Abstract
This study was conducted to evidence the dissemination potential of antibiotic resistance genetic elements in E. coli isolates of Costa Rican domestic wastewater treatment plants (WWTPs). Few studies have addressed this phenomenon in WWTPs in Central America. Phenotypical resistance profiles to β-lactams, quinolones, aminoglycosides, phenicols, tetracyclines, and folate pathway inhibitors of 133 Escherichia coli isolates from the influent and effluent of two urban WWTPs located in the Greater Metropolitan Area of Costa Rica were described. Thirty multidrug-resistant profiles were identified and grouped into 15 genetic clones by ERIC-PCR; 6 of 15 genetic clones were from effluents. Six of the seven examined genes (sulI, sulII, intI1, intI2, blaTEM, and tetA) were found in multidrug-resistant isolates, whereas blaOXA was absent. The horizontal gene conjugation test confirmed the gene transfer capacity of all tested isolates n = 8. Multidrug-resistant isolates in effluents with horizontal gene transfer capacity suggest that Costa Rican WWTPs represent spots related to antibiotic resistance spread to the environment. In domestic WWTPs, we found that nearly 22% of E. coli isolates presented a multidrug-resistant phenotype capable of transferring their resistance determinants by conjugation processes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Antimicrobial resistance remains a global health threat. By 2050, it is estimated that up to 10 million deaths associated with antimicrobial resistance will occur (O’Neill et al., 2016). The World Health Organization has described the multifactorial causes of antimicrobial resistance (The Interagency Coordination Group on Antimicrobial Resistance (IACG), 2019). Misuse in clinical contexts (Goossens et al., 2005) and even expected antibiotic use in dense populations exert a selective pressure on bacteria to maintain antibiotic resistance elements, among other factors (Levy & Bonnie, 2004). In addition, the prophylactic use of antibiotics as a growth factor in animal breeding and agricultural processes liberates these molecules into the environment (Davies & Davies, 2010). Likewise, the industrial production of antibiotics releases high concentrations of antimicrobial drugs into the environment through wastewater (Akhil et al., 2021).
Wastewater treatment plants (WWTPs) have been described as hot spots for the dispersion and evolution of antibiotic-resistant bacteria as they are antibiotic accumulation sites and generally have low antimicrobial compound removal efficiencies where resistance selection pressures exist (Michael et al., 2013). Water used in industrial processes, domestic households, and healthcare facilities is discarded and directed to wastewater treatment systems. Both organic and inorganic compounds, such as nutrients, drugs — including antibiotics — and heavy metals, are combined with a rich diversity of microbes (Hubeny et al., 2021). In Costa Rica, 53% of installed WWTPs use activated sludge as a treatment technology. Regarding end-use, 77% are disposed in a receiving water body, and 18% are reused (Centeno Mora & Murillo Marín, 2019).
Multiple bacterial species are considered reservoirs of resistance in WWTPs (Everage et al., 2014). For example, Escherichia coli is a well-known biomarker for contamination associated with human activity, as it is a constituent part of the intestinal microbiome. In addition, multidrug-resistant E. coli strains are present in influents, effluents, and sludge of WWTPs (Garcia et al., 2007), and horizontal gene transfer mechanisms have been identified (Silva et al., 2006), making it a good model to study resistance determinants in WWTPs and their dissemination. Furthermore, Escherichia coli removal by activated sludge technology is estimated between 1 and 2 log10 (Barrios-Hernández et al., 2020), which can lead to high concentrations of E. coli being discharged into receiving water bodies. Also, there is limited information regarding WWTPs’ role in resistance dissemination in Central America (D. C. Domínguez et al., 2021).
This study aimed to investigate the dissemination potential of resistance determinants of Costa Rican WWTPs. Phenotypical resistance profiles of E. coli isolates from the influent and effluent of two urban activated sludge-based WWTPs located in the Greater Metropolitan Area of Costa Rica were described. From multidrug-resistant isolates, clonal diversity and specific resistance genes were determined. Furthermore, phenotype changes and molecular analysis were used to assess the horizontal resistance-related gene transfer capacity of E. coli multidrug-resistant isolates.
2 Materials and Methods
2.1 Sample Collection
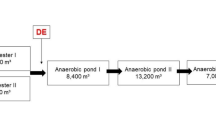
Wastewater samples were collected from two urban WWTPs located in the Greater Metropolitan Area of Costa Rica (WWTP1: 9°56ʹ26.0159ʺ N, 84°16ʹ36.3719ʺ W; WWTP2: 9°55ʹ14.9880ʺ N, 84°14ʹ35.9519ʺ W, coordinate system WGS84). Both WWTPs are considered small (i.e., treating waste from less than 4000 inhabitants) and only receive domestic wastewater. They consist of primary treatment followed by secondary treatment via conventional activated sludge processes. No further disinfection processes are used. The effluents are discharged into river surface waters that were also source water for agricultural irrigation.
Samples from the influent (250 mL) and effluent (250 mL) of both WWTPs were collected on three consecutive days between 9:00 a.m. and 12:00 p.m. Six samples (three influent and three effluent) for each WWTP were collected. Sample collection occurred in May (samples from A642 to A655), October (samples from A1156 to A1176), and December (samples from A1516 to A1539) 2013. In total, 36 samples (18 influent and 18 effluent) were collected in sterile amber bottles and maintained at 4 °C until further analysis.
2.2 Escherichia coli Quantification
All samples were analyzed for Escherichia coli using Standard Methods for the Examination Water and Wastewater 9221E Method (most probable number method) within eight hours of collection (American Public Health Association (APHA) et al., 2017). Briefly, influent and effluent samples were serially diluted to 1:1,000,000 and 1:100,000, respectively. Dilutions were inoculated in a series of five tubes with lauryl sulfate broth and incubated at 35 °C for 48 ± 4 h. After this enrichment step, an inoculum of each positive tube (bacterial growth and gas) was transferred to EC medium with methylumbelliferyl-β-glucuronide (EC-MUG) broth and were incubated for 24 ± 2 h at 44.5 °C; positive samples for Escherichia coli had bacterial growth, gas, and fluorescence characteristics (Chacón et al., 2020). A positive control (E. coli ATCC 25922), a negative control (S. enterica serovar Enteritidis ATCC 13076), and a blank (containing the dilution buffer as the inoculum) were analyzed alongside all samples. No contamination was observed, and all positive and negative controls generated positive and negative results, respectively.
2.3 Escherichia coli Isolation
All positive tubes of the most concentrated dilution from the enrichment phase (turbidity and gas) were pooled. Each pool was inoculated in MacConkey (Oxoid®) following Chacón et al. (2012). After 24 ± 2 h of incubation at 35 ± 0.5 °C, 15 lactose-positive and five lactose-negative colonies — a total of 20 per sample — were inoculated in trypticase soy broth with 20% of glycerol (Oxoid®). Lastly, all broths were incubated for 24 ± 2 h at 35 ± 0.5 °C and then stored at − 70 °C for further analyses. Our study included 36 wastewater samples (18 influent and 18 effluent) from which 720 isolates were isolated.
2.4 Escherichia coli Biochemistry Characterization and Isolation
Each frozen broth was inoculated onto Levine EMB agar (Oxoid®). After 18–24 h of incubation at 35 ± 0.5 °C, colonies with typical E. coli phenotype (greenish metallic sheen by reflected light) were selected for further phenotypical confirmation using EC medium supplemented with methylumbelliferyl-β-glucuronide (EC-MUG broth, (Oxoid®)) and incubated for 24 ± 2 h at 44.5 °C. Those isolates displaying gas production and fluorescence at 295 nm were considered E. coli. E. coli ATCC 25922 was used as a control, both in the Levine EMB agar and CEC-MUG broth.
2.5 Antibiotic Susceptibility Testing
Kirby-Bauer disc diffusion method was performed using Müller Hinton agar (Oxoid®) (Hudzicki, 2009). CLSI thresholds were used to classify isolates as susceptible or resistant (CLSI, 2018). The following antimicrobial susceptibility discs (OXOID®) were used: penicillin (amoxicillin (AML 10 µg)), first-generation cephalosporins (cephalothin (KF 30 µg), cefazolin (KZ 30 µg)), third-generation cephalosporins (ceftazidime (CAZ 30 µg) and cefotaxime (CTX 30 µg)), aminoglycosides (gentamicin (CN 10 µg) and amikacin (AK 30 µg)), quinolones (nalidixic acid (NA 30 µg), fluoroquinolones (norfloxacin (NOR 10 µg)), tetracycline (TE 30 µg), chloramphenicol (C 30 µg), and folate pathway inhibitors (trimethoprim-sulfamethoxazole (SXT 25 µg)). E. coli ATCC 2592 was used as a reference strain. Those isolates resistant to three or more antibiotics from different categories were classified as multidrug-resistant strains (Magiorakos et al., 2012).
2.6 Molecular Analysis
2.6.1 DNA Extraction
DNA extraction from multidrug-resistant isolates was executed according to Barrantes et al. (2010) using the phenol-chloroform method. First, each isolate was inoculated in 1.5-mL trypticase soy broth and incubated overnight at 35 ± 0.5 °C. After overnight growth, they were centrifuged at 6000 rpm (Eppendorf® 5417C) for 6 min. The resultant pellets were resuspended in 560 µL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Thirty microliters of SDS (10% v/v, Sigma®) and 70 µg of proteinase K (Thermo Fisher Scientific®) were added. Once homogenized, this mixture was incubated for 1 h at 38 °C ± 0.5 °C. Afterward, 100 µL of NaCl 5 M (Fermont®) and 80 µL of a mixture of CTAB (0.28 M)/NaCl (0.7 M) (Merck® and Fermont®) were added and incubated for 10 min at 65 °C ± 0.5 °C.
After this incubation, 650 µL of chloroform/isoamyl alcohol (24:1) (Sigma-Aldrich®) was added and mixed. Then, to separate the phases, the mixture was centrifuged for 10 min at 10,000 rpm (Eppendorf® 5417C). Next, the supernatant was transferred and mixed with 650 µL of a mixture of phenol/chloroform/isoamyl alcohol (25:24:1) (Sigma-Aldrich®) was added. Then, it was centrifuged for 10 min at 10,000 rpm (Eppendorf® 5417C). Later, this supernatant was recovered. Then 450 µL of ice-cold isopropanol (Sigma-Aldrich®) was added and incubated overnight at 4 °C. Microtubes were centrifuged at 12,000 rpm for 10 min (Eppendorf® 5417C). The resultant pellets were washed with 70% ice-cold ethanol twice. Then, each pellet was resuspended in 250 µL of TE buffer. After 24 h, purity and concentration were measured using BioPhotometer Plus (Eppendorf ®). DNA was stored at − 80 °C until further analysis.
2.6.2 Escherichia coli 16SrRNA Gene
The presence of the specific 16S rRNA gene from E. coli was confirmed for each bacterial isolate. Briefly, a 25-µL reaction was prepared using DreamTaq 2X Master Mix (Thermo Fischer Scientific®), 0.8 µM of each primer (detailed primers and cycling conditions are shown in Table 1), and 200 ng of DNA. A Veriti thermal cycler system (Applied Biosystems®) was used to perform PCR. E. coli ATCC 25922 was used as a positive control.
2.6.3 Enterobacterial Repetitive Intergenic Consensus Sequences (ERIC)
The ERIC-PCR method proposed by Versalovic et al. (1991) and the GelJ software (Heras et al., 2015) were used to analyze the clonal relationship of all E. coli isolates. Twenty-five microliter reaction volumes were prepared using GoTaq® 5 × Master Mix (Promega®), 1.75 mM MgCl2, 0.2 mM of a dNTPs mix (Promega®), 1.2 µM of each primer (detailed primers and cycling conditions are shown in Table 1), and two units of GoTaq® DNA polymerase (Promega®). Two hundred nanogram of isolate DNA was used as a template. Thus, a clonal relationship between isolates was established. A Pearson correlation matrix from densitometric band analysis was calculated to obtain a similarity index between isolates using the software GelJ (Heras et al., 2015). The unweighted pair group method with arithmetic mean (UPGMA) linkage method was followed to build the dendrogram (Heras et al., 2016). The threshold to consider isolates as the same clone was 0.90 (Alsultan & Elhadi, 2022).
2.6.4 Detection of Antibiotic Resistance Genes
Antibiotic resistance genes (blaOXA, blaTEM, intlI, intlII, sulI, sulII) were amplified using Barrantes et al. (2010) methods. In addition, the protocol from Maynard et al. (2003) was used to amplify the tetA gene. The expected amplicon size for each set of primers and cycling conditions are shown in Table 1. The total volume for each reaction was 25 µL, of which 200 ng was isolated DNA. E. coli ATCC 25922 was used as a negative control. INISA 03 strain, E. coli A653-2 strain (accession number: GCA_008806725.1), and Aeromonas hydrophila INISA 09 strain were used as positive controls.
2.7 Resistance Transfer Determination
During the experiment, E. coli JM107 strain was used as a gene acceptor strain to test horizontal gene transfer by conjugation between MDR E. coli isolates. This strain is resistant to nalidixic acid (NA) and susceptible to ampicillin (AMP). Eight selected isolates from this study (susceptible to NA and resistant to AMP) were the donor strains.
First, each donor strain was inoculated onto Luria Bertani (LB) agar plates supplemented with ampicillin (100 µg mL−1). Then, acceptor strain E. coli JM107 was cultivated on LB agar plates supplemented with nalidixic acid (25 µg mL−1) during 18–24 h at 35 ± 2 °C. After incubation, E. coli JM107 (acceptor strain) was mixed with a donor strain using a 5:1 recipient–donor ratio for each cross. Next, this mix was single-streaked on LB agar without antibiotics for 18–24 h at 35 ± 2 °C. Later, an inoculum from the mixture was subcultured for 18–24 h at 35 ± 2 °C on LB plates supplemented with nalidixic acid and ampicillin. The presence of resistance (blaTEM, sulI, sulII) and integron (intI1, intI2) genes was determined in transconjugant bacteria using the PCR as above-mentioned methods. Additionally, resistance phenotype to SXT and AML was performed using the Kirby-Bauer disc diffusion method (Hudzicki, 2009). Antibiotic discs used were SXT (25 µg) and AML (10 µg) (OXOID ®). The interpretation was made following the CLSI (2018) thresholds.
2.8 Statistical Analysis
Differences in the prevalence of antibiotic resistance between influent and effluents of WWTPs were analyzed by using non-parametric test: chi-square, using the software SPSS Statistic 20® software.
3 Results
3.1 Escherichia coli Quantification and Isolation
E. coli counts are shown in the Supplementary Material (Table S1). Removal percentages are shown in Table S1. Influent counts range from 7.9 × 105 to > 1.6 × 107 MPN/100 mL. Effluent counts range from 1.3 × 103 to > 1.6 × 106 MPN/100 mL. Furthermore, both WWTPs discharged over the maximum permitted value into the receiving water body which is 3 log10/100 mL according to national legislation (Ministerio de Ambiente, E. y T., & Ministerio de Salud, 2007). From the 36 wastewater samples, a total of 140 suspected E. coli isolates were recovered from Levine’s agar. According to biochemical criteria, 133 isolates out of 140 were considered E. coli using CEC-MUG broth (OXOID®).
3.2 Antibiotic Susceptibility Testing
Ninety-five percent (n = 126) of isolates with biochemical characteristics of E. coli were considered resistant to at least one of the antibiotics tested. All 133 resistance profiles of the isolates are shown in the Supplementary Material (Table S2). In terms of the frequency of resistance phenotypes, considering both WWTP influent and effluent together, 91% of isolates (n = 121) were amoxicillin resistant, followed by cephalothin (38.3%, n = 51/133), tetracycline (22.6%, n = 30), and nalidixic acid (15.8%, n = 21) (Table 2). Notably, only a single sample was resistant to third-generation cephalosporin. There was a significant increase (χ2 = 6,813, df = 1, p value = 0.014) in the prevalence of sulfonamide resistance between influents and effluents (7.4% vs. 24.6%) as can be seen in Table 2. With other resistance determinants, statistical tests showed no difference between influents and effluents (p > 0.05) (Table 2). Furthermore, we determined that 22.6% of the isolates (n = 30) were classified as multidrug-resistant strains showing resistance from three up to seven different antibiotics. The most common phenotype in effluents was combined resistance to AML-NA-SXT. The most complex resistance phenotype in effluents was AML-KF-KZ-CTX-TE-CN-STX phenotype; meanwhile, in influents, it was AML-KF-TE-NOR-NA-AK-SXT. In total, 17 different multidrug-resistant profiles were described, as shown in Table S3.
3.3 Detection of Antibiotic Resistance Genes
All 30 multidrug-resistant isolates were confirmed as E. coli by 16S rRNA. According to the ERIC-PCR method, these isolates were clustered into 15 clonal groups (Fig. 1). Complete ERIC-PCR electrophoresis results can be seen in Supplementary Material (Fig. S1). The presence of class 1 and 2 integrons was confirmed (Table 3). Also, four out of five studied antibiotic resistance genes were detected, except the blaOXA gene. It should be noted that the phenotype and genotype profiles for isolates belonging to the same clone may differ, as seen in Table 3, except for 4 isolates. Also, the genotypic profiles are shown in Table S3, where the blaTEM, sulI, sulII, intI1, and tetA genotype was the most common in effluents of the 18 different profiles found in multi-resistant bacteria. This genotype was mainly present in effluents of WWTP 1.
Of the thirty multi-resistant isolates, 26 were unique regarding phenotype and genotype. blaTEM was present in 19 isolates, of which 16 also had sulI, 16 sulII, and 14 tetA, and 15 had at least one intI gene. Five isolates show a sul gene without intI. There are no differences in the presence of these between influents or effluents.
3.4 Resistance Transfer Determination
Of the donor strains assessed, 100% generated successful transconjugants with NA and AMP resistance phenotype. Resistance to AML (8/8) and SXT (4/4) was identified in all cases according to the donor strain (Table 4). Furthermore, it was possible to identify the resistance genes present in the donor strain in the transconjugant bacteria.
4 Discussion
The role of WWTPs in multidrug-resistant bacteria and resistance genetic determinant dissemination and their impact on the environment of Central America has yet to be well studied, with just a handful of studies carried out in the region (Amaya et al., 2012). In this study, the frequency of resistance phenotypes observed was consistent with the use of antibiotics in Costa Rica. Particularly, penicillin resistance was found in 88.2% of influent isolates. Penicillins are nowadays the most used antibiotics by the Costa Rican population, followed by other beta-lactam molecules, tetracyclines, and sulfonamides (World Health Organization, 2018). Other studies in the Latin American region show lower resistance rates in domestic sewage were reported, but high rates were found in hospital sewage systems from León, Nicaragua (Amaya et al., 2012). In Mexico, lower penicillin resistance rates (lower than 25%, n = 200) have been found in wastewater plant effluents, while this study found 93.8% (n = 133) of resistance in the same matrix. This is despite the fact that in both countries, beta-lactams are the most prescribed antibiotics. These differences could be due to decreased selective pressure, poor resolution of the methods, or dilution effects (Rosas et al., 2015).
Regarding molecular characterization within clonal groups, E. coli clones may have different phenotypes but similar genotypes (Aristizábal-Hoyos et al., 2019). Therefore, it is necessary to study both clonal relationships and resistance profiles to observe these events. Six of the 15 different multidrug-resistant clones (with one or more isolates) identified in this study were isolated from WWTP effluents. Also, even in the same clone, different genotypes and phenotypes were identified in our study, as seen in Table 3, where different highly related strains isolated from the same sample presented a core of antibiotic resistance genes and phenotypes. For instance, clone 1.3 isolated from WWTP 1 in different months shares blaTEM, sulII, and intI1 genes and AML, KF, and NA resistance. However, despite their close genetic relationships within the clone, each strain can gain or lose resistance determinants and change its phenotype. This event suggests a dynamic transfer environment within WWTPs and the presence of mobile genetic elements both in influents and effluents, as we confirmed with the conjugation test. Therefore, as both effluents of the WWTPs discharge in surface water bodies without further disinfection processes, in this scenario, they can contribute to the spread of transferable resistant determinants into aquatic environments.
Even though this study’s genotyping method was not extensive, this phenomenon was still observed. Methods, such as multilocus sequence typing (MLST), are recommended and widely used for this purpose, but ERIC-PCR assays were compatible with both the aim of this study and our available resources. Therefore, future studies should use MLST-based analyses to standardize strain identification whenever possible.
Further analysis of the genotypical results obtained from the thirty multidrug-resistant isolates, all possessed a gene associated with resistance to sulfonamides: 12% carried only sulI, 20% carried only sulII, and 68% carried both. Using the CARD database (Alcock et al., 2022), of the WGS sequences deposited at NCBI for E. coli, the sulI gene is present in 17% and sulII in 23%. This study’s overall frequencies in multidrug-resistant strains were 80% and 88%, respectively. Regarding the other genes studied, blaTEM-1 is present in 22% and tetA in 21% of the E. coli reported in the CARD database; meanwhile, in this study, those genes were present in 72% and 68% of the multi-resistant isolates, respectively. When interpreting these data, as they are multi-resistant, they are expected to have higher frequencies of resistance and more mechanisms of horizontal gene transfer.
All isolates from effluents were STXR and had functional sul genes. Isolates from influents also were positive for sul genes, but 66% of the multi-resistant isolates from influents were susceptible to SXT despite carrying one or both of these markers. Previous reports confirmed the presence of sulfonamides in both WWTP influents and effluents from other wastewater plants in Costa Rica (Ramírez-Morales et al., 2020). Sulfonamides or similar molecules may have a selective pressure for functional genes in WWTPs. Fazel et al. (2019) also reported a higher prevalence of sulfonamide-resistant genes than phenotypic resistance. sulI genes are generally associated with class I integrons by their presence as part of a conserved 3’-CS region (Deng et al., 2015; Jiang et al., 2019), but not exclusively, as Table 3 shows. The observed antibiotic susceptibility in these isolates is possible when the promoter variant located at the 5ʹCS position of the integron is weakly expressed and mediates poor transcription of the cassettes (Moura et al., 2012). Also, when the cassettes are far from intI, there is a lower transcription and translation rate (Boucher et al., 2007). Moreover, mutated genes encoding no functional proteins do not confer resistance to SXT, (Homma et al., 2002), reversing the resistant phenotype.
On the other hand, sulfonamide resistance genes were transferred successfully even in the absence of the intlI gene (M. Domínguez et al., 2019). This result has also been described previously in clinical isolates (Gündoǧdu et al., 2011). However, it is more likely that sulI resides on other mobile genetic elements because of that conjugation capacity. In our study, 16 of 18 isolates with the tetA gene also had sulII, which aligns with the association between sulI and ISCR elements and co-transference between tetA and sulII genes (Jiang et al., 2019). Interestingly in this study, even strains without resistance to SXT but with sulI or sulII genes could transfer these genes, and those transconjugant strains were also susceptible, supporting previous suggestions. Similar differential expression occurred with two blaTEM-positive isolates (A642-2 and A1167-1), with no consistent phenotype with beta-lactam resistance.
Multidrug-resistant enterobacteria isolated from WWTPs have also been identified as carrying β-lactam resistance determinant genes like blaOXA, blaTEM, blaSHV, and/or blaCTX-M, indicating that these sites are conducive to their storage, release, and lateral transmission. The blaTEM gene is easily disseminated among enterobacteria and constitutes a risk of horizontal transmission in WWTPs (Korzeniewska & Harnisz, 2013). In this study, 19 of 26 isolates with unique genotype–phenotype were positive for this extended-spectrum beta-lactamase gene. It was also possible to demonstrate the bacterial capacity to carry this out with the blaTEM gene (Table 4).
Similar phenotypes and genotypes (Table S3) were found in both influents and effluents within WWTP, with significant enrichment just in the phenotypic sulfonamide resistance and a tendency to increase for beta-lactamics in effluents at phenotypical level (Tables 2 and 3). It is reported that the activated sludge WWTPs have limited effect in removing ARGs (Uluseker et al., 2021) and poor removal efficiencies for fecal indicators (Table S1). This suggests that WWTPs can be reservoirs and hotspots for the enrichment and potential transference of functional genes that enter the wastewater system.
These results show that multidrug-resistant microorganisms can be released from WWTP effluents. As mentioned, both WWTP’s effluents discharge in surface water bodies without further disinfection steps, which contributes to spreading resistant determinants in the environment. Furthermore, we found functional elements of mobile resistance in conjugation assays and differential expression and genotype of resistance genetic determinants even in closely related strains, suggesting a highly dynamic genetic pool within WWTPs that is released into the surrounding natural environment. Knitting these biological phenomena with removal efficiencies of conventional activated sludge technology, the disposal of effluents in receiving water bodies or their reuse, and the lack of legislation regarding ARGs present in discharges (Mohammadali & Davies, 2017) presents a worrisome scenario for the spread of resistance in developing economies such as Costa Rica and other countries which are not contemplating this emerging pollutant and its impact in the environment and thus, human health.
Data Availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Akhil, D., Lakshmi, D., Senthil Kumar, P., Vo, D.-V.N., & Kartik, A. (2021). Occurrence and removal of antibiotics from industrial wastewater. Environmental Chemistry Letters, 19(2), 1477–1507. https://doi.org/10.1007/s10311-020-01152-0
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., Edalatmand, A., Petkau, A., Syed, S. A., Tsang, K. K., Baker, S. J. C., Dave, M., McCarthy, M. C., Mukiri, K. M., Nasir, J. A., Golbon, B., Imtiaz, H., Jiang, X., Kaur, K., … McArthur, A. G. (2022). CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research. https://doi.org/10.1093/NAR/GKAC920
Alsultan, A., & Elhadi, N. (2022). Evaluation of ERIC-PCR method for determining genetic diversity among Escherichia coli isolated from human and retail imported frozen shrimp and beef. International Journal of Food Contamination, 9(1), 1–12. https://doi.org/10.1186/S40550-022-00098-1/TABLES/2
Amaya, E., Reyes, D., Paniagua, M., Calderón, S., Rashid, M. U., Colque, P., Kühn, I., Möllby, R., Weintraub, A., & Nord, C. E. (2012). Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in León, Nicaragua. Clinical Microbiology and Infection, 18(9). https://doi.org/10.1111/j.1469-0691.2012.03930.x
American Public Health Association (APHA), American Water Works Association (AWWA), & Water Environment Environment (WEF). (2017). Standard Methods for Examination of Water and Wastewater. (R. Baird, A. Eaton, & E. Rice, Eds.; 23rd ed.). American Public Health Association Inc.
Aristizábal-Hoyos, A. M., Rodríguez, E. A., Arias, L., & Jiménez, J. N. (2019). High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. Journal of Environmental Management, 245(May), 37–47. https://doi.org/10.1016/j.jenvman.2019.05.073
Barrantes, K., McCoy, C., & Achí, R. (2010). Detection ofShigella in lettuce by the use of a rapid molecular assaywith increased sensitivity. Brazilian Journal of Microbiology, 41(4), 993–1000.
Barrantes, K., Chacón, L., Solano, M., & Achí, R. (2014). Class 1 integrase and genetic cassettes bla oxa and bla tem among multi-drug resistant Shigella isolates in Costa Rica. International Journal of Biological Sciences and Applications, 1(1), 24–27.
Barrios-Hernández, M. L., Pronk, M., Garcia, H., Boersma, A., Brdjanovic, D., van Loosdrecht, M. C. M., & Hooijmans, C. M. (2020). Removal of bacterial and viral indicator organisms in full-scale aerobic granular sludge and conventional activated sludge systems. Water Research X, 6. https://doi.org/10.1016/j.wroa.2019.100040
Boucher, Y., Labbate, M., Koenig, J. E., & Stokes, H. W. (2007). Integrons: Mobilizable platforms that promote genetic diversity in bacteria. Trends in Microbiology, 15(7), 301–309. https://doi.org/10.1016/j.tim.2007.05.004
Centeno Mora, E., & Murillo Marín, A. (2019). Tipología de las tecnologías de tratamiento de aguas residuales ordinarias instaladas en Costa Rica. Revista de Ciencias Ambientales, 53(2), 97–110. https://doi.org/10.15359/rca.53-2.5
Chacón, L., Taylor, L., Valiente, C., Alvarado, I., & Cortés, X. (2012). A DNA pooling based system to detect Escherichia coli virulence factors in fecal and wastewater samples. Brazilian Journal of Microbiology, 43(4), 1319–1326.
Chacón, L., Arias-Andres, M., Mena, F., Rivera, L., Hernández, L., Achi, R., Garcia, F., & Rojas-Jimenez, K. (2021). Short-term exposure to benzalkonium chloride in bacteria from activated sludge alters the community diversity and the antibiotic resistance profile. Journal of Water and Health, 19(6), 895–906. https://doi.org/10.2166/wh.2021.171
Chacón, L., Barrantes, K., Santamaría-Ulloa, C., Solano, M., Reyes, L., Taylor, L., Valiente, C., Symonds, E. M., & Achí, R. (2020). A somatic coliphage threshold approach to improve the management of activated sludge wastewater treatment plant effluents in resource-limited regions. Applied and Environmental Microbiology, 86(17). https://doi.org/10.1128/AEM.00616-20
CLSI. (2018). Performance standards for antimicrobial susceptibility testing. (28th ed. C). Clinical and Laboratory Standards Institute.
Davies, J., & Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews, 74(3), 417–433. https://doi.org/10.1128/MMBR.00016-10
Deng, Y., Bao, X., Ji, L., Chen, L., Liu, J., Miao, J., Chen, D., Bian, H., Li, Y., & Yu, G. (2015). Resistance integrons: Class 1, 2 and 3 integrons. Annals of Clinical Microbiology and Antimicrobials, 14, 45. https://doi.org/10.1186/s12941-015-0100-6
Domínguez, M., Miranda, C. D., Fuentes, O., de La Fuente, M., Godoy, F. A., Bello-Toledo, H., & González-Rocha, G. (2019). Occurrence of transferable integrons and suland dfrgenes among sulfonamide-and/or trimethoprim-resistant bacteria isolated from Chilean salmonid farms. Frontiers in Microbiology, 10(APR), 1–14. https://doi.org/10.3389/fmicb.2019.00748
Domínguez, D. C., Chacón, L. M., & Wallace, D. (2021). Anthropogenic activities and the problem of antibiotic resistance in Latin America: A water issue. Water (Switzerland), 13(19). https://doi.org/10.3390/w13192693
Everage, T. J., Boopathy, R., Nathaniel, R., LaFleur, G., & Doucet, J. (2014). A survey of antibiotic-resistant bacteria in a sewage treatment plant in Thibodaux, Louisiana, USA. International Biodeterioration and Biodegradation, 95(PA), 2–10. https://doi.org/10.1016/j.ibiod.2014.05.028
Fazel, F., Jamshidi, A., & Khoramian, B. (2019). Phenotypic and genotypic study on antimicrobial resistance patterns of E. coli isolates from bovine mastitis. Microbial Pathogenesis, 132(August 2018), 355–361. https://doi.org/10.1016/j.micpath.2019.05.018
Garcia, S., Wade, B., Bauer, C., Craig, C., Nakaoka, K., & Lorowitz, W. (2007). The effect of wastewater treatment on antibiotic resistance in Escherichia coli and Enterococcus sp. Water Environment Research, 79(12), 2387–2395. https://doi.org/10.2175/106143007x183826
Goossens, H., Ferech, M., Vanderstichele, R., & Elserviers, M. (2005). Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. The Lancet, 365(9459), 579–587. https://doi.org/10.1016/S0140-6736(05)70799-6
Gündoǧdu, A., Long, Y. B., Vollmerhausen, T. L., & Katouli, M. (2011). Antimicrobial resistance and distribution of sul genes and integron-associated inti genes among uropathogenic Escherichia coli in Queensland, Australia. Journal of Medical Microbiology, 60(11), 1633–1642. https://doi.org/10.1099/jmm.0.034140-0
Heras, J., Domínguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., & Zarazaga, M. (2015). GelJ - A tool for analyzing DNA fingerprint gel images. BMC Bioinformatics, 16(1), 1–8. https://doi.org/10.1186/s12859-015-0703-0
Heras, J., Domínguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., & Zarazaga, M. (2016). A survey of tools for analysing DNA fingerprints. Briefings in Bioinformatics, 17(6), 903–911. https://doi.org/10.1093/bib/bbv016
Homma, K., Fukuchi, S., Kawabata, T., Ota, M., & Nishikawa, K. (2002). A systematic investigation identifies a significant number of probable pseudogenes in the Escherichia coli genome. Gene, 294(1–2), 25–33. https://doi.org/10.1016/S0378-1119(02)00794-1
Hu, R. M., Huang, K. J., Wu, L. T., Hsiao, Y. J., & Yang, T. C. (2008). Induction of L1 and L2 β-Lactamases of Stenotrophomonas maltophilia. Antimicrobial Agents and Chemotherapy, 52(3), 1198–1200. https://doi.org/10.1128/AAC.00682-07
Hubeny, J., Harnisz, M., Korzeniewska, E., Buta, M., Zieliński, W., Rolbiecki, D., Giebułtowicz, J., Nałęcz-Jawecki, G., & Płaza, G. (2021). Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PLOS ONE, 16(6), e0252691. https://doi.org/10.1371/journal.pone.0252691
Hudzicki, J. (2009). Kirby-Bauer disk diffusion susceptibility test protocol. American Society for Microbiology.
Jiang, H., Cheng, H., Liang, Y., Yu, S., Yu, T., Fang, J., & Zhu, C. (2019). Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Frontiers in Microbiology, 10(August). https://doi.org/10.3389/fmicb.2019.01787
Kerrn, M. B., Klemmensen, T., Frimodt-Møller, N., & Espersen, F. (2002). Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. Journal of Antimicrobial Chemotherapy, 50(4), 513–516. https://doi.org/10.1093/jac/dkf164
Korzeniewska, E., & Harnisz, M. (2013). Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. Journal of Environmental Management, 128, 904–911. https://doi.org/10.1016/j.jenvman.2013.06.051
Levy, S. B., & Bonnie, M. (2004). Antibacterial resistance worldwide: Causes, challenges and responses. Nature Medicine, 10(12S), S122–S129. https://doi.org/10.1038/nm1145
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., & Monnet, D. L. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18(3), 268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Maynard, C., Fairbrother, J. M., Bekal, S., Sanschagrin, F., Levesque, R. C., Brousseau, R., Masson, L., Larivière, S., & Harel, J. (2003). Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149: K91 isolates obtained over a 23-year period from pigs. Antimicrobial Agents and Chemotherapy, 47(10), 3214–3221. https://doi.org/10.1128/AAC.47.10.3214-3221.2003
Mazel, D., Dychinco, B., Webb, V. A., & Davies, J. (2000). Antibiotic Resistance in the ECOR Collection: Integrons and Identification of a Novel aad Gene. Antimicrobial Agents and Chemotherapy, 44(6), 1568–1574. https://doi.org/10.1128/AAC.44.6.1568-1574.2000
Michael, I., Rizzo, L., McArdell, C. S., Manaia, C. M., Merlin, C., Schwartz, T., Dagot, C., & Fatta-Kassinos, D. (2013). Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Research, 47(3), 957–995. https://doi.org/10.1016/J.WATRES.2012.11.027
Ministerio de Ambiente, E. y T., & Ministerio de Salud. (2007). Reglamento de Vertido y Reuso de Aguas Residuales. No 33601-MINAE-S.
Mohammadali, M., & Davies, J. (2017). Antimicrobial resistance genes and wastewater treatment. In Antimicrobial Resistance in Wastewater Treatment Processes (pp. 1–13). John Wiley & Sons, Inc. https://doi.org/10.1002/9781119192428.ch1
Moura, A., Jové, T., Ploy, M. C., Henriques, I., & Correia, A. (2012). Diversity of gene cassette promoters in class 1 integrons from wastewater environments. Applied and Environmental Microbiology, 78(15), 5413–5416. https://doi.org/10.1128/AEM.00042-12
O'Neill, J. (2016). Tackling drug-resistant infections globally: final report and recommendations. Wellcome Collection. Attribution 4.0 International (CC BY 4.0).
Ramírez-Morales, D., Masís-Mora, M., Montiel-Mora, J. R., Cambronero-Heinrichs, J. C., Briceño-Guevara, S., Rojas-Sánchez, C. E., Méndez-Rivera, M., Arias-Mora, V., Tormo-Budowski, R., Brenes-Alfaro, L., & Rodríguez-Rodríguez, C. E. (2020). Occurrence of pharmaceuticals, hazard assessment and ecotoxicological evaluation of wastewater treatment plants in Costa Rica. Science of The Total Environment, 746, 141200. https://doi.org/10.1016/j.scitotenv.2020.141200
Rosas, I., Salinas, E., Martínez, L., Cruz-Córdova, A., González-Pedrajo, B., Espinosa, N., & Amábile-Cuevas, C. F. (2015). Characterization of Escherichia coli isolates from an urban lake receiving water from a wastewater treatment plant in Mexico City: Fecal pollution and antibiotic resistance. Current Microbiology, 71(4), 490–495. https://doi.org/10.1007/s00284-015-0877-8
Silva, J., Castillo, G., Callejas, L., López, H., & Olmos, J. (2006). Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile. Electronic Journal of Biotechnology, 9(5), 533–540. https://doi.org/10.2225/vol9-issue5-fulltext-7
The Interagency Coordination Group on Antimicrobial Resistance (IACG). (2019). No time to wait: securing the future from drug-resistant infections. Report to the Secretary General of the United Nations.
Uluseker, C., Kaster, K. M., Thorsen, K., Basiry, D., Shobana, S., Jain, M., Kumar, G., Kommedal, R., & Pala-Ozkok, I. (2021). A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: Mechanisms and perspectives. Frontiers in Microbiology, 12, 3003. https://doi.org/10.3389/FMICB.2021.717809/BIBTEX
Versalovic, J., Koeuth, T., & Lupski, R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Research, 19(24), 6823–6831. https://doi.org/10.1093/nar/19.24.6823
World Health Organization. (2018). WHO report on surveillance of antibiotic consumption: 2016–2018 early implementation.
Author information
Authors and Affiliations
Contributions
Luis Rivera-Montero: investigation, formal analysis, visualization, methodology, and writing — original draft preparation. Gabriel Acuña-Espinola: investigation, formal analysis, and writing — original draft preparation. Kenia Barrantes: supervision, formal analysis, and writing — review and editing. Keilor Rojas: conceptualization and writing — review and editing. Luz Chacón: conceptualization, funding acquisition, formal analysis, supervision, project administration, and writing — review and editing.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rivera-Montero, L., Acuña, G., Barrantes, K. et al. Multidrug-Resistant Escherichia coli in Costa Rican Domestic Wastewater Treatment Plants Maintains Horizontal Transfer Capacity of Resistance Determinants in Effluents. Water Air Soil Pollut 234, 397 (2023). https://doi.org/10.1007/s11270-023-06401-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-023-06401-w