Dierckx RA, Zaidi H (2016) Generic and robust method for automatic segmentation of PET images using an active contour model. Med Phys 43:4483–4494
Foster B, Bagci U, Ziyue X et al (2014) Segmentation of PET images for computer-aided functional quantification of tuberculosis in small animal models. IEEE Trans Biomed Eng 61:711–724
Zou KH, Warfield SK, Bharatha A, Tempany CMC, Kaus MR, Haker SJ, Wells WM III, Jolesz FA, Kikinis R (2004) Statistical validation of image segmentation quality based on a spatial overlap index—scientific reports. Acad Radiol 11:178–189
Hatt M, le Rest CC, Descourt P et al (2010) Accurate automatic delineation of heterogeneous functional volumes in positron emission tomography for oncology applications. Int J Radiat Oncol Biol Phys 77:301–308
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Frigge M, Hoaglin DC, Iglewicz B (1989) Some implementations of the boxplot. Am Stat 43:50–54
Karakatsanis NA, Lodge MA, Zhou Y, Wahl RL, Rahmim A (2013) Dynamic whole-body PET parametric imaging: II. Task-oriented statistical estimation. Phys Med Biol 58:7419–7445
Chen HW, Jiang JZ, Gao JL, Liu D, Axelsson J, Cui M, Gong NJ, Feng ST, Luo L, Huang B (2014) Tumor volumes measured from static and dynamic F-18-fluoro-2-deoxy-D-glucose positron emission tomography-computed tomography scan: comparison of different methods using magnetic resonance imaging as the criterion standard. J Comput Assist Tomogr 38:209–215
Ilan E, Sandstrom M, Velikyan I, Sundin A, Eriksson B, Lubberink M (2017) Parametric net influx rate images of 68Ga-DOTATOC and 68Ga-DOTATATE: quantitative accuracy and improved image contrast. J Nucl Med 58:744–749
Wangerin KA, Muzi M, Peterson LM, Linden HM, Novakova A, Mankoff DA, Kinahan PE (2017) A virtual clinical trial comparing static versus dynamic PET imaging in measuring response to breast cancer therapy. Phys Med Biol 62:3639–3655
Karakatsanis N, Lodge M, Fahrni G et al (2016) Simultaneous SUV/patlak-4D whole-body PET: a multi-parametric 4D imaging framework for routine clinical application. J Nucl Med 57(Suppl. 2):367
Karakatsanis N, Lodge M, Zhou Y et al (2015) Novel multi-parametric SUV/Patlak FDG-PET whole-body imaging framework for routine application to clinical oncology. J Nucl Med 56(Suppl. 3):625
Funding
This work was supported by the Swiss National Science Foundation under grant SNSF 320030_176052, the Swiss Cancer Research Foundation under Grant KFS-3855-02-2016, and an Open Grant (2014GDDSIPL-06) from the Key Laboratory of Digital Signal and Image Processing of Guangdong Province, Shantou University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 248 kb)
Rights and permissions
About this article
Cite this article
Zhuang, M., Karakatsanis, N.A., Dierckx, R.A.J.O. et al. Quantitative Analysis of Heterogeneous [18F]FDG Static (SUV) vs. Patlak (Ki) Whole-body PET Imaging Using Different Segmentation Methods: a Simulation Study. Mol Imaging Biol 21, 317–327 (2019). https://doi.org/10.1007/s11307-018-1241-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-018-1241-8