Abstract
We have made and partially sequenced two subtracted cDNA libraries, one representing genes predominantly expressed in a tree from an early-flushing family of Norway spruce (early-flushing library; EFL) and the second from a late flushing family (late flushing library; LFL), during 4 weeks before bud burst. In the EFL, expressed sequence tags (ESTs) encoding proteins of the photosynthetic apparatus and energy metabolism and proteins related to stress (abiotic and biotic) and senescence were abundant. ESTs encoding metallothionein-like and histone proteins as well as transcription factors were abundant in the LFL. We used quantitative real-time reverse transcription polymerase chain reaction to study the expression patterns of 25 chosen genes and observed that the highest levels of activity for most genes were present when plants were still ecodormant. The results indicate that the late flushing is not a result of a delay in gene activity, but is rather associated with an active transcriptional process. Accordingly, certain metabolic processes may be turned on in order to prevent premature flushing. We discuss the putative role of the studied genes in regulation of bud burst timing. Among the candidate genes found, the most interesting ones were the DNA-binding proteins, water-stress-related genes and metallothioneins. Expression patterns of some genes involved in chemical modification of DNA and histones indicate that epigenetic factors are involved in the timing of bud burst. In the obtained transcriptomes, we could not find genes commonly believed to be involved in dormancy and bud set regulation (PHY, CRY, ABI etc.) in angiosperm plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Synchronization of active growth and dormancy with seasonal changes in temperature and day length is necessary for survival and competitive success of boreal forest tree species. Adapted populations tune timing of dehardening and bud burst in the spring and of growth cessation, dormancy and cold acclimation in autumn in proper synchrony with seasonal variation in temperature. The timing of bud burst in conifers correlates well with spring frost hardiness, and the trait is an important component of adaptability [1, 25]. Norway spruce (Picea abies [L.] Karst), like other boreal forest trees, expresses substantial variation in the timing of bud burst in spring, which is correlated with the latitudinal and altitudinal origin of the seed sources [4]. The trait is under strong genetic control with narrow-sense heritability estimates of 0.8 [2] and is one of the most important traits affecting mortality, growth and quality [51]. Progeny tests show that early-flushing Norwegian families of P. abies contain more individuals with defects and injuries and with reduced growth compared to late-flushing families (Skrøppa et al., unpublished data). Temperature is the main regulator of the timing of bud burst [18], but long days may modify its timing as well [44]. After the bud burst, the new shoots become very susceptible to freezing injury [12]. Hence, the idea to delay the timing of bud burst in spring through breeding is a feasible option to increase survival and decrease the frequency of defects [21].
Modern genomic tools have not been widely used to study bud burst regulation. Most efforts have been made to study the autumn processes: cold hardiness and dormancy [13], and leaf senescence [5, 8, 19]. The recent development of genomic analyses makes it possible to identify genes that regulate bud burst in Norway spruce. Our primary goal is to understand the genetic basis for bud burst timing. We describe here the sequencing and analysis of Norway spruce expressed sequence tags (ESTs) to identify candidate genes for regulation of this process, and we characterize expression profiles of some of them. For gene identification, we used the suppressive subtraction hybridization (SSH) technique. This powerful method enables us to compare two populations of mRNA and obtain clones of genes expressed in one population but not in the other. One of the advantages of this technique is the enrichment of rare transcripts, and it reduces redundancy in the library. In recent years, SSH has been used to study gene expression and identify transcripts involved in important processes in many plant and woody species, e.g. [19, 20, 46, 47].
For expression profiling, we used quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR), which is rapidly being adopted as a standard method for in-depth expression studies and offers a robust method for precisely quantifying changes in gene expression over a wide dynamic range [9].
Here, we present the first analyses of two different sets of Norway spruce ESTs and use gene expression data to propose that delayed bud burst in Norway spruce is associated with active transcription. Identification of genes specifically expressed when trees prepare for spring and early summer could be the initial step in exploring the co-regulating genetic network during spring dehardening and growth initiation. Such knowledge could be used in breeding and gene conservation by improving our understanding of local adaptation and micro-evolution under global warming.
Materials and methods
Plant material and RNA isolation
Samples were collected at the trial plot in Skiptvet (Norway) once per week from 16 April 2004 until 10 June 2004, from five trees within each of two families (79 and 95) expressing largest differences in timing of bud burst (Table 1). Based on phenological observation, we selected samples from the earliest flushing tress in the early family and from the latest flushing tree in the late family. The early-flushing family 79 (EF) is the full-sib progeny of control cross between trees both originated from Norway (mother, #1589 and father, #39). Tree #5 was selected as the earliest flushing within the family (E79-5). The late-flushing family 95 (LF) is the full-sib progeny of control cross between trees originated from Poland (mother, #5460) and Ukraine (father, #5466). In the family 95, we chose tree #25 as having the latest time of bud burst within the family (L95-25). The difference in bud burst time between selected trees was nearly 3 weeks; within-family difference did not exceed 10 days for early and 1 week for late families. Bud burst was defined as the day when bud scales were broken and green needles became visible.
Twigs (the last node, nearly 15 cm) from each tree were cut in the afternoon, from the south side and the middle part of the crown, to avoid within-tree variation in the bud burst timing. They were immediately frozen in liquid nitrogen to keep intact transcriptome and avoid forming artefact stress genes. Total RNA was extracted from buds (bud scales removed) or from juvenile needles from flushing buds using the RNAqueous-Midi kit (Ambion #1911). Contaminating DNA was removed from total RNA samples using the DNA-Free kit (Ambion #1906) according to the manufacturer’s protocol. RNA was quantified using spectrophotometric OD260 measurements, and quality was assessed by OD260/OD280 ratios and by electrophoresis on 1% formaldehyde agarose gels followed by ethidium bromide staining. Total RNA was stored at −80°C.
Subtractive cDNA libraries construction
Equal amounts of 1 μg of total RNA from two samples, collected at 4 weeks before bud burst (E79-5, date 16 April 2004; L95-25, date 07 May 2004), were used for subtractive hybridization. SSH was performed using the Clontech PCR-Select cDNA Subtraction Kit (BD Bioscience Clontech #K1804-1) and the SuperSMART cDNA Synthesis Kit (BD Bioscience Clontech #K1054-1) according to the manufacturer’s protocols. We synthesized two (forward and reverse) subtracted arrays. In the early-subtracted array, we used sample E79-5 as tester and L95-25 as driver. It represents transcripts predominantly expressed in the early-flushing tree 4 weeks before bud burst. In the late-subtracted array, we made the reverse subtraction. As tester, we used the sample L95-25, and E79-5 was used as driver. It represents transcripts predominantly expressed in the late-flushing tree 4 weeks before bud burst. Obtained arrays were non-directionally cloned in a pDrive vector using PCR Cloning Kit (QIAGEN #231224) according to the manufacturer’s protocols. Two libraries with the titers 5×108 and 3.5×108 cfu/ml were used for partial sequencing.
cDNA sequencing and EST analysis
The two obtained subtracted libraries [early-flushing library (EFL) and late-flushing library (LFL)] were partially sequenced at the DNA Sequencing Laboratory at the Institute of Biotechnology (University of Helsinki, Finland). In total, 4,010 sequences from 3,406 randomly picked colonies were obtained. We re-sequenced all 3′-end sequences from the 5′-end due to non-directional cloning of transcripts into the vector and analysed them together with the 5′ sequences obtained in the first sequencing run. All sequence processing and contigs assemblies were made with SecMan II sequence analysis software (DNAStar Inc., WI). The obtained nucleotide sequences (all EST singletons and the longest sequence from each contig) were manually scrutinized for open reading frames (ORFs) using NCBI ORF-finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and the BLASTP analysis of the largest ORFs against the National Center of Biotechnology Information (NCBI) database (NIH, USA) was performed. In parallel, TBLASTX and BLASTN analyses of some of the nucleotide sequences against the NCBI database were performed. Any similarities with a score more than 46 or an e-value of less than 10−4 were considered a hit.
Multiple sequence alignment was made using WWW resources: Multiple Sequence Alignment (MultAlign) by Florence Corpet (http://prodes.toulouse.inra.fr/multalin/multalin.html) [15] and CLUSTALW: Multiple Sequence Alignment (http://clustalw.genome.jp). We grouped the ESTs into functional categories based on a literature search.
Relative quantitative real-time RT-PCR
cDNA was synthesized from 150 ng of total RNA using the SuperSMART cDNA Synthesis Kit for reverse transcription in a 40-μl reaction volume. After heat-deactivation of the enzyme, obtained ss cDNA was diluted twice. Real-time PCR reactions were performed in a 25-μl volume containing 250 nM of each primer, 4 μl of cDNA sample and 1× SYBR Green PCR master mix (Applied Biosystems #4309155). For internal control gene (Actin 2/7) amplification, primer concentration was 150 nM. Real-time PCR was performed on the 7500 Fast Real-Time PCR System (Applied Biosystems #4351106) in a 96-well reaction plate using the parameters recommended by the manufacturer (2 min at 50°C, 10 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min). Each PCR reaction was duplicated and no-template controls were included. We verified the specificity of the amplifications at the end of the PCR run using 7500-system SDS Dissociation Curve Analysis Software. Description of selected Norway spruce ESTs for genes with known or putative functions and primers for real-time PCR are given in Supplementary Table 1. The entire experiment, including both the RT and real-time PCR steps, was repeated twice, giving two experimental replications. Results were then averaged because the expression patterns were nearly equal. All the gene expression levels were normalized to Actin gene expression. Actin was chosen as internal control among other tested genes, i.e. α-tubulin (PaαTub), glyceraldehyde-3-phosphate dehydrogenase (PaGAPDH) and polyubiquitin (PaUbq), based on the lowest estimates of the stability index ([7]; for details see Supplementary Tables 2.1 and 2.2). Analysis of data was done using 7500-system SDS software for absolute quantification and MS Excel software.
Results
Characterization of subtractive cDNA libraries and EST sequences
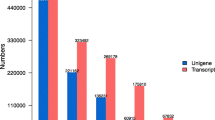
Both subtracted libraries from the EFL and LFL trees were partially sequenced and processed separately. From the EFL, 1,263 colonies were sequenced and analysed and 1,585 EST sequences were obtained (plasmids from 322 colonies were re-sequenced from the 5′-end). After pDrive-vector and quality end trimming and removal of all ambiguous short (<200 bp) sequence fragments, we revealed 1,234 meaningful clone sequences. A total of 831 ESTs (67.3% of the total number of ESTs) were assembled into 270 contigs composed of 2–54 sequences. The remaining 403 ESTs formed contigs containing only single ESTs (singletons). Overall, the current subtraction set consisted of 673 contigs, with an average length of 471 bp, potentially enriched with early-bud-flushing regulating genes.
From the LFL, 2,143 colonies were sequenced and analysed and 2,425 EST sequences were obtained (plasmids from 282 colonies were sequenced from both ends). Following pDrive-vector and quality end trimming, we removed all short (<200 bp) fragments and found 1,862 meaningful sequences. Out of these 1,862 ESTs, we obtained 789 contigs potentially enriched with late bud-flushing regulating genes, with an average length of 533 bp. They consist of 361 contigs assembled from 1,434 ESTs (77% of the total number of ESTs) composed of 2–75 sequences, and the remaining 428 contigs consist of singletons.
Comparison of the unique sequences obtained from each library against the GenBank databases revealed that 416 contigs (61.8%) in the EFL and 450 contigs in the LFL (57.0%) had a high degree of sequence similarity to genes from other organisms. Fifty-four contigs in EFL (8.0% of the total ESTs) and 64 contigs in LFL (8.1%) showed a high degree of sequence similarity to submitted genes from other plants with unknown functions. Sixteen contigs in the EFL (2.3% of the total ESTs) and 102 contigs in the LFL (12.9%) were found to have ORFs with no significant match in the databases, and we considered these as true unknown. In addition, we found 187 contigs in the EFL (27.8% of the total ESTs) and 173 contigs in the LFL (21.9%) as having no good ORF or consisting of just 3′-end of mRNA.
A snapshot of transcriptomes in the trees from the early- and the late bud-flushing trees
Despite the fact that the relatively low numbers of 4,010 clones from the subtractive libraries were sequenced, we obtained a diverse group of ESTs, matching to nearly 617 putative genes in the public databases. We classified these sequences according to their putative functional role in spruce development (Table 2). Based on similarity to the genes with annotated functions, we compared the gene homologues across the two libraries. The comparison is based entirely on the gene homologue names, so it gives only a preliminary evaluation of similarity between the two libraries.
Most of the identified transcripts appeared to be genes related to metabolism, photosynthesis, energy and red/ox reactions (including thioredoxins), protein biosynthesis and degradation (Table 2). The most frequently found hits in both libraries were genes encoding metallothionein-like proteins (three types) and Rubisco small subunit. In addition, we found several other genes related to stress (abiotic and biotic) and protein folding (e.g. chaperons), regulation and signaling in abundance (Table 2, Supplementary Table 3).
In the EFL, ESTs encoding proteins of the photosynthetic apparatus and energy metabolism and proteins related to stress (abiotic and biotic) and senescence were relatively more abundant. These genes are responsible for supporting maintenance and photosynthesis under unfavourable conditions in early spring (Table 2). In fact, from the 20 most abundant genes, six were related to photosynthesis (Table 3): photosystem I and II proteins, small unit of Rubisco, and thioredoxin-H-like proteins. Among the most abundant sequences selected in the EFL, we found two putative stress-related proteins (dehydrin 1 and universal stress protein) and a senescence-associated protein, as well as several ribosomal proteins (acidic r-protein P1a, chloroplast r-proteins and precursors CL35a and CL27). None of these sequences represented more than 1% of the clones.
In the LFL, we found abundant ESTs that encoded histone proteins as well as transcription factors, i.e. genes responsible for early stages of gene expression (Table 2). We identified ESTs encoding some related to photosynthesis proteins, histones H2A and H2B and High Mobility Group (HMG) box proteins in great abundance. Among the most frequently found ESTs were several ribosomal proteins (e.g. 60S r-proteins L17 and L36) and ESTs encoding Ca2+-binding proteins such as calmodulin-like (CaM1), and EF-hand calcium-binding motif proteins (Table 4).
Transcription patterns of candidate genes
We decided to study the expression patterns of 25 genes in detail by qRT-PCR. They were chosen based on their putative function and on our research interest in epigenetic regulation in Norway spruce [28]. We suspected that stress-related proteins and dehydrins, signal transduction proteins (including temperature- and light-sensitive proteins), photosynthesis and respiratory proteins, and proteins that activate the cell division machinery might govern bud burst. Bud burst in Norway spruce is probably regulated epigenetically through temperature influence during sexual reproduction [27–29]; therefore, genes involved in regulation by chemical modification of DNA and chromatin modification are of high interest. In addition, we have also focused our attention on genes that regulate expression of other genes. Detailed analysis of expression could allow us to estimate the efficiency of subtraction and relate expression pattern to the presence of genes in subtracted libraries.
First, we analysed the expression patterns of several DNA-binding proteins and eukaryotic translation initiation factor 5A (PaeIF5A). In both libraries, we found a large number of ESTs having a Zn-finger DNA-binding motif. From the list of differently expressed genes, we selected PaZfPDOF (DNA binding one finger), zinc finger PaC 3 HC 4 and PaC 2 H 2 type proteins. The expression patterns of PaC 2 H 2 and PaeIF5A, both reported to be involved in senescence, looked very similar. For the late-flushing tree (LF), these four genes were up-regulated during the early deacclimation period (April to early May) and down-regulated after bud burst (Fig. 1). The early-flushing pattern (EF) was completely different. The expressions of these genes were down-regulated all the time, and the reduction in transcript level was most prominent during and after bud burst (Fig. 1).
Expression profiles of PaZfPDOF (DNA binding one finger), PaZfC 3 HC 4 and PaZfC 2 H 2 type proteins and translation initiation factor 5A(PaeIF5a) genes in the trees from early- (E) and late- (L) bud-flushing families of Norway spruce at different calendar dates (a) and developmental stages before and after bud burst (b). Expression was normalized to PaAct. Obtained relative values were transformed into log2. Each unit line indicates the twofold (corresponding to log2=1) difference in expression, which is considered important. L and E indicate dates of bud burst for earliest and latest trees from corresponding flushing families in a
The transcript level of PaC 3 HC 4 between EF and LF correlate well with the calendar dates. The gene expression was up-regulated in April and down-regulated in May (Fig. 1a). However, relative to the developmental stage before bud burst, PaC 3 HC 4 was expressed in an opposite manner between the EF and LF (Fig. 1b). Expression of PaZfPDOF showed an opposite pattern between EF and LF with respect to both calendar dates and developmental stages. Having distinctly different expression patterns between the EF and LF, we consider that these four genes are of high interest for further study.
We also included another group of genes putatively involved in epigenetic gene regulation through DNA methylation and histone modifications (acetylation, methylation, phosphorylation, ubiquitination or ADP-ribosylation). Histone acetylation is reversible; the modification could be reverted by histone deacetylation [3, 35]. We could not find any ESTs related to cytosine methyltransferases; instead, one EST of Arabidopsis thaliana (adenosine-N6-)-methyltransferase (DAM) homologue was found. DAM methylates the deoxyadenine residues in 5′-GATC-3′ sequences and plays an important role in the global regulation of genes in Escherichia coli [41].
We identified three contigs related to histone acetylation. The expression patterns of two of them—GCN5-related N-acetyltransferase (PaGNAT) and histone deacetylase (PaHDAC)—were further analysed by qRT-PCR. GNATs are reported to be involved in histone acetylation [6, 43], and HDACs revert histones to the inactive state. An EST for a SET-domain-containing protein was found in the EFL. The SET domain is the integral part of histone methyltransferases and is required for its enzymatic activity [56]. As methylation of histones occurs on histones H3 and H4, we also chose the corresponding ESTs for expression analysis.
Expression of four out of the six analysed genes—PaGNAT, PaDAM, PaSET and PaH3a—correlate better with the calendar date than with the developmental stage (Fig. 2). Expression of these genes was decreasing gradually from the start until May 7, followed by an increase in transcription level. For most transcripts, expression was higher for LF than that for EF. However, considering the transcript levels at the same developmental stages, the expression patterns between LF and EF were nearly opposite and the levels of gene expression differed considerably for these four genes. The expression patterns of PaHDAC and PaH4 on the other hand appeared to be independent of calendar date and more dependent on developmental stage (Fig. 2).
Expression profiles of putative epigenetic regulation and development regulation genes in the trees from early- (E) and late- (L) bud-flushing families of Norway spruce. For further description, see Fig. 1. PaDAM (adenosine-N6-)-methyltransferase, PaGNAT GCN5-related N-acetyltransferase, PaSET SET-domain-containing protein, PaHDAC histone deacetylase family, PaH3a histone H3a, PaH4 histone H4, PaNLY NEEDLY/FLORICAULA (FLO)/LEAFY (LFY), PaSCF/UIP2 E3 ubiquitin ligase SCF complex subunit SKP1/ASK1/UFO-binding protein (UIP2)
We analysed only two (PaSCF/UIP2 and PaNLY) of the genes that are putatively involved in organ development. E3 protein ligases are components of the ubiquitination mechanism in cells, ensuring substrate specificity and a fine degree of regulation over diverse cellular processes [52]. EST encoding the SCF complex subunit SKP1/ASK1/UFO-binding protein (PaSCF/UIP2), reported to play a role in the regulation of floral organ development [54], was identified in the LFL. It is believed to facilitate the degradation (i.e. control the level) of a negative modulator (X) of LFY protein activity [58]. NEEDLY (PaNLY), which is a homologue of flower meristem-identity genes FLORICAULA (FLO)/LEAFY (LFY), have been identified in Pinus radiata [37, 38] and Picea abies [10] earlier.
The PaNLY transcript level had a clear decreasing trend, reaching a minimum during and after bud burst. It was always higher for LF and was not correlated with calendar date but with internal developmental stage (Fig. 2). The expression pattern of PaSCF/UIP2 was less clear and more closely correlated with developmental stage than with calendar date. For both LF and EF, the PaSCF/UIP2 transcript level was elevated during the first three developmental stages. However, for EF the PaSCF/UIP2 expression level was decreasing over the later stages of the studied period, while for LF it was increasing. The lowest transcript level was at 1 week before bud burst for both LF and EF (Fig. 2).
During spring, before bud burst, trees may be subjected to unfavourable abiotic factors (low temperatures/high light intensities, droughts, large day and night temperature fluctuations etc.). We thus considered the network of stress response genes reported for rice [14] and Arabidopsis [31, 36, 46] as a basis for looking for genes involved in dehardening and timing of bud break in spruce. Among the 100 ESTs of abiotic stress- or senescence-related genes, identified in both libraries, we selected two dehydrins, one metallothionein, early responsive to dehydration and aquaporin (AQP). The expression patterns of the chosen stress-related genes are shown in Figs. 3 and 4. In addition, we looked at the transcript level of a cytokinin-dependent light-regulated protein and two protein kinases.
Expression profiles of stress-related genes in the trees from early- (E) and late- (L) bud-flushing families of Norway spruce. For further description, see Fig. 1. PaMtl(Fs) metallothionein-like Fagus sylvatica type, PaDhnX dehydrin 1 and dehydrin 2, PaERD early responsive to dehydration
Expression profiles of genes putatively involved in bud burst regulation in the trees from early- (E) and late- (L) bud-flushing families of Norway spruce. For further description, see Fig. 1. PaLir1 putative cytokinin-repressed, light-regulated protein Lir1; PaL13a 60s ribosomal L13a protein; PaAqp aquaporin; PaSK5 SHAGGY-related protein kinase 5; PaPRKase phosphoribulokinase
Three types of dehydrin ESTs were identified in both libraries. Two of the dehydrin genes most divergent at the amino acid level, PaDhn1 and PaDhn2, were analysed by qRT-PCR. Both genes showed similar pattern of down-regulation. PaDhn1 had a larger amplitude of expression (up to ninefold) than that of PaDhn2 (up to sixfold). In all cases, the expression level of both PaDhn1 and PaDhn2 was higher for LF than that for EF (Fig. 3).
At the same time, we noticed differences in the expression pattern for PaAqp between EF and LF (Fig. 4). For EF, expression of PaAqp was clearly down-regulated all the time, reaching a minimum level just prior to bud burst. For LF, the clear down-regulated expression trend is supplemented by a drastic increasing of expression starting at 2 weeks before bud burst (Fig. 4). The expression pattern of PaERD (early responsive to dehydration stress) gene differed only slightly between EF and LF, showing common decreasing levels until and after the time of bud burst (Fig. 3).
From the three different types of metallothionein-like proteins, we selected PaMtl(Fs)—metallothionein-like protein class II (Fagus sylvatica type)—as less abundant and more similar within group. For LF, PaMtl(Fs) was up-regulated prior to the bud burst and down-regulated thereafter. For EF, on the contrary, expression of PaMtl(Fs) showed nearly the opposite pattern. However, on average, the difference in PaMtl(Fs) expression between EF and LF rarely exceeded twofold and the maximum difference in expression occurred at 1 week before bud burst and reached 18-fold. The fall in expression of PaMtl(Fs) appeared to be closely correlated with calendar date, but not with developmental stage (Fig. 3).
Expression patterns of phosphoribulokinase (PaPRKase) differed between EF and LF. For LF, PaPRKase transcription was up-regulated during the early period (until mid-May), reaching its peak 3 weeks before bud burst; thereafter, the transcription level was decreasing to basal level around the time of bud burst. For EF, the expression level of PaPRKase varied only up to twofold around basal level and with an irregular pattern. The difference in expression between EF and LF reached more than 90-fold at 3 weeks before bud burst (Fig. 4).
The expression pattern of SHAGGY-related protein kinase SK5 (PaSK5), with reduction in transcript levels prior and immediately after bud burst in both LF and EF, is very similar to that of PaNLY. Therefore, we suspect that similar factors regulate the activities of PaSK5 and PaNLY during the spring. The activity of PaSK5 for LF was always higher than for EF at the same calendar date (Fig. 4).
Among the putative light-regulated proteins, the expression of cytokinin-repressed/light-regulated protein Lir1—PaLir1—was studied. For EF, the transcript level of PaLir1 was gradually increasing, reaching a maximum first after bud burst. For LF, gene expression was more or less stable and increased considerably (up to eightfold) in the week before bud burst, followed by a gradual decreasing. Therefore, the period before bud burst coincide with low levels of PaLir1 transcription, while initiation of bud burst coincides with an increased transcript level (Fig. 4).
The expression pattern of 60s ribosomal L13a protein—PaL13a—dropped before bud burst and correlated better with calendar date rather than with developmental stage related to bud burst. When comparing LF and EF at the same developmental stages, the expression pattern of PaL13a showed opposite trends without any obvious relationships with bud burst.
We also analysed the expression of genes with unknown functions (Fig. 5). The expression pattern of PaUnk18 differed significantly between EF and LF, with a nearly opposite pattern in a manner reminiscent of the expression pattern for DNA-binding proteins (Fig. 1). The expression patterns of PaUnk24 and PaUnk35 looked similar and seemed to correlate with calendar date and not with developmental stage. Their expression patterns resembled that of PaNLY and PaL13a, with higher transcript level of EF in comparison with LF before bud burst and lower after. Both genes had highest expression in April.
Expression profiles of four unknown genes putatively involved in bud burst regulation in the trees from early- (E) and late- (L) bud-flushing families of Norway spruce. For further description, see Fig. 1
The expression pattern of the fourth gene (PaUnk44) resembled the pattern for PaHDAC, with gradual increasing in the LF at around the time of bud burst. Therefore, we suggest it could be involved in regulation of bud burst timing in the late-flushing spruce.
Discussion
We have presented our transcriptional data in two different ways, namely, by calendar date and by developmental stage relative to bud burst (Figs. 1–5; panels a and b). When we compare EF and LF trees (and families) at identical time points, we measure the approximate genetic difference between the contrasting trees in response to the same sum of accumulated environmental conditions. This is because their contrasting behaviour is due to a genetic identification based on the covariance among their relatives, their full siblings (Table 1). Moreover, we further increased the contrast between the two trees (used for constructing the subtractive cDNA libraries) by selecting them in opposite directions; the late tree was the latest among their late family members, and the early tree was the earliest within the early family. However, the late tree was in a very different phenological state compared to the early tree. To correct for phenotypic difference, we also compared the genotypes when they approximately attained the same developmental stages. In this comparison, we cannot distinguish between genotypic and environmental factors because they are confounded. We have done this because the phenotypic comparisons could generate interesting hypotheses for further research.
In general, the analysis of gene expression, most notably for LF, which started 7 weeks before bud burst, suggested that flushing is the final event of extensive changes in the expression of a great number of genes that occur in buds before the needles emerge from the bud scales. The period of ecodormancy (no visible traces of growth following dormancy release after chilling), especially at the earliest period, was characterized by very high levels of gene activity during the time when buds prepare to flush. Based on the high expression levels seen even at the start of this experiment, we propose that the starting point for sampling may well be shifted to time points 7–8 weeks or earlier before the bud burst of the early-flushing trees in future experiments. This is to further elucidate the expression of these early stages in ecodormancy.
The patterns of gene expressions between the two ecotypes and the distribution of ESTs in the two libraries were noticeably different. The expression profiles that we report for 25 genes showed that LF expressed a number of genes at higher levels than EF during the whole period. This means that being a late flusher is not a result of a simple delay in gene activity until the necessary temperature sum or day-length are reached. Being late is rather a complex process, involving many genes that may actively delay the time of flushing for 2–3 weeks. We thus propose that being a late flusher is a product of an active process and that specific metabolic processes, developed through evolution, may function as a safeguard against injury from frost. Such genotypes have a selective advantage in areas where late spring frosts would otherwise cause high mortality.
Our results support the assumption that DNA-binding proteins affecting transcription and proteins involved in regulating translation should be involved in bud burst regulation. They are known to be involved in regulation of different processes in plants, e.g. the ZnfC2H2 protein could be related to leaf senescence [33] or floral organogenesis, leaf initiation and seed development [11]. ZfPDOF has been shown to be involved in stress response and seed development [14], and eIF5A has been shown to facilitate translation of a subset of mRNAs required for cell division and to participate in many late developmental processes like senescence and fruit ripening [39].
The expression profiles of genes participating in histone acetylation (PaGNAT) and deacetylation (PaHDAC) suggest that preparation for bud burst involves chromatin remodelling by both acetylation and deacetylation in a complex way. Transcription of sequences putatively involved in acetylation seems to have a nearly opposite direction in EF compared to LF. At the same time, the sequences involved in deacetylation seem to be transcribed at a low level for EF during the entire period, but for LF, the increase is most pronounced during bud swelling, returning later to basal level. PaSET expression (involved in histone methylation) looked very similar to that of PaGNAT, indicating that the chromatin opens up, and allowing for transcriptional activity to start. Methylation of histones correlates with both transcriptional repression and activation, even when it occurs at the same site. This might depend on the particular gene, the type of the functionally redundant HMTs involved, and the level of methylation [35]. Therefore, we suspect that genes that modify histones might play an important regulatory role in the timing of bud burst. We did not find any literature data about the possible role of adenosine methylation in gene regulation and participation in epigenetic regulation in plants. The expression pattern of PaDAM could be the first implication of epigenetic regulation by DAM genes in spruce.
We compared our data with the results of gene expression analysis of several biological processes in plants. These are salt stress and recovery [47], cold acclimation [46, 48], leaf senescence [5], dormancy release [42] and abiotic stresses in rice [14] and Arabidopsis [31, 36]. We found a considerable part of homologues participating in these processes in our subtracted libraries. When trees prepare for spring and summer, they obviously need to protect themselves from unfavourable abiotic factors. They activate stress-related signaling repertoire of genes available.
The expression patterns of water-stress-related genes, the dehydrins and aquaporins, were very interesting. It is generally observed that the levels of dehydrins are lowest during the active growth period and highest during the winter months [30, 55]. The abrupt decrease and low levels of dehydrin transcripts for EF indicate that EF trees had a lower level of freezing tolerance. Low freezing temperatures could damage EF buds during swelling and bursting. For LF, the level of dehydrin transcription decreased gradually, which could mean that freezing tolerance was kept higher during this period of the spring [22]. AQPs are integral membrane proteins, which facilitate water transport across membranes. They are regulated by water, abiotic stresses and ABA treatment [26]. We saw quite different expression patterns between EF and LF for PaAqp. Up-regulation of the PaAqp gene during bud swelling may help increase membrane water permeability and improve water transport (and maybe some other substances) for LF. So water-transport proteins are thus related to the timing of bud burst.
The most abundant ESTs in both libraries were the metallothionein-like proteins. The MTLs have been reported as one of the highly expressed genes in leaf senescence and cell death [5, 40], fruit ripening [34, 39] and defence mechanisms. We presume that some cellular events occur in the LF during the time before bud burst, which demands high level of MTLs activity and increased levels of other stress-related genes.
Our data indicate that some of the protein kinases may play an important role for LF bud burst regulation. SHAGGY-related kinase genes (ASK) play a role in floral meristem patterning [16], suggesting that they may perhaps be involved in vegetative meristem activity regulations as well. PaSK5 could analogously regulate the very early, post-endodormant stages before bud burst, depending more on photoperiod than on temperature. PRKase expression is regulated by light under the control of the circadian clock and is down-regulated during senescence [32, 45]. Similarly, PaPRKase expression for LF bud burst could be triggered by long days and could participate in photoperiodic signaling. A possible light-dependant regulation of bud burst for LF is supported by the expression pattern of PaLir1. The Lir1 homologue PaLir1 seems to be under the control of circadian clocks [24] controlling the timekeeping mechanism of angiosperms and could be involved in mechanisms in the photoperiodic regulation of bud burst as well. We do not find any clear expression pattern indicating the involvement of histone (PaH3a and PaH4) and ribosomal protein (PaL13a) genes in regulation of bud burst, but some noticeable differences in expression were found.
From the sequencing effort on both LFL and EFL, we did not obtain transcripts (ESTs), which are commonly recognized as being involved in terminal bud formation and dormancy in the autumn. Most notably among these presupposed (but not obtained) transcripts are the light-regulated and diurnal clock-controlled genes, which are responsible for photocontrol of development and the circadian clock in Arabidopsis and rice: the phytochromes (PHY), cryptochromes (CRY), CO, LHY, TOC1, ZTL and others (see reviews: [23, 50]). We identified just a few ESTs corresponding to putative light-regulated proteins. This could happen if these genes were eliminated during the subtraction procedure, indicating that circadian clock genes and genes involved in light-regulated development are commonly expressed in both flushing trees or expressed at low levels, and thus, a much larger number of clones would have been needed to be sequenced in order to be able to obtain these. On the other hand, the light-regulated pathway may start to function considerably later, just during bud burst and after, and not been obtained from our libraries for that reason.
Another group of genes which we did not find in our ESTs were the abscisic acid (ABA)-related genes. The QTL approach shows that phytochromes (PHYB1 and PHYB2) and abscisic acid insensitive genes (ABI1B) affect bud set and bud break in Populus [17]. The participation of ABA and other plant hormones in dormancy regulation and outgrowth of buds is documented [49, 57]. On the other hand, we found many ESTs involved in the Ca2+ signaling pathway, which seems to be involved in temperature-dependent signal transduction [53]. These results may indicate that bud burst is mostly temperature-regulated [18]. Nevertheless, the expression profiles of some genes [e.g. PaMtl(Fs), PaLir1, PaAqp] do also indicate that other environmental factors (e.g. photoperiod) may be involved in regulating the timing of bud burst in late-flushing trees of spruce.
The LFL had a higher percentage of ESTs containing good ORFs, but showing no significant similarity to genes with known function (37%). Real-time PCR analysis of four of these unknown genes showed that they have noticeably different expression patterns for EF and LF, similar to the expression patterns of genes with known function. Therefore, we have in fact traced potentially new genes that could be involved in bud burst regulation.
In conclusion, the present study demonstrated significant differences in gene expression between an early- and a late-flushing tree of Norway spruce. Timing of bud burst is a complex character involving function and regulation of multiple genes. Several of the studied genes are putative candidates as markers of timing of bud burst, and they provide a good starting point for further research. Those studies should cover several contrasting genotypes. Further, genetic and relevant physiological studies under more controlled conditions are also needed to investigate the interaction of environmental and genetic regulation of bud burst. Understanding this regulation is a key to identifying genotypes with preferable phenotypic characteristics for future climatic conditions in the temperate and boreal areas.
References
Aitken SN, Adams WT (1997) Spring cold hardiness under strong genetic control in Oregon populations of Pseudotsuga menziesii var. menziesii. Can J For Res 27:1773–1780
Aitken SN, Hannerz M (2001) Genecology and gene resource management strategies for conifer cold hardiness. In: Bigras FJ, Colombo SJ (eds) Conifer cold hardiness. Series: tree physiology, vol 1. Kluwer, Dordrecht, pp 23–53
Berger SL (2002) Histone modifications in transcriptional regulation. Curr Opin Genet Dev 12:142–148
Beuker E (1994) Adaptation to climatic changes of timing of bud burst in populations of Pinus sylvestris L. and Picea abies (L.) karst. Tree Physiol 14:961–970
Bhalerao R, Keskitalo J, Sterky F Erlandsson R, Bjorkbacka H, Birve SJ, Karlsson J, Gardestrom P, Gustafsson P, Lundeberg J, Jansson S (2003) Gene expression in autumn leaves. Plant Physiol 131:430–442
Bhat RA, Riehl M, Santandrea G, Velasco R, Slocombe S, Donn G, Steinbiss H-H, Thompson RD, Becker H-A (2003) Alteration of GCN5 levels in maize reveals dynamic responses to manipulating histone acetylation. Plant J 33:455–469
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Buchanan-Wollaston V, Ainsworth C (1997) Leaf senescence in Brassica napus, cloning of senescence related genes by subtractive hybridization. Plant Mol Biol 33:821–834
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR):trends and problems. J Mol Endocrinol 29:23–39
Carlsbecker A, Tandre K, Johanson U, Englund M, Engstrom P (2004) The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J 40:546–557
Chrispeels HE, Oettinger H, Janvier N, Tague BW (2000) AtZFP1, encoding Arabidopsis thaliana C2H2 zinc-finger protein 1, is expressed downstream of photomorphogenic activation. Plant Mol Biol 42:279–290
Christersson L (1978) The influence of photoperiod and temperature on the development of frost hardiness in seedling of Pinus sylvestris and Picea abies. Physiol Plant 44:288–294
Clapman D, Ekberg I, Little CHA, Savolainen O (2001) Molecular biology of conifer frost tolerance and potential application to tree breeding. In: Bigras FJ, Colombo SJ (eds) Conifer cold hardiness. Series: tree physiology, vol 1. Kluwer, Dordrecht, pp 187–219
Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbuhl P, Ellero C, Goff SA, Glazebrook J (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100:4945–4950
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Dornelas MC, van Lammeren AAM, Kreis M (2000) Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J 21:419–429
Frewen BE, Chen THH, Howe GT, Davis J, Rohde A, Boerjan W, Bradshaw HD Jr (2000) Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154:837–845
Fuchigami LH, Weiser CJ, Kobayashi K, Timmis R, Gusta LV (1982) A degree growth stage (°GS) model and cold acclimation in temperate woody plants. In: Li PH, Sakai A (eds) Plant cold hardiness and freezing stress mechanisms and crop implication. Academic, New York, pp 93–116
Gepstein S, Sabehi G, Carp M-J, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36:629–642
Gu R, Fonseca S, Puskás LG (2004) Transcript identification and profiling during salt stress and recovery of Populus euphratica. Tree Physiol 24:265–276
Hannerz M (1999) Evaluation of temperature models for predicting bud burst in Norway spruce. Can J For Res 29:9–19
Haug H (2002) Phenological differences in Norway spruce (Picea abies) and the ability to meet the changes in climate. Ph.D. thesis, The Agricultural University of Norway
Hayama R, Coupland G (2004) The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol 135:677–684
Hayama R, Izawa T, Shimamoto K (2002) Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol 43:494–504
Howe GT, Aitken SN, Neale DB, Jermstad KD, Wheeler NC, Chen THH (2003) From genotype to phenotype, unraveling the complexities of cold adaptation in forest trees. Can J Bot 81:1247–1266
Jang JY, Kim DG, Kim YO, Kim JS, Kang H (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis Thaliana. Plant Mol Biol 54:713–725
Johnsen Ø (1989) Phenotypic changes in progenies of northern clones of Picea abies (L) Karst grown in a southern seed orchard. II Seasonal growth rhythm and height in field trials. Scan J For Res 4:331–341
Johnsen Ø, Fossdal CG, Nagy N, Mølmann J, Dæhlen OG, Skrøppa T (2005) Climatic adaptation in Picea abies progenies is affected by the temperature during zygotic embryogenesis and seed maturation. Plant Cell Environ. DOI 10.1111/j.1365-3040.2005.01356.x
Johnsen Ø, Dæhlen OG, Østreng G, Skrøppa T (2005) Day length and temperature during seed production interactively affect adaptive performance of Picea abies progenies. New Phytol 168(3):589–596
Kontunen-Soppela S, Laine K (2001) Seasonal fluctuations of dehydrins is related to osmotic status in Scots pine seedlings. Trees, Structure and Function 15:425–430
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol 130(4):2129–2141
Lemaire S, Stein M, Issakidis-Bourguet E, Keryer E, Benoit V, Pineau B, Gerard-Hirne C, Miginiac-Maslow M, Jacquot J-P (1999) The complex regulation of ferredoxin/thioredoxin-related genes by light and the circadian clock. Planta 209:221–229
Lin JF, Wu SH (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39(4):612–628
Liu P, Goh CJ, Loh CS, Pua EC (2002) Differential expression and characterization of three metallothionein-like genes in Cavendish banana (Musa acuminata). Physiol Plant 114(2):241–250
Loidl P (2004) A plant dialect of the histone language. Trends Plant Sci 9(2):84–90
Mahalingam R, Gomez-Buitrago A, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4(3):R20
Mellerowicz EJ, Kathryn H, Adrian W, Astrid C, Christian W (1998) PRFLL, a Pinus radiata homologue of FLORICAULA and LEAFY, is expressed in buds containing vegetative shoot and undifferentiated male cone primordial. Planta 206:619–629
Mouradov A, Glassick T, Hamdorf B, Murphy L, Fowler B, Marla S, Teasdale RD (1998) NEEDLY, a Pinus radiata ortholog of FLORICAULA/LEAFY genes, expressed in both reproductive and vegetative meristems. Proc Natl Acad Sci USA 95:6537–6542
Moyle R, Fairbairn DJ, Ripi J, Crowe M, Botella JR (2005) Developing pineapple fruit has a small transcriptome dominated by metallothionein. J Exp Bot 56:101–112
Navabpour S, Morris K, Allen R, Harrison E, A-H-Mackerness S, Buchanan-Wollaston V (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54:2285–2292
Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, Masuda Y, Hiraga S, Mori H (2002) Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol Microbiol 45:673–695
Pacey-Miller T, Kirsten S, Effie A, Scott T, Ada C, Robert H (2003) Genes associated with the end of dormancy in grapes. Funct Integr Genomics 3:144–152
Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30:5036–5055
Partanen J, Koski V, Hänninen H (1998) Effects of photoperiod and temperature on the timing of bud burst in Norway spruce (Picea abies). Tree Physiol 18:811–816
Raines CA, Longstaff M, Lloyd JC, Dyer TA (1989) Complete coding sequence of wheat phosphoribulokinase, developmental and light-dependent expression of the mRNA. Mol Gen Genetics 220:43–48
Reyes BG, Morsy M, Gibbons J, Varma TSN, Antoine W, McGrath JM, Halgren R, Redus M (2003) A snapshot of the low temperature stress transcriptome of developing rice seedlings (Oryza sativa L) via ESTs from subtracted cDNA library. Theor Appl Genet 107:1071–1082
Sahi C, Agarwal M, Reddy M, Sopory S, Grover A (2003) Isolation and expression analysis of salt stress-associated ESTs from contrasting rice cultivars using a PCR-based subtraction method. Theor Appl Genet 106:620–628
Sanchez-Ballesta MT, Lluch Y, Gosalbes MJ, Zacarias L, Granell A, Lafuente M (2003) A survey of genes differentially expressed during long-term heat-induced chilling tolerance in citrus fruit. Planta 218:65–70
Shimizu-Sato S, Mori H (2001) Control of outgrowth and dormancy in axillary buds. Plant Physiol 127:1405–1413
Spalding EP, Folta KM (2005) Illuminating topics in plant photobiology. Plant Cell Environ 28:39–53
Timmis R, Flewelling J, Talbert C (1994) Frost injury prediction model for Douglas-fir seedlings in the Pacific Northwest. Tree Physiol 14:855–869
VanDemark AP, Hill CP (2002) Structural basis of ubiquitylation. Curr Opin Struct Biol 12:822–830
Viswanathan C, Zhu JK (2002) Molecular genetic analysis of cold-regulated gene transcription. Philos Trans R Soc Biol Sci 357(1423):877–886
Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N (2003) The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15(5):1071–1082
Welling A, Rinne P, Vihera-Aarnio A, Kontunen-Soppela S, Heino P, Palva ET (2004) Photoperiod and temperature differentially regulate the expression of two dehydrin genes during overwintering of birch (Betula pubescens Ehrh). J Exp Bot 55:507–516
Yu Y, Dong A, Shen W-H (2004) Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding and segregation. Plant J 40:699–711
Zeng Y, Nancy R, Allison RK (2003) Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant Mol Biol 51:39–49
Zhao D, Yu Q, Chen M, Ma H (2001) The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128:2735–2746
Acknowledgements
We wish to thanks Lejla Ljevo for her help during RNA extraction and subtractive hybridization, and Lars Paulin for quick and high quality sequencing. The project was supported by the Research Council of Norway (Grant No. 143276/140).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yakovlev, I.A., Fossdal, CG., Johnsen, Ø. et al. Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genetics & Genomes 2, 39–52 (2006). https://doi.org/10.1007/s11295-005-0031-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-005-0031-z