Abstract
Despite the notable success of combination antiretroviral therapy, how to eradicate latent HIV-1 from reservoirs poses a challenge. The Tat protein plays an indispensable role in HIV reactivation and histone demethylase LSD1 promotes Tat-mediated long terminal repeats (LTR) activation. However, the role of LSD1 in remodeling chromatin and the role of its component BHC80 in activation of latent HIV-1 in T cells are unknown. Our findings indicate that LSD1 could decrease the level of histone H3 lysine 4 trimethylation (H3K4me3) at the HIV-1 promoter by recruiting histone lysine demethylase 5A (KDM5A) and preventing histone methyltransferase Set1A and WD-40 repeat protein 5 (WDR5) from binding to LTR. Moreover, BHC80 is necessary for LSD1-triggered LTR activation and assists LSD1 in activating LTR by binding to nucleotides 305–631 of LTR. In activated J-Lat-A2 cells, BHC80 expression was elevated and its isoform BHC80-6 promoted the association of BHC80 with LSD1. These results suggest that the LSD1–BHC80 complex enhances HIV-1 transcription by a decrease of H3K4me3 level at the viral promoter. Therefore, it might be used as a new drug target to reactivate latent HIV-1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Combination antiretroviral therapy (cART) and regimens to block mother-to-child transmission of human immunodeficiency virus (HIV) have led to a steady decrease in death due to AIDS during the past decade [1]. The major obstacle results from latently infected CD4+ T cells and other viral tissue reservoirs which are not completely characterized, which are not eliminated by antiretroviral drugs and the host immune system because the viral DNA is integrated into the host cellular genome and therefore unrecognized [2]. It is particularly important to understand the precise molecular mechanisms underlying HIV-1 transcription and latency.
HIV transcription consists of transcriptional initiation and transcriptional elongation. Initiation depends on transcription factors such as Sp1 and TATA–box binding protein (TBP) that bind to the core promoter region of long terminal repeats (LTR) [3]. However, efficient initiation is strongly induced by binding of NF-κB to the enhancer region. The transcriptional coactivator p300, which interacts with NF-κB, can also be recruited to the LTR promoter and acetylate histones H3 and H4 to activate LTR transcription [4,5,6,7]. In the absence of the transactivator Tat, HIV transcription pauses, because DRB sensitivity-inducing factor (DSIF), and negative elongation factor (NELF) interact with RNA polymerase II (RNAP II) to repress elongation [8]. When presenting, Tat binds to the bulge region of the transactivation-responsive region (TAR) RNA element and recruits the positive transcriptional elongation factor (P-TEFb) and other elongation factors to the elongation complex [9, 10]. The subunit CDK9 phosphorylates NELF, DSIF, and the C-terminal repeat domain (CTD) of RNAP II, and the phosphorylated NELF and DISF are released from RNAP II. Then, the hyperphosphorylated RNAP II continues HIV transcription [11, 12].
Besides the Tat–TAR mechanism, epigenetic regulation, which includes diverse modifications of histones, plays a key role in HIV transcriptional control. The histone methyltransferases SUV39H1 and G9a are recruited to the viral promoter and are responsible for synthesizing methylated H3K9 to maintain HIV-1 latency [13, 14]. The HKMT enhancer of Zeste 2 (EZH2), a key component of the polycomb repressive complex 2 (PRC2), accelerates HIV-1 silencing by increasing the amount of H3K27me3 at the LTR [15]. The co-repressor COUP-TF-interacting protein 2 (CTIP2) recruits the histone deacetylases HDAC1 and HDAC2 to promote histone H3 deacetylation at the HIV-1 promoter region, which maintains silencing of transcription [16]. The trimethylation status of H3K4 (H3K4me3) plays an essential role in the reactivation of latent HIV proviruses. Phorbol-12-myristate 13-acetate (PMA) has been reported to activate HIV-1 in latently infected U1 cells, with a dramatic decrease in H3K4me3 [17]. However, another recent study showed that Tat can increase the amount of H3K4me3 at the LTR in TZM-bl cells [18]. Therefore, the role of the H3K4 trimethylation state in activating HIV-1 transcription needs to be more elucidated.
Lysine-specific demethylase 1 (LSD1) is the first discovered FAD-dependent demethylase, and its primary function is to remove mono- or di-methylation modifications by Lys4 or Lys9 of histone H3, as well as lysine residues of some non-histone proteins [19,20,21]. LSD1-mediated demethylation depends on components in this complex, including co-repressor of RE1 silencing transcription factor (CoREST), histone deacetylases HDAC1/2, and BRAF–histone deacetylase complex 80 (BHC80) [22, 23]. Among these regulatory factors, CoREST stimulates LSD1 demethylase activity and BHC80 negatively regulates LSD1 function in vitro [22]. LSD1’s role in modulating gene transcription is related to cofactors bound to it. Several studies have shown that when associated with coactivators such as Pit1 and FOXA1, LSD1 can promote transcriptional activation, but when linked to corepressors such as deacetylase SIRT1, it tends to silence Notch target gene expression [24,25,26]. For instance, in an α-herpesviruses infection, LSD1 couples with the Set1/MLL1 complex and thus promotes the expression of viral immediate early (IE) genes by reducing H3K9 methylation levels and increasing H3K4me3 [27]. There are currently two different perspectives on the impact of LSD1 on HIV-1 replication. On one hand, LSD1 can activate HIV-1 gene transcription in latently infected T cells by removing mono-methylation modifications at Tat K51 [28]. On the other hand, in microglial cells with integrated proviruses, LSD1 can inhibit HIV-1 gene transcription by reducing the acetylation of H3 and increasing the methylation at both H3K4 and H3K9 [17].
To elucidate the role of the LSD1 complex in the reactivation of HIV-1 transcription from latency, we first measured the luciferase activity of HIV LTR in NH1 cells with integrated LTR when LSD1 was either overexpressed or knocked down. Then, we investigated whether LSD1 binds to the HIV-1 LTR and affects the amount of H3K4me3 by ChIP assay. In addition, we examined the role of BHC80 in LSD1-modulated HIV-1 transcription via Co-IP and ChIP assays.
Materials and methods
Plasmid constructs
The mammalian expression plasmids were prepared as follows: pCMV-myc-LSD1 was generated by PCR; pcDNA3.1-Tat K50A-flag, pcDNA3.1-Tat K51A-flag, and pcDNA3.1-Tat K50/51A-flag were generated by site-directed mutagenesis; pGL3-NL-4.3-LTR ΔSp1 mutation was generated by deletion of Sp1-binding site nucleotides via site-directed mutagenesis (Thermo Fisher); pLKO.1-shLSD1 targeting LSD1 (CCACGAGTCAAACCTTTATTT) was cloned into the pLKO.1 vector; and pcDNA3.1-Tat-flag, pRL-TK, and pGL3-NL-4.3-LTR were prepared as previously described [29].
Cell culture and transfection
HEK 293T (ATCC) and HeLa-derived NH1 cells (kindly provided by the Qiang Zhou Lab [30]) were maintained in Dulbecco’s modified Eagle medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS, BI), 50 U/ml penicillin, and 50 µg/ml streptomycin (Gibco). Jurkat-derived J-Lat-A2 cells (kindly provided by the Qiang Zhou Lab [30]) which express Tat-flag and GFP from the integrated HIV LTR upon exposure to the protein kinase C agonist prostratin were maintained in RPMI 1640 medium (Corning) supplemented with 10% FBS (Biological Industries), 50 U/ml penicillin, and 50 µg/ml streptomycin (Gibco).
Co-immunoprecipitation and western blot analysis
HEK 293T cells transfected with Tat-expressing vectors were cultured for 48 h, lysed with lysis buffer (50 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail), and then the lysates were incubated with Anti-Flag M2 Affinity Gel (Sigma) for 2 h at 4 °C. The resin was washed and the samples were analyzed by immunoblotting.
J-Lat-A2 cells were treated with 5 µM prostratin (Sigma) overnight. Then, the cells were collected, lysis buffer was added as described above, and the protein extracts were gently shaken with anti-LSD1 antibody for 2 h at 4 °C. Protein A/G Plus-Agarose (Santa Cruz Biotechnology) was incubated with the mixture for 1 h. After a wash, the pellets were assessed by western blot analysis. The primary antibodies used were anti-LSD1 (ab17721, Abcam), anti-BHC80 (A303-603A, Bethyl), anti-CoREST (ab32631, Abcam), anti-Flag (ab1162, Abcam), and anti-Actin (sc-58673, Santa Cruz Biotechnology). Secondary antibodies (anti-rabbit HRP, anti-mouse HRP, and anti-donkey HRP) were purchased from Promega.
Chromatin immunoprecipitation and quantitative real-time PCR (qRT-PCR)
Following the instructions of the Magna ChIP A/G kit (Millipore), cells were fixed in 1% formaldehyde for 10 min at room temperature. Then, the cells were lysed and the nuclear extracts sonicated for 20 min. Sheared cross-linked chromatin was incubated with 1–4 µg of the indicated antibody and 20 µl of fully resuspended protein A/G beads for 3 h at 4 °C with rotation. The beads were washed and incubated with elution buffer at 62 °C overnight. Free DNA was obtained and analyzed using qRT-PCR equipment. Antibodies used in the ChIP analysis were as follows: anti-Sp1 antibody (ab13370, Abcam), anti-H3K4me3 antibody (ab8580, Abcam), anti-Pol II antibody (Millipore), anti-H3 antibody (Millipore), anti-LSD1 antibody (ab17721, Abcam), anti-Set1A antibody (Millipore), anti-WDR5 antibody (Millipore), anti-KDM5A antibody (Millipore), and anti-BHC80 antibody (A303-603A, Bethyl). Primer sequences are listed in Table S1.
Total RNA was extracted from J-Lat-A2 cells using Trizol reagent (Invitrogen) and cDNA was synthesized using a PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). Quantitative PCR was performed using SYBR Green reagent (Roche). Primer sequences are listed in Table S2.
Luciferase assays
To detect luciferase activity, NH1 or HEK 293T cells were transfected with the specified vectors using PEI (Polyscience) and incubated for 48 h. Then, cells were collected and luciferase activity was measured using the Steady-Glo Luciferase Assay System or Dual-Luciferase Reporter Assay System (Promega). Briefly, NH1 cells were transfected with 100 pmol siRNA (LSD1 siRNA, sc-60970, Santa Cruz; NC siRNA, sc-37007, Santa Cruz; BHC80 siRNA, Ribobio) using Lipofectamine RNAiMAX reagent (#13778, Invitrogen). After 48 h, cells were re-transfected with the Tat-expressing vectors (50 ng) and corresponding amounts of empty vector using Lipofectamine 3000 reagent (#L3000, Invitrogen). After 24 h, cell lysates were analyzed for luciferase activity using the Steady-Glo Luciferase Assay System (Promega). HEK 293T cells were transfected with 100 pmol siRNA (LSD1 siRNA, sc-60970, Santa Cruz; NC siRNA, sc-37007, Santa Cruz; BHC80 siRNA, Ribobio) using Lipofectamine RNAiMAX reagent (#13778, Invitrogen). After 48 h, cells were re-transfected with pGL3-NL4.3LTR (100 ng), pRL-TK (5 ng) and corresponding amounts of empty vector using Lipofectamine 3000 reagent (#L3000, Invitrogen). After 24 h, cell lysates were analyzed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
The statistical significance of differences between the two groups was assessed using a Student’s t test. All data on luciferase activity and the qRT-PCR were presented as the mean ± SEM of at least three independent experiments and differences were considered statistically significant at p < 0.05.
Results
LSD1 enhances Tat-induced HIV-1 transcriptional activity
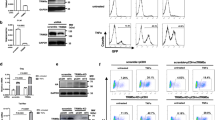
We used NH1 cells harboring a luciferase-tagged HIV-1 LTR DNA sequence as the indicator to test the effect of LSD1 on HIV-1 transcription. Through overexpression of Myc-LSD1, we found that LSD1 could enhance Tat-mediated LTR transcription (Fig. 1a). In contrast, HIV-1 LTR luciferase activity strongly decreased in NH1 cells with small interfering RNA (siRNA)-mediated depletion of LSD1 (Fig. 1b). Sakane et al. reported that LSD1 activates HIV-1 transcription through demethylation of Tat K51me [28]. To investigate whether LSD1 regulation during HIV-1 transactivation is correlated with Tat K50/51, amino acids K50/51 in the Tat protein were substituted with alanine. These mutations led to an obvious decline in HIV-1 LTR transcription when compared with that of wild-type Tat in previous reports. When LSD1 was knocked down, an individual Tat K50A or Tat K51A mutation led to a reduction in LTR luciferase activity compared to that in the corresponding negative control group, but LSD1 had no significant effect on activation by the K50/51A mutated Tat (Fig. 1c). To confirm the relationship between LSD1 and Tat K50/51, we analyzed the LSD1 content in the Flag-Tat immunoprecipitate by western blotting. LSD1/CoREST complex may be recruited to the HIV-1 promoter by Tat [28]. Consistent with this, our experiment revealed that the LSD1/CoREST complex interacted with Tat (Fig. 1d). Moreover, we found that a Tat K50A, K51A, or K50/51A mutation weakened the interaction with LSD1/CoREST (Fig. 1d). These results suggest that Tat K50 and K51 play a vital role in LSD1’s efficient stimulation of Tat-dependent HIV-1 LTR transactivation.
LSD1 enhances Tat-induced HIV-1 transcriptional activity. a LSD1 promotes Tat-mediated transactivation. Schematic representation of HIV-1 LTR and the firefly luciferase reporter gene integrated into NH1 cells (top). Co-transfection of Tat (50 ng) and increasing amounts of a myc-tagged LSD1-expressing vector (0, 0.5, and 1 µg) in NH1 cells. After 48 h, a luciferase activity assay was performed. The relative differences in luciferase activity were compared to empty vector transactivation (bottom). Expression levels of myc-LSD1 and Tat-flag in corresponding NH1 lysates were detected by immunoblot, with actin used as a control (middle). Data are shown as the mean ± SEM of three independent experiments. **p < 0.005, ***p < 0.0001. b Tat-mediated transactivation is repressed in the presence of LSD1 siRNA. LSD1 siRNA or NC siRNA was transfected into NH1 cells, after 48 h Tat (50 ng) was re-transfected, luciferase activity was detected 24 h later. The relative differences in luciferase activity compared to that in empty vector transactivation were calculated (bottom). Immunoblot analysis of endogenous LSD1 and Tat-flag in corresponding NH1 lysates, with actin used as a control (top). Data are shown as the mean ± SEM of three independent experiments. **p < 0.005. c LSD1 affects Tat-mediated transactivation by Tat K50/51. Luciferase activity of NH1 cells transfected with si-NC or si-LSD1 followed by transfection with expression vectors for wild-type or mutant Tat (50 ng) (bottom). Immunoblot analysis of endogenous LSD1 and Tat-flag in corresponding NH1 lysates, with actin used as a control (top). Data are shown as the mean ± SEM of three independent experiments. ***p < 0.0001. d Protein immunoprecipitated from HEK 293T cells expressing wild-type or mutant Tat-flag compared to that from cells expressing the empty vector control
LSD1 is associated with the Sp1-binding sites in the HIV-1 LTR
The core promoter in HIV-1 LTR consists of three Sp1-binding sites and a TATA box; Sp1 can bind to these sites. We analyzed nucleotides 380–400 in LTR of HIV-1 NL-4.3 using AliBaba2.1 online software and found that these nucleotides could bind AP-2, Egr-1, or Sp1. In addition, by blasting sequences between LSD1 and Sp1, we found the sequence in Sp1 that had a high similarity with LSD1. Thus, we hypothesized that LSD1 might bind to the Sp1-binding sites of the HIV-1 LTR to regulate HIV transcription. To test this hypothesis, we co-transfected HEK 293T cells with both Myc-LSD1 and wild-type LTR or LTRΔSp1 (Sp1-binding sites in NL-4.3 LTR were deleted). Our results showed that the overexpression of Myc-LSD1 upregulated wild-type LTR activity, but did not affect LTRΔSp1 function (Fig. 2a). However, when the LSD1 protein was decreased following shRNA transfection, LTR activity was reduced substantially. The transcriptional activity of the LTR without Sp1-binding sites remained almost unchanged, regardless of LSD1 (Fig. 2b). Next, we performed ChIP experiments using HEK 293T cells transfected with a wild-type LTR or LTRΔSp1 plasmid to determine whether LSD1 binds to Sp1-binding sites in the LTR. Accumulation of LSD1 in the NL-4.3 promoter region was abolished in the cells with Sp1-binding sites deleted, showing that LSD1 binds to the Sp1-binding sites in LTR (Fig. 2c). Taken together, our data established that LSD1 induces HIV-1 LTR activity by binding to the Sp1-binding sites.
LSD1 is associated with Sp1-binding sites in HIV-1 LTR. a LSD1 promotes HIV-1 transcription. Schematic representation of HIV-1 LTR (wild-type) and ΔSp1 LTR (deleted Sp1-binding sites) (top left). HEK 293T cells were co-transfected with the pGL3-LTR or pGL3-ΔSp1 LTR (100 ng), pRL-TK (5 ng) and pmyc-LSD1 (1 µg) vectors. Firefly luciferase activity was normalized to Renilla luciferase activity. The relative differences in luciferase activity were compared to that with pGL3-LTR vector transactivation (right). Expression levels of myc-LSD1 in corresponding HEK 293T lysates were detected by immunoblot, with actin used as a control (bottom left). Data are shown as the mean ± SEM of three independent experiments. ***p < 0.0001. b Knockdown of LSD1 reduces HIV-1 transcription. LSD1 shRNA vector (1 µg) was transfected into HEK 293T cells and, after 48 h, pGL3-LTR vector or pGL3-ΔSp1 LTR vector (100 ng) and pRL-TK (5 ng) were also transfected. Luciferase activity was detected 24 h later. Firefly luciferase activity was normalized to Renilla luciferase activity. The relative differences in luciferase activity compared to that with pGL3-ΔSp1 LTR vector transactivation were calculated (bottom). Immunoblot analysis of endogenous LSD1 in corresponding HEK 293T lysates, with actin used as a control (top). Data are shown as the mean ± SEM of three independent experiments. **p < 0.005. c ChIP analysis of LSD1, Sp1, and Pol II enrichment at HIV-1 LTR in HEK 293T cells transfected with the pGL3-LTR or pGL3-ΔSp1 LTR vector
LSD1 promotes HIV-1 transcription while reducing the level of H3K4me3
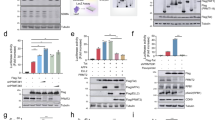
ChIP-Seq studies have shown that H3K4me3 is present at transcriptional start sites (TSS) in active genes in humans [31, 32]. Given that H3K4me3 is a general indicator of transcriptional activation, we next investigated whether LSD1 affected H3K4me3 modifications during Tat-induced transactivation. In a ChIP assay with anti-H3K4me3 antibody, we found unexpectedly that the amount of H3K4me3 in the nuc-1 region of LTR decreased with the overexpression of LSD1 (Fig. 3a), whereas the level of H3K4me3 upregulated when LSD1 was knocked down (Fig. 3b). This suggests that LSD1 promotes Tat-modulated LTR activity by reducing H3K4me3 level. In humans, H3K4 trimethylation is specifically performed by histone lysine methyltransferase complexes Set1A and Set1B, which include three conserved factors: WD-40 repeat protein 5 (WDR5), absent small homeotic-2-like (ASH2L), and retinoblastoma-binding protein 5 (RBBP5) [33,34,35]. Histone methylation is reversible and the tri-methyl group of H3K4 can be removed by histone lysine demethylase KDM5A [36, 37]. To determine which factors participate in the regulation of H3K4me3 in NH1 cells, we analyzed whether Set1A and WDR5 in histone methyltransferase complexes and demethylase KDM5A were recruited to the viral promoter using a ChIP assay. As anticipated, overexpression of LSD1 induced binding of KDM5A to the HIV-1 LTR and weakened the binding of Set1A and WDR5 to the LTR (Fig. 3c). However, when LSD1 was knocked down, Set1A and WDR5 binding to the LTR increased and KDM5A binding to the LTR decreased (Fig. 3d). Collectively, these results demonstrate that LSD1 reduced the amount of histone H3K4me3 during HIV-1 transcription by recruiting KDM5A and preventing the binding of Set1A and WDR5 to the HIV-1 LTR.
LSD1 promotes HIV-1 transcription by decreasing the amount of H3K4me3. a ChIP analysis of H3 and H3K4me3 enrichment at LTR in NH1 cells transfected with the pCMV-tag3B (empty vector) or pCMV-myc-LSD1 vector and the expression vector for Tat. b ChIP analysis of H3 and H3K4me3 enrichment at LTR in NH1 cells transfected with the negative control shRNA or LSD1 shRNA vector and the expression vector for Tat. c ChIP analysis of Set1A, WDR5, and KDM5A enrichment at LTR in NH1 cells transfected with the pCMV-tag3B or pCMV-myc-LSD1 vector and the expression vector for Tat. d ChIP analysis of Set1A, WDR5, and KDM5A enrichment at LTR in NH1 cells transfected with the negative control shRNA or LSD1 shRNA vector and the expression vector for Tat
LSD1 associated with BHC80 enhances Tat-dependent HIV-1 transcription
BHC80, a member of the LSD1 complex, can inhibit the demethylase activity of LSD1. While, the role of BHC80 in LSD1 inducing Tat-mediated transactivation remains unknown. First, as shown in Fig. 4a, the expression of BHC80 remarkably increased in Tat-transfected HEK 293T cells as compared to that in control vector-transfected cells. To reactivate J-Lat-A2 cells, we performed flow cytometry to seek the suitable treatment time and concentration of prostratin which is a protein kinase C agonist and activator of latent HIV-1 (Fig. S1). Further, we used 5 µM prostratin to activate J-Lat-A2 cells and then measured the level of BHC80 mRNA in these activated cells by qRT-PCR. The analysis showed that the level of BHC80 mRNA in prostratin-treated cells was twice that in the untreated cells (Fig. 4b). These data indicated that Tat can induce the expression of BHC80. Next, we asked whether BHC80 stimulated Tat-dependent HIV-1 transcription. To answer the question, we performed luciferase assays in NH1 cells with BHC80 siRNA. We found that after BHC80 was knocked down, LTR luciferase activity was reduced greatly, indicating that BHC80 induced LTR transcription. Importantly, with BHC80 expression knocked down, LSD1 expression did not increase the Tat-dependent luciferase activity (Fig. 4c). LSD1 lost the ability to enhance HIV-1 transcription in the absence of BHC80, indicating that BHC80 plays an essential role in LSD1 induction of Tat-dependent HIV-1 transactivation. BHC80 has two differentially spliced forms, BHC80-4 (NM_016621) and BHC80-6 (NM_001101802), in human cells [38]. We used LSD1 antibody to carry out co-immunoprecipitation in J-Lat-A2 cells during latent and reactivated periods to evaluate the binding of BHC80 isoforms with LSD1. Unexpectedly, in the J-Lat-A2 cells activated by prostratin, LSD1 tended to bind with BHC80-6 instead of BHC80-4 (Fig. 4d). We did obviously not observe two bands in the input samples. This may be due to BHC80 expression in J-Lat-A2 cells being lower, but being enriched in the IP samples, suggesting that BHC80 might mainly bind to LSD1 in J-Lat-A2 cells. By analyzing the differences in the amino acids sequence between in BHC80-6 and BHC80-4, we found that BHC80-6 had an AT-hook DNA binding motif other than BHC80-4 (Fig. 4d). Because this motif is capable of binding AT-rich DNA, we designed five primer pairs based on the HIV-1 LTR sequence. By ChIP analysis with anti-BHC80 antibody, we found that BHC80 selectively bound to nucleotides 305–631 of the viral promoter in activated J-Lat-A2 cells (Fig. 4e). Together, these data indicated that BHC80 is required for LSD1-mediated Tat transactivation. HIV-1 Tat can prompt the expression of BHC80 and LSD1 might enhance the interaction of Tat with BHC80-6 in activated J-Lat-A2 cells; in addition, BHC80 can bind the LTR to modulate HIV-1 transcription.
LSD1 associated with BHC80 enhances Tat-dependent HIV-1 transcription. a Immunoblot analysis of BHC80 in HEK 293T cells transfected with Tat-expressing vector for 48 h. b qRT-PCR analysis of BHC80 expression in J-Lat-A2 cells treated with DMSO (D) or 5 µM prostratin (P) overnight. GAPDH was used for normalization. Data are shown as the mean ± SEM of three independent experiments. ***p < 0.0001. c Luciferase activity analysis of Tat-mediated transactivation in the presence of BHC80 siRNA and the pCMV-myc-LSD1 vector. BHC80 siRNA was transfected into NH1 cells and, after 48 h, pTat-flag (20 ng) and the empty or pLSD1-myc vector (1 µg) were transfected also. Luciferase activity was detected 24 h later. The relative differences in luciferase activity compared to that with empty vector transactivation were calculated (bottom). Immunoblot analysis of endogenous BHC80 and myc-LSD1 in corresponding NH1 lysates, with actin used as a control (top). Data are shown as the mean ± SEM of three independent experiments. ***p < 0.0001. d Co-immunoprecipitation of BHC80, CoREST, or Tat-flag with anti-LSD1 in J-Lat-A2 cells treated with either DMSO (D) or 5 µM prostratin (P) overnight (top). Differences in the amino acid sequences are shown as follows: the BHC80-6 sequence (NM_001101802) is depicted in red, the BHC80-4 sequence (NM_016621) is depicted in blue, and the AT-hook is depicted in a purple rectangular box (bottom). The percentages of BHC80 isoforms derived from immunoblots of BHC80 (middle) are indicated. e Schematic representation of HIV-1 LTR divided into five regions (right). ChIP analysis of BHC80 enrichment in different (1–5) regions of LTR in J-Lat-A2 cells treated with DMSO (D) or 5 µM prostratin (P) overnight (left). (Color figure online)
Discussion
To eradicate HIV-1 proviral latency and cure HIV infection, Richman et al. proposed a “shock and kill” strategy [39]. Latent proviruses can be reactivated using multiple approaches, including the use of HDAC inhibitors, Tat, P-TEFb agonists, NF-κB inducers, and DNA methyltransferase inhibitors [40]. Among these factors, Tat plays a central role in virus reactivation because it is indispensable for efficient HIV transcription. Therefore, it is extremely important to elucidate the molecular mechanism of Tat-mediated reactivation of latent HIV. In this report, we found that, in HeLa-derived NH1 cells, LSD1 could bind to the Sp1-binding sites in the LTR and promote Tat-mediated HIV transcription through Tat K50/51. This is in concordance with prior data [28]. We also found that BHC80, which is a member of the LSD1 complex, was vital for HIV transcription. The association between the LSD1 complex and the LTR was stabilized when BHC80 bound to the R and U5 regions of the LTR. Moreover, our results revealed that LSD1 could reduce the amount of H3K4me3 by recruiting KDM5A and preventing the Set1/MLL complex from binding to the LTR and that BHC80 was crucial for LSD1-induced activation of the LTR by binding to demethylated H3K4.
The luciferase assay revealed that LSD1 promoted HIV-1 Tat transactivation in NH1 cells with integrated HIV, which agrees with the results of Sakane et al. who demonstrated that LSD1 activates HIV transcription [28]. LSD1 specifically removes mono- or di-methyl groups from H3K4 and represses transcription [19]. On the other hand, in latently infected microglial cells, LSD1 can be recruited to the HIV proximal promoter to repress viral transcription while increasing the amount of H3K4me3 [17]. Because different cell types may be associated with different mechanisms underlying HIV latency, we focused our research on whether LSD1 can induce H3K4me3 in latent T cells. ChIP assays revealed that LSD1 decreased the level of H3K4me3 binding to the viral promotor in NH1 cells. Although our results show a role for LSD1 that is different from that in microglial cells with latent HIV, we speculate that this difference is associated with the different host cell types. In addition, our ChIP experiment showed that LSD1 could bind to Sp1-binding sites in HIV-1 LTR, consistent with a report from Douce et al. [17]. Meanwhile, Tat interacted physically with LSD1 in J-Lat-A2 cells during exposure to prostratin, suggesting that LSD1 plays an essential role in HIV-1 transcription.
We next evaluated which components of the LSD1 complex participated in Tat-mediated transactivation. We found that LSD1 interacted with CoREST and BHC80 in J-Lat-A2 cells. BHC80 can associate with unmethylated H3K4 and prevent re-methylation of unmodified H3K4 [41]. Our data revealed that Tat could associate with the LSD1–BHC80 complex and promote the expression of BHC80 in J-Lat-A2 cells during exposure to the protein kinase C agonist prostratin. Tat was necessary for binding of BHC80 to H3K4 at the proximal viral promoter. In addition, the luciferase activity assay confirmed that BHC80 could enhance Tat transactivation and was required for LSD1-mediated HIV transcription. BHC80 seems to play a more critical role in promoting HIV-1 transcription than LSD1, but the precise mechanism underlying this function remains unclear. Moreover, LSD1 tends to associate with BHC80-6 rather than BHC80-4 because BHC80-6 can bind to the HIV promoter.
Moreover, LSD1 interactions with CTIP2 promote recruitment of Set1A and WDR5 to the viral promoter in microglial cells with latent HIV-1 [17]. However, we revealed that LSD1 reduced the binding between Set1A/WDR5 and the viral promoter, as well as decreased H3K4me3 levels, suggesting that whether LSD1 recruits the Set1A complex is determined by its cofactors. In our study, LSD1 associated with BHC80 that binds to unmodified H3K4, which is why LSD1 leads to a reduction in binding of the Set1A complex with the viral promoter. We next investigated which factor was anchored to the HIV-1 promoter. The ARID domain of KDM5A can bind to promoters via the CCGCCC motif or close variants with a single-base deviation and this domain is necessary for KDM5A’s demethylase activity [42]. We also found a CCGCCT sequence similar to the CCGCCC viral promoter motif in J-Lat-A2 and NH1 cells by sequence blasting. We speculate that KDM5A might be recruited to the HIV-1 promoter region and reduce the level of trimethylated H3K4. Undoubtedly, we confirmed via ChIP assay that LSD1 favored the recruitment of KDM5A to the viral promoter. Nevertheless, we did not detect LSD1 association with KDM5A in an immunoprecipitation experiment (Fig. S2), suggesting that the association of these two enzymes may depend on one or more bridging factors.
H3K4me3 is generally considered a mark of transcriptional activation. However, several studies have shown that reduced H3K4me3 is accompanied by active transcription. For example, when genes associated with estrogen-modulated cellular proliferation were activated, the PHD domain of TRIM24 recognized and interacted with unmodified H3K4, preventing methylation of H3K4 and ensuring the transcription of related genes [43]. Moreover, the autoimmune regulator AIRE promoted the gene expression of thymic medullary epithelial cells when its PHD finger bound to hypomethylated H3K4 [44]. These findings indicate that the transcription of some genes is indeed accompanied by H3K4 hypomethylation. Our experiments also showed that the amount of H3K4me3 decreased as LSD1 stimulated the activation of HIV-1 LTR. Consequently, a lower level of H3K4me3 is compatible with the transcription of active genes. However, it remains unclear whether a decrease in H3K4me3 is accompanied by the change of other lysine modifications in histone H3 during HIV-1 transcription in infected T cells.
New anti-latency drugs such as HDAC inhibitors (SAHA, vorinostat, and panobinostat) are currently in clinical trials [45,46,47]. Unfortunately, these drugs can rapidly increase HIV-1 RNA expression in resting T cells, but fail to decrease plasma HIV RNA and the number of latent T cells [48, 49]. Hence, new drug targets to reactivate latent HIV will be needed and combined anti-latency therapies may be essential for eliminating latently infected cells. Here, we revealed that the LSD1–BHC80 complex promoted Tat-mediated HIV reactivation from latency. We anticipate that a better understanding of how the LSD1/BHC80 complex modulates HIV-1 transcription will shed new light on the eradication of latent virus reservoirs.
References
Wang H, Wolock TM, Carter A et al (2016) Estimates of global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2015: the Global Burden of Disease Study 2015. Lancet HIV 3(8):e361–e387
Maartens G, Celum C, Lewin SR (2014) HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet 384(9939):258–271
Rittner K, Churcher MJ, Gait MJ et al (1995) The human immunodeficiency virus long terminal repeat includes a specialised initiator element which is required for Tat-responsive transcription. J Mol Biol 248(3):562–580
Nabel G, Baltimore D (1987) An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326(6114):711–713
Perkins ND, Felzien LK, Betts JC et al (1997) Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275(5299):523–527
Ogryzko VV, Schiltz RL, Russanova V et al (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87(5):953–959
Lusic M, Marcello A, Cereseto A et al (2003) Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22(24):6550–6561
Yamaguchi Y, Takagi T, Wada T et al (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97(1):41–51
Lu H, Li Z, Xue Y et al (2013) Viral-host interactions that control HIV-1 transcriptional elongation. Chem Rev 113(11):8567–8582
Karn J, Stoltzfus CM (2012) Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2(2):a006916
Fujinaga K, Irwin D, Huang Y et al (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24(2):787–795
Phatnani HP, Greenleaf AL (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20(21):2922–2936
du Chene I, Basyuk E, Lin YL et al (2007) Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 26(2):424–435
Imai K, Togami H, Okamoto T (2010) Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem 285(22):16538–16545
Friedman J, Cho WK, Chu CK et al (2011) Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol 85(17):9078–9089
Marban C, Suzanne S, Dequiedt F et al (2007) Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 26(2):412–423
Le Douce V, Colin L, Redel L et al (2012) LSD1 cooperates with CTIP2 to promote HIV-1 transcriptional silencing. Nucleic Acids Res 40(5):1904–1915
Zhang HS, Du GY, Liu Y et al (2016) UTX-1 regulates Tat-induced HIV-1 transactivation via changing the methylated status of histone H3. Int J Biochem Cell Biol 80:51–56
Shi Y, Lan F, Matson C et al (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119(7):941–953
Metzger E, Wissmann M, Yin N et al (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437(7057):436–439
Boehm D, Ott M (2017) Host methyltransferases and demethylases: potential new epigenetic targets for HIV cure strategies and beyond. AIDS Res Hum Retroviruses 33(S1):S8–S22
Shi YJ, Matson C, Lan F et al (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19(6):857–864
Hakimi MA, Bochar DA, Chenoweth J et al (2002) A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA 99(11):7420–7425
Wang J, Scully K, Zhu X et al (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446(7138):882–887
Mulligan P, Yang F, Di Stefano L et al (2011) A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 42(5):689–699
Cai C, He HH, Gao S et al (2014) Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep 9(5):1618–1627
Liang Y, Vogel JL, Narayanan A et al (2009) Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15(11):1312–1317
Sakane N, Kwon HS, Pagans S et al (2011) Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog 7(8):e1002184
Zhao X, Qian L, Qi D et al (2016) The 57th amino acid conveys the differential subcellular localization of human immunodeficiency virus-1 Tat derived from subtype B and C. Virus Genes 52(2):179–188
Wu J, Ao MT, Shao R et al (2017) A chalcone derivative reactivates latent HIV-1 transcription through activating P-TEFb and promoting Tat-SEC interaction on viral promoter. Sci Rep 7(1):10657
Bernstein BE, Kamal M, Lindblad-Toh K et al (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120(2):169–181
Barski A, Cuddapah S, Cui K et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129(4):823–837
Wu M, Wang PF, Lee JS et al (2008) Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28(24):7337–7344
Li Y, Han J, Zhang Y et al (2016) Structural basis for activity regulation of MLL family methyltransferases. Nature 530(7591):447–452
Shinsky SA, Monteith KE, Viggiano S et al (2015) Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem 290(10):6361–6375
Tumber A, Nuzzi A, Hookway ES et al (2017) Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem Biol 24(3):371–380
Horton JR, Engstrom A, Zoeller EL et al (2016) Characterization of a linked Jumonji domain of the KDM5/JARID1 family of histone H3 lysine 4 demethylases. J Biol Chem 291(6):2631–2646
Iwase S, Januma A, Miyamoto K et al (2004) Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem Biophys Res Commun 322(2):601–608
Richman DD, Margolis DM, Delaney M et al (2009) The challenge of finding a cure for HIV infection. Science 323(5919):1304–1307
Ay E, Banati F, Mezei M et al (2013) Epigenetics of HIV infection: promising research areas and implications for therapy. AIDS Rev 15(3):181–188
Lan F, Collins RE, De Cegli R et al (2007) Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448(7154):718–722
Tu S, Teng YC, Yuan C et al (2008) The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Biol 15(4):419–421
Tsai WW, Wang Z, Yiu TT et al (2010) TRIM24 links a non-canonical histone signature to breast cancer. Nature 468(7326):927–932
Org T, Chignola F, Hetenyi C et al (2008) The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep 9(4):370–376
Rasmussen TA, Tolstrup M, Brinkmann CR et al (2014) Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1(1):e13–e21
Desimio MG, Giuliani E, Doria M (2017) The histone deacetylase inhibitor SAHA simultaneously reactivates HIV-1 from latency and up-regulates NKG2D ligands sensitizing for natural killer cell cytotoxicity. Virology 510:9–21
Archin NM, Liberty AL, Kashuba AD et al (2012) Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487(7408):482–485
Elliott JH, Wightman F, Solomon A et al (2014) Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10(10):e1004473
Archin NM, Bateson R, Tripathy MK et al (2014) HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 210(5):728–735
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81571987, 81371820). We thank Dr. Qiang Zhou and Dr. Yuhua Xue for providing NH1 and J-Lat-A2 cells.
Author information
Authors and Affiliations
Contributions
CL and XK designed the experiments. YL, DZ, DQ, JF, ZL, YH, and WS performed the experiments. YL and CL analyzed the data and designed the figures. YL and XK wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
All authors read and approved the manuscript.
Additional information
Edited by Wolfram Gerlich.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Y., Zhou, D., Qi, D. et al. Lysine-specific demethylase 1 cooperates with BRAF–histone deacetylase complex 80 to enhance HIV-1 Tat-mediated transactivation. Virus Genes 54, 662–671 (2018). https://doi.org/10.1007/s11262-018-1589-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-018-1589-5