Abstract
The diversity of cyanobacteria in the North-Eastern region of India has not been studied except for a few sporadic and inconclusive reports. Loktak Lake is a huge reservoir for various kinds of organisms, including cyanobacteria. The present study describes the isolation and molecular diversity of 72 filamentous, heterocystous cyanobacterial strains isolated from samples collected from Loktak Lake, its adjoining rice fields and rice fields in Indian Council of Agricultural Research (ICAR) complex, Shillong, Meghalaya, India. The isolated strains belonged to the genera Anabaena, Nostoc, Calothrix, Cylindrospermum and Mastigocladus. The molecular analysis of isolates revealed the occurrence of certain strains being present in the sample collected from the rice fields falling in the catchment area of Loktak Lake, Manipur and rice fields in ICAR complex, Shillong, Meghalaya both. A polyphasic approach based on morphological features and PCR based molecular polymorphism revealed enormous level of molecular diversity. Out of three primers targeted regions used for determining genetic polymorphism, STRR1A produced best fingerprint profile of cyanobacterial strains. The morphological diversity of isolates was assured by light microscope whereas PCR based multiple fingerprint profile was used for molecular characterization. Molecular typing using short tandemly repeated repetitive STRR1A sequences as primer provided strain specific fingerprint profiles of the isolates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyanobacteria constitute an ancient group of morphologically diverse phototrophic gram-negative bacteria. Most of them can fix molecular nitrogen to meet their cellular nitrogen requirements. These mutually exclusive properties have made them a very fascinating and ecologically significant group. Cyanobacteria can be found in terrestrial and aquatic environment as well as in symbiosis with Bryophyta, Pteridophyta, Gymnosperm, Angiosperm and fungi (Rai 1990). Identification and characterization of cyanobacterial strains from habitats like Loktak Lake, a wetland of international importance under Ramsar convention (1990) is extremely important as they affect humans and economically important animals and may provide background information concerning cyanobacterial toxins and toxicology with emphasis on ecological, morphological and physiological aspects. A study by Komarek and Anagnostidis (1989) shows that more than 50% strains in culture collections are misidentified. Lack of enough taxonomic criteria has led to incorrect assignment of cyanobacteria in different groups. Morphologically similar strains differ greatly at molecular level and vice versa. Such changes become evident in field isolates maintained under artificial culture conditions. This include morphological features and physiological characteristics e.g. pigment composition, variation in vacuole formation, akinete production etc. Hence use of phenotypic characters in combination with molecular markers as part of a robust approach seems to be a better method to understand molecular affiliation and systematics of cyanobacteria and the composition of natural cyanobacterial communities (Garcia Pichel et al. 1998).
Repeated sequences constitute an important part of the prokaryotic genome. Various categories of repetitive DNA sequences were initially reported in the genome of Calothrix sp. (Mazel et al. 1990) and then in Anabaena sp strain PCC 7120 Masepohl et al. (1996). These were also later identified in other heterocystous cyanobacterial genera (Prasanna et al. 2006; Rasmussen and Svenning 1998). The short repeated sequences dispersed in the genome of the bacterial species have been used as primer target sites for developing their DNA fingerprint profile. Similarly, unique distribution of repetitive sequences has been used as primer targets for generating PCR based specific DNA fingerprint profile of cyanobacterial strains (Prasanna et al. 2006; Rasmussen and Svenning 1998). Cyanobacteria being a highly diverse group of prokaryotes, need isolation and characterization of more genera using molecular methods. The diversity of cyanobacteria has focussed attention of researchers to realize their untapped applied and environmental potential. This attains added significance as many cyanobacteria are known to produce bioactive compounds e.g. toxins (Halinen et al. 2007; Namikoshi and Rinehart 1996; Sivonen et al. 1992) in addition to being useful sources of pharmaceutical, cosmetics, protein, biofertilizer etc. The North-East region of India has been described as a biodiversity hot spot harbouring different kinds of flora and fauna unique to this region. The semitropical climatic condition augmented with high annual rainfall has played a crucial role enhancing the biodiversity richness that supports luxuriant growth of cyanobacteria as well. However, little information is available about occurrences of cyanobacteria in Loktak Lake and adjoining areas (Tiwari and Singh 2005). The present study describes for the first time the molecular diversity of cyanobacteria in Loktak Lake, its adjoining rice fields and further isolation of some of those genotypes from the rice fields in Indian Council of Agricultural Reserach (ICAR) complex, Shillong, Meghalaya as well.
Materials and methods
Sample collection sites
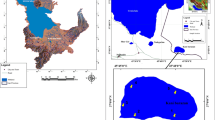
Loktak Lake is the largest freshwater wetland in the North-Eastern region of India situated between 24° 25′ to 24° 42′ N latitudes and 93° 46′ to 93° 55′ E longitudes. It is a shallow wetland with depth varying from 0.5 m to 10 m. This lake was designated as wetland of international importance under Ramsar convention in 1990 because of its biological richness where naturally occurring Phumdis cover the lake extensively and provide a specialized habitat for many Biotas besides being useful to the local people in many ways. The direct catchment area of the lake is 1040 sq km (Devi et al. http://www.gisdevelopment.net/application/nrm/water/overview/ma0790htm). Samples for isolation of cyanobacteria were collected from a wide range of locations in Loktak Lake and from rice fields in ICAR complex, Shillong, Meghalaya. This included cyanobacteria growing as thin film or mat on surface water or from a depth of about 50 cm to 100 cm of water, and from soil surface covered with few centimetres of water, from benthic and epiphytic substrata and from exposed surface of roots and stems of some angiospermic plants. In addition, isolates were also obtained from rice fields falling in the catchment area of the lake. All the samples were collected by using clean sterilized implements and stored at 4°C until further use. The samples collected were brought to the laboratory at North-Eastern Hill University, Meghalaya, India and stored at 4°C in the cold room.
Strain isolation and cultivation
Environmental samples were washed twice in sterilized BG110 (N2 medium) media and suspended in the same. A small portion (50 μl) of the suspension was spread on nutrient agar BG110 plates without combined nitrogen source and incubated at 22 ±2°C with a photon fluence rate of 30 μmol.photons m−2 s−1 of standard culture conditions for 2 weeks (Rippka et al. 1979). From these plates, cyanobacterial strains were isolated to their axenic forms as described in Packer and Glazer (1988). Final purification of individual strains was achieved by subjecting to cycloheximide and polymyxin B sulphate treatment in nutrient agar BG110 medium. Identification of clonal strains was perfomed according to Desikachary (1959) and clonal cyanobacterial isolates were grown and maintained at 22 ±2°C under the light intensity of 30 μmol photons.m−2 s−1 in BG110 media or in media supplemented with nitrogen sources as 5 mM NaNO3 (NO3 media).
DNA fingerprint profile analysis
For molecular characterization, 6 days old axenic culture of cyanobacterial strains were washed twice and suspended in an appropriate volume of sterile milli Q water to obtain few filaments in every 1 μl suspension. 1 μl from such suspension was used directly as template in Rep-PCRs with STRR1A (5′-CCARTCCCCARTCCCC-3′) or STRR1B (5′-GGGGAYTGGGGAYTGG-3′) as described by Rasmussen and Svenning (1998) or with ERIC (forward primer ERIC1R: 5′ATGTAAGCTCCTGGGGATTCAC-3′ and reverse primer ERIC2:5′-AAGTAAGTGA CTGGGGTGAGCG-3′) primers as described by Bruijn et al. (1992). The PCRs were performed in 50 μl reaction volume containing 100 pmol of each primer, 1.25 mM deoxynucleoside triphosphate, and 1 μl of cyanobacterial filaments pre-treated at 100°C for 10 min as template and 2 U of DNA polymerase (Bioline) using Applied Biosystems 2720 thermal cycler. The buffer supplied with Taq pol enzymes was used according to the manufacturer’s instruction. For the STRR primers, the cycles used were as follows: 1 cycle at 95°C for 6 min, 35 cycles at 94°C for 1 min, 56°C for 1 min, 65°C for 5 min, 1 cycle at 65°C for 16 min and a final soak step at 4°C. For the ERIC primers, the first cycle at 95°C for 7 min was followed by 30 cycles at 94°C for 1 min, at 52°C for 1 min, and at 65°C for 8 min; 1 cycle at 65°C for 16 min; and a final soak step at 4°C. After the reaction, 6 μl of DNA amplicon was electrophorsed at 80 V in 1.5% agarose gels in 1xTAE buffer (0.04 M Tris–acetate, 0.001 M EDTA, pH 8.0) for 1.5 h. The PCR amplicons thus separated were visualized on a transilluminator under UV light after staining with ethidium bromide by using a CCD camera (Olympus Camedia C4000).
Results
Strain isolation and identification
The designations of 72 strains of filamentous heterocystous cyanobacteria isolated from different sites and habitats in Loktak Lake and 15 Strains of Anabaena spp and Nostoc spp isolated from rice fields falling in the catchment area of Loktak Lake in Manipur and from rice fields in ICAR complex, Shillong, Meghalaya used in this study are given in Table 1. The isolates were identified morphologically by microscopic observation under light microscope. They belonged to the following genera: Anabaena, Nostoc, Calothrix, Cylindrospermum and Mastigocladus.
Distribution pattern of cyanobacterial isolates
Cyanobacterial strains, either planktonic or benthic or epilithic or epiphytic, were isolated from four distinct sites (Table 2) during September 2007 which are as follows: Open water (A), Shore of the lake (B), Phumdis (C) and aquaponds at the shore of the lake (D). Occurrence pattern of individual strains was analyzed by enumerating individual strains in different habitats. Out of the four habitats, phumdis and lake shore were found to be supporting maximum number of cyanobacterial species whereas aquaponds supported the least. This might be because of nutrients availability in phumdis and shore of the lake. A total of 30 taxa dominated by Anabaena spp were present on phumdis occurring mainly as epiphytes on the surfaces of annual angiospermic plants. STRR1A based analysis of cyanobacterial strains isolated from site C samples collected during September 2008 reconfirmed the prevalence of previously isolated identical strains during September 2007. Numbers of cyanobacterial strains isolated from the other three sites were 13 strains from site A, 24 strains from site B, and 5 strains from site D. Strains of Anabaena and Nostoc were found to occur in samples from all the 4 sites whereas Calothrix strains could be found only in samples from site B. Cylindrospermum spp were found to occur in samples from all the sites except for site A. Mastigocladus laminosus occurred in samples from site B and C.
Strain differentiation by Rep-PCR generated fingerprint profile
The use of STRR1A target sequence in PCR were found to be more efficient generating strain specific DNA fingerprint profile of cyanobacterial strains isolated from different sites and habitats of Loktak lake and its adjoining rice fields as compared to when STRR1B and ERIC primers (data not shown) were used in Rep-PCR. DNA template from all isolated strains revealed the capacity to produce PCR based multiple DNA amplicons with primer STRR1A. Figs.1a, 1b, and 1c represent STRR1A DNA fingerprint profile of strains of Anabaena oscillarioides, Anabaena fertilissima and Anabaena variabilis respectively. A. oscillarioides isolates A26, A27 and A28, out of their five isolates, generated identical multiple DNA fingerprint profile indicating their similarity at molecular level (Fig 1a). Similarly DNA templates from four isolates (strain numbers A23, A24, A38 and A39) of A. fertilissima produced similar multiple fingerprint profiles (Fig 1b). Further, STRR1A based molecular analysis of 15 Anabaena variabilis strains generated 12 distinct DNA fingerprint profiles with strain numbers A4 and A5 generating identical DNA profile. Also, strain number A13 and A14 generated identical DNA profile. The remaining 11 strains produced unique multiple DNA profiles (Fig. 1c). DNA templates from three strains each of Anabaena naviculoides (strain numbers A35, A36 and A37) and Anabaena oryzae (strain numbers A32, A33 and A34) produced unique DNA fingerprint profiles with STRR1A primer. Similarly DNA template from Anabaena vaginicola isolates from phumdis (strain number A30) and from open water (strain number A31) produced distinct fingerprint profile. All strains of Nostoc punctiforme, Nostoc spongiaeforme, Nostoc piscinale, Nostoc linckia, Cylindrospermum spp and Mastigocladus laminosus produced unique DNA fingerprint profiles with STRR1A primer. Five cyanobacterial strains belonging to the genera Anabaena (RM11,RM21,RM22,) and Nostoc (RM12,RM13) isolated from the rice plant rhizosphere from rice fields falling under the catchment area of Loktak lake were also found to occur in rice fields in ICAR complex, Shillong, Meghalaya as revealed by STRR1A fingerprinting profile (Fig. 2a, b). Nostoc strain RS12 isolated from the rice fields in Shillong produced identical DNA fingerprint profile as that of Nostoc linckia Strain N15 isolated from the phumdis in Loktak Lake (Fig 2c). The STRR1A based DNA fingerprint profiles were, therefore, able to distinguish and differentiate closely related strains and were also specific enough to relate strains isolated from distinct locations.
Discussion
To the best of our knowledge there has been only a single report on cyanobacterial diversity in Loktak Lake based on morphological attributes (Tiwari O and Singh 2005). Therefore objectives of the present study were to isolate heterocystous cyanobacterial strains from Loktak Lake and analyse molecular diversity based on their multiple DNA fingerprint profile.
The biochemical plasticity and diversity of cyanobacteria has enabled them to occupy almost every conceivable habitat on the earth. The present study describes the distribution pattern and diversity of filamentous cyanobacterial strains in Loktak Lake, categorize them in different morphological groups followed by comparing such isolates on the basis of their multiple DNA fingerprint profile. Distribution pattern indicates the presence and dominance in all the samples collected from different sites and habitats by Anabaena and Nostoc strains emphasizing their ability to adapt to a wide range of ecological niche. Calothrix species were found to occur near the shore of the lake on benthic habitat or as free floating form near the shore. The absence of Calothrix spp in samples from other site might be due to the isolation biases. Calothrix strains formed brown patches on soil or rock surfaces. Alternatively, they were deep blue green in colour in their free floating forms. Mastigocladus laminosus seems to prefer to grow on the soil surface, rock surface or surface of small plants as their abundances were maximum in soil samples from phumdis and shore of the lake. Cylindrospermum spp. forms free floating scum or mat with Anabaena/Nostoc or other non-heterocystous cyanobacteria such as Phormidium. While growing on the soil surface it forms patches which remain dark blue green in colour.
Analysis of genotypic diversity of cyanobacterial strains using tandemly repeated repetitive genomic DNA sequences has been reported (Prasanna et al. 2006; Rasmussen and Svenning 1998; Zheng et al. 1999). Number of short tandemly repeated sequences has been found to be about 100 per genome in Calothrix sp by hybridization experiment (Mazel et al. 1990). The specificity of genotyping of microbes using STRR1A and STRR1B primers varies. Compared to STRR1A primer, STRR1B primer is less specific for cyanobacteria as it produces amplicons with other bacterial genomes as template (Rasmussen and Svenning 1998). In addition, many cyanobacterial isolates failed to produce or yielded few PCR amplicons with STRR1B primer as compared to when STRR1A primer was used for generating strain specific fingerprint profile. A good correlation between identification of cyanobacterial strains using STRR and LTRR based fingerprint pattern and 16S rRNA gene sequence has been reported (Valério et al. 2009). Use of STRR1A primer for genotypic cyanobacterial diversity analysis appears to be an appropriate approach (Nayak et al. 2009). In the present study, use of STRR1A primer for developing strain specific DNA fingerprint profile could efficiently distinguish closely related cyanobacterial strains. Some of the isolates from different sites produced similar/identical DNA fingerprint profile indicating their similarity at molecular level e.g. group of strains A4 & A5; strains A26, A27 & A28; strains A23, A24, A38 & A39 and strains A13 & A14 (Table 2 and Fig 1). Although, the use of ERIC primers in Rep-PCR was also able to generate strain specific reproducible PCR profile, the generated profile appears to be complicated. Moreover, the presence of ERIC sequences among eubacteria, primarily in the gram-negative group, requires the use of axenic culture which is often difficult and time consuming (Rasmussen and Svenning 1998).
STRR1A primer was able to establish the similarity of cyanobacterial diversity and the similarity of certain strains in different Gunnera species originating from different geographical areas (Nilsson et al. 2000). In the present study the occurrence of certain strains of Anabaena spp and Nostoc spp revealed by using STRR1A primer based PCR fingerprinting in the rice field isolates of Manipur and Shillong signifies the efficiency of STRR1A primer in determining the similarity of cyanobacterial diversity in various rice fields. The presence of these cyanobacterial strains in distinctly located rice fields highlights their ability to adapt in rice fields. Such strains could be exploited as potential candidate for their use as biofertilizer.
In conclusion, Loktak Lake shows a high level of cyanobacterial diversity dominated by Anabaena and Nostoc. Further, a combination of morphological and PCR based molecular method was able to differentiate closely related strains with some of the isolates being present in the rice field in Manipur and Shillong both.
References
de Bruijn FJ (1992) Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ Microbiol 58:2180–2187
Desikachary TV (1959) Cyanophyta. ICAR Monograph on Algae. ICAR, New Delhi, India
Devi R S, Srivastava P, Gupta A. Catchments characterization of Loktak lake using remote sensing and geographical information system (GIS) techniques. (http://www.gisdevelopment.net/application/nrm/water/overview/ma0790.htm)
Garcia Pichel F, Nübel U, Muyzer G (1998) The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol 169:469–482
Halinen K, Jokela J, Fewer DP, Wahlsten M, Sivonen K (2007) Direct evidence for production of microcystins by Anabaena strains from the Baltic Sea. Appl Environ Microbiol 73:6543–6550
Komarek J, and Anagnostidis K (1989) Modern approach to the classification system of cyanophytes. 4. Nostocales. Arch Hydrobiol (Suppl. 83) (Algol Stud) 56: 247–345
Masepohl B, Görlitz K, Böhmen H (1996) Long tandemly repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim Biophy Acta 1307:26–30
Mazel D, Houmard J, Castets AM, de Marsac NT (1990) Highly repetitive DNA sequences in cyanobacterial genomes. J Bacteriol 172:2755–2761
Namikoshi M, Rinehart KL (1996) Bioactive compounds produced by cyanobacteria. J Industrial Microbiol 17:373–384
Nayak S, Prasanna R, Prasanna BP, Sahoo D (2009) Genotypic and phenotypic diversity of Anabaena isolates from diverse rice agroecologies of India. J Basic Microbiol 49:165–177
Nilsson M, Bergman B, Rasmussen U (2000) Cyanobacterial diversity in geographically related and distant host plants of the genus gunnera. Arch Microbiol 173:97–102
Packer L, Glazer A N (1988) Cyanobacteria. Methods in Enzymology; Vol.167; Academic press London
Prasanna R, Kumar R, Sood A, Prasanna BM, Singh PK (2006) Morphological, physiochemical and molecular characterization of Anabaena strains. Microbiol Res 161:187–202
Rai AN (1990) In Handbook of symbiotic cyanobacteria. A. N. Rai (ed) CRC Press, Boca Raton, Fla
Rasmussen U, Svenning MM (1998) Fingerprinting of cyanobacteria based on PCR with primers derived from short and long tandemly repeated repetitive sequences. Appl Environ Microbiol 64:265–272
Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Genetic assignment, strain histories and properties of pure culture of cyanobacteria. J Gen Microbiol 111:1–61
Sivonen K, Namikoshi M, Evans WR, Carmichael WW, Sun F, Rouhiainen L, Luukkainen R, Rinehart KL (1992) Isolation and characterization of a variety of microcystins from seven strains of the cyanobacterial genus Anabaena. Appl Environ Microbiol 58:2495–2500
Tiwari O N, H Tombi Singh (2005) Biodiversity of Cyanobacteria in Loktak Lake and rice fields of Manipur, India having acidic properties. Proc Nat Acad Sci India 75 (B) III: pp 209–213
Valério E, Chambel L, Paulino S, Faria N, Pereira P, Tenreiro R (2009) Molecular identification, typing and traceability of cyanobacteria from fresh water reservoirs. Microbiol 155:642–656
Zheng WW, Nilsson M, Bergman B, Rasmussen U (1999) Genetic diversity and classification of cyanobacteria in different Azolla species by the use of PCR fingerprinting. Theor Appl Genet 99:1187–1193
Acknowledgments
Financial assistance from Department of Science & Technology, Government of India vide project number SP/SO/PS-56/2003 is gratefully acknowledged. Authors would like to thank Dr KD Pandey, Botany Department, BHU, Varanasi, India for assistance in strain identification and Akoijam Rina for checking the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akoijam, C., Singh, A.K. Molecular typing and distribution of filamentous heterocystous cyanobacteria isolated from two distinctly located regions in North-Eastern India. World J Microbiol Biotechnol 27, 2187–2194 (2011). https://doi.org/10.1007/s11274-011-0684-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0684-8