Abstract
Yellow vein mosaic disease of okra is a whitefly transmitted begomovirus causing heavy economic loss in different parts of India. The okra isolate (OY131) of this virus from a bhendi plant [(Abelmoschus esculentus L.) Moench] showing yellow vein mosaic, vein twisting, reduced leaves, and a bushy appearance in the Palem region, New Delhi, India, was characterized in the present study. The complete DNA-A and DNA-B sequences have been determined and are comprised of 2,746 and 2,703 nucleotides, respectively. The betasatellite (DNA-β) component was absent in the sample. The genome organization was typically of biparite begomoviruses, which were characterized earlier. Comparison of DNA-A component with other known begomoviruses suggest that this virus, being only distantly related (<85.9% similarity with its nearest relative, BYVMV) to other known begomoviruses, is a new species. We have tentatively assigned the genome to a novel geminivirus species Bhendi yellow vein mosaic Delhi virus [BYVDV-IN (India: Delhi: okra)]. DNA-B showed highest sequence identity (87.8% identical) to that of a ToLCNDV (AY158080). The phylogenetic analysis of the present isolate is distinct from all other viruses; however clusters with ToLCNDV group infect different crops. The recombination analysis revealed that this isolate has sequences originated from ToLCNDV. This is the first known bhendi yellow vein mosaic disease associated bipartite begomovirus from India.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Geminiviruses are small single-stranded circular DNA plant viruses with distinctive twinned isometric particle morphology of approximately 20 × 30 nm in size. Viruses in the Begomovirus genus of the family Geminiviridae are transmitted by the whitefly Bemisia tabaci (Gennadius), and represent a major threat to many cultivated dicotyledonous plants. Begomoviruses can either have monopartite or bipartite genomes. Bipartite begomovirus genomes consist of two ssDNA molecules of approximately 2.6 kb, referred to as DNA-A and DNA-B, with each component being responsible for different functions in the infection process. The genes on the A component are involved in encapsidation and replication, whereas the genes on the B component are involved in viral systemic movement, host range determination, and symptom expression [1]. Recently, some satellite DNA molecules, known as DNA-β or betasatellites, have been found associated with monopartite begomoviruses. Like DNA-B, these satellites depend upon the DNA-A component for encapsidation, replication, and insect-transmission [2–5].
Bhendi [Abelmoschus esculentus (L.) Moench], also called as okra or lady’s finger, belongs to the family Malvaceae. Despite being an important Indian vegetable crop that is grown extensively throughout the year in all parts of the country, bhendi yields are quite low due to infection by a number of diseases, of which viral diseases are particularly important [6]. Among these viral diseases, Yellow vein mosaic disease of bhendi (YVMD) (Fig. 1), characterized by veinal clearing, chlorosis and swelling coupled with slight downward curling of leaf margins, twisting of petioles, and retardation of growth [7] has been consistently reported in past few years from different parts of India. The rise in importance of this disease has emphasized the need, to properly determine its etiology.
Although an earlier report has indicated that YVMD is possibly caused by betasatellit associated monopartite begomoviruses [8], the begomoviruses associated with YVMD of bhendi across large parts of northern India have never been properly characterized. Cataloging the full complement of species associated with a disease such as YVMD is important, because in the past few years it has become apparent that in many parts of the world such disease are in fact caused by a diverse complex of different begomovirus species [2, 9–17]. Such complexes and the opportunities for mixed infections that afford a crucial feature of begomovirus ecology for the inter-species recombination seems to be a major driver of begomovirus evolution. It was quite probable that, recombination during the last 50 years has directly contributed to the emergence of multiple new begomovirus diseases [18–22].
In the present study, we have characterized for the first time a new recombinant bhendi bipartite begomovirus species infecting bhendi which is different from previously characterized monopartite begomoviral pathogens of bhendi from India.
Materials and methods
Virus isolate and its maintenance
The leaf sample from bhendi plant exhibiting YVMD with yellow mosaic and vein thickening symptoms (Fig. 1) was collected from the Palem region, New Delhi, India, and designated as Bhendi yellow vein mosaic virus (BYVMV)-isolate OY131. The virus culture of this isolate was established through grafting and whitefly transmission and maintained on susceptible bhendi cultivar (cv. 1685) by periodic transfer. The B. tabaci adults were used for the transmission experiments which were initially collected from horsegram [Macrotyloma uniflorum (Lam.) Verdc.] from the Talagavara village, Kolar district, India, and maintained in the laboratory on healthy cotton plants (Gossypium hirsutum L. cv. Laxmi), for long enough to insure they carried no unknown begomoviruses, after which eggs were collected. The collected eggs were allowed to hatch and a virus free colony was obtained and maintained on healthy cotton plants in large wooden cages (45 × 45 × 30 cm), covered with insect proof, galvanized wire mesh (40-mesh size) at 30°C and 60% RH in a controlled glasshouse. Adult non-viruliferous whiteflies were given an acquisition access period (AAP) of 24 h on leaves of infected plants and were then released onto healthy test plants for an inoculation access period (IAP) of 24 h unless otherwise specified. After this, plants were sprayed with an insecticide and maintained under insect-proof glasshouse conditions until symptoms were evaluated. Samples from plants showing symptoms along with infected samples collected from the field were utilized for further analysis.
PCR and cloning of viral genome
Total nucleic acids were extracted from both healthy and infected leaf tissues from whitefly inoculated samples from the glasshouse and field collected samples by the cetyl trimethyl ammonium bromide method [23]. Full length genomes BYVMV-OY131 isolate were amplified using the three sets of overlapping primers OY2395F 5′-GCTCCCTGAATGTTCGGATGGA-3′/OY680R5′-GTTCTCRTCCATCCATATCTTAC-3′; MKBEGF4 5′-ATATCTGCAGGGNAARATHTGGATGGA-3/MKBEGR5 5′-TGGAC TGCAGACNGGNAARACNATGTGGGC-3′; and GEMA1223F:5′-GTCGGAGGSTGTAAGG TCGTCCAG-3′/GEMA2454R:5′-CTCACWTAYCHCAARTGCTCTC-3′specifically designed to DNA-A components and the primers ToLCBD2081F 5′-GCGTACTCWACGC GCTCAGATTG-3′/ToLCBD656R 5′-GTGTTTCACAGATTTCCTTACGCG-3′, ToLCBD971F 5′-GTGGCAGAACGCCACCATGAACG-3′,ToLCBD2142R 5′-GCTGCGCG GCCAATATGTCAATAG-3′/ToLCBD971R 5′-GTTCATGGTGGCGTTCTGCAAC-3′, ToLCBD656F 5′-CGCGTAAGGAA ATCTGTGAAACAC-3′ designed to DNA-B by utilizing the sequences of BYVMV, ToLCNDV, ACMV, and other begomoviruses available in NCBI data bank. The PCR reactions were carried out in a Gene Amp PCR system 9700 (PE Applied Biosystems, Foster City, CA, USA) thermocycler. The total volume of the PCR reaction in each case was 25 μl (100 pmol DNA template, 1.5 U Taq DNA polymerase, 25 mM Mgcl2, 2 mM dNTPs, and 25 pmol of each primer).
The total number of amplification cycles given was 35 with initial denaturation at 94°C for 3 min and final extension at 72°C for 15 min. The cycling conditions were denaturation at 94°C for 1 min, annealing at 55–58°C for 45 s, and extension at 72°C for 90 s. Similarly, attempts were made to amplify betasatellite by PCR using universal primers (β01/β02) [24]. PCR products were electrophoresed (1 h at 80 volts) on 0.8% agarose gel and viewed on a Gel documentation system (Alpha Innotech, USA). Potentially full-length amplification products of DNA-A and DNA-B were cloned into the pTZ57R/T vector (Fermentas, Germany) according to the manufacturer's instructions. The complete nucleotide sequence of DNA clones were determined using automated ABI PRISM 3730 DNA sequencer, (Applied Biosystems) at Anshul Biotechnologies DNA Sequencing facility, Hyderabad, Andhra Pradesh, India.
Detection for the presence of betasatellite DNA by dot blot analysis
Betasatellite DNA of YVMD (Isolate OYnun acc.no GU111991) was oligonucleotide primed and labeled with DIG dUTP. The labeling was done in a total volume of 20 μl comprising of 12 μl of betasatellite DNA clone, 2 μl of exo-resistant random hexanucleotide (Fermentas), 2 μl of 10× Klenow buffer (Fermentas), 2 μl of dNTP (Fermentas), 1 μl of digoxinin dUTP alkali stable (Roche diagnostics), and 2 μl Klenow enzyme, E. coli (Fermentas).
Genomic DNA was spotted onto nylon membranes as described by Boulton et al. [25]. Full length DNA-β probes were used to detect betasatellite in DNA samples isolated from filed samples and whitefly inoculated plants displaying typical YVMD symptoms. 5 μl of the DNA solutions were spotted directly onto nitrocellulose membranes and the DNA was fixed on to the membranes by ultraviolet cross linking for 3 min. The pre-hybridization, hybridization, and detection procedures were carried out according to the protocol given in DIG High Prime DNA labeling and detection starter kit II (Roche diagnostics). Nitroblue tetrazolium (NBT) and X-phosphate based colorimetric detection were used to visualize labeled spots.
Sequence analysis
The DNA-A and DNA-B component of nucleotide sequences from bhendi were compared with selected begomovirus sequences retrieved from GenBank (Table 1). Pair wise sequence similarity matrixes were generated using Bioedit Sequence Alignment Editor (version 5.0.9) [26] and neighbor joining phylogenetic trees (with 1,000 bootstrap replicates and the P-distance nucleotide substitution model) were generated using MEGA 5.0 [27].
Detection of recombination events
This was carried out using selected begomovirus sequences previously reported from the Indian subcontinent. The conflicting phylogenetics with signals' potential indication of recombination were detected using the program split tree version 4.11.3 with default settings [28]. Further recombination analysis to identify recombinants and their recombination breakpoints was carried out using the RDP, GENECOV, Bootscan, Max Chi, Chimara, SiScan, and 3Seq methods implemented in the program RDP 3 [29]. Default RDP3 settings with a 0.05 P value cutoff with standard Bonferroni correction for multiple testing were used throughout.
Results
Transmission and maintenance
YVMD was successfully transmitted by whitefly B. tabaci (Fig 2a) and grafting (Fig. 2b) from the field-collected bhendi samples to bhendi cultivar (cv. 1685) rised in glasshouse and achieved 100 transmission. The infected plants start producing symptoms within 10 days after inoculation, which were similar to those observed in the field infected plants (i.e., yellow vein symptoms and failed to produce fruits).
Genome amplification, detection cloning, and genomic organization of BYVDV
The complete sequence of begomovirus DNA-A and DNA-B components from field samples as well whitefly inoculated samples were amplified using six primer sets as described above. This confirmed that BYVDV is a bipartite begomovirus. However, attempts to amplify betasatellite in both samples failed confirming its non-association with the present virus isolate. In order to provide further evidence for this, DNA blotting with DIG dUTP-labeled probes specific to betasatellite was carried out. The results were negative for the association of betasatellite in the infected sample, and a strong positive signal was observed in the +ve betasatellite clone (S1). Non-symptomatic samples were negative for amplification of DNA-A, DNA-B, and betasatellite components by PCR, and betasatellite in dot blot hybridization.
Genome organization and sequence and phylogenetic analysis of BYVDV DNA-A
The PCR amplified products of DNA-A and DNA-B were cloned into pTZ57R/T vector and sequenced. At first, we compared the sequences of field infected sample and whitefly inoculated sample. The sequences obtained in both the cases were identical with respect to both DNA-A and DNA-B components of the virus, and therefore we continued with the representative sequences from these samples for further analysis. The sequence of the 2,746 nt long DNA-A component was determined in both orientations and deposited in GenBank under the accession number FJ515747. Analysis of the sequence indicates that the organization of genes within the BYVDV is typically of old world bipartite begomoviruses (Table 2). When individual predicted amino acid sequences for the different inferred proteins of BYVDV were compared with other begomoviruses, ORFs AV2 and AV1 were most similar to those of ToLCNDV-Luffa (AM292302) and ToLCNDV-AVT1 (AY428769), respectively, ORF AC1was most similar to that of CLCuBV, and ORFs AC2 and AC3 were most similar to those of Okra leaf curl virus (GQ245760) and BYVMV-PK (AJ002453) (Table 3), respectively. Overall the 227 nucleotide long IR of the BYVDV shared <63.7% with its nearest relatives BYVMV and Bhendi yellow vein India virus, 75.75 to 85.7% with Tomato leaf curl new Delhi virus (ToLCNDV), which are infecting okra in India (the sequences are available in the data base). Within the intergenic region, incomplete direct repeats of an iteron sequence, GGTGT, was detected adjacent to the TATA box, which is presumably the AC1 (or replication associated gene) promoter.
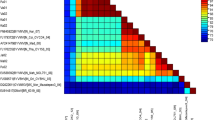
When compared with homologous sequences from selected Indian begomoviruses it was found to be <85.9% identical with any other previously characterized DNA-A component begomoviruses so for (Table 3). Phylogenetically, the DNA-A component is on a well-supported isolated branch distinct from other known Indian begomoviruses (Fig. 3a). Based on the present begomovirus species demarcation criterion of <89% nucleotide sequence identity [30], the BYVDV from bhendi is a distinct species, for which we propose the name Bhendi yellow vein Delhi virus (BYVDV). Based on its sampling location and host the isolate described here has been given the additional descriptor [India: Delhi: Okra].
Phylogeny of complete nucleotide sequences of DNA-A (a) and DNA-B (b) of Bhendi yellow vein delhi virus (OY131 isolate) with other related begomoviruses. The tree was generated using the Neighbor-joining method in MEGA5. Horizontal distances are proportional to sequence distances, vertical distances are arbitrary. The trees are unrooted. A bootstrap analysis with 1,000 replicates was performed and the bootstrap percent values more than 50 are numbered along branches
Genome organization and sequence and phylogenetic analysis of BYVDV DNA-B
A compete nucleotide sequence of DNA-B was determined to be 2,703, and submitted to GenBank under the accession number HQ542082. The genome organization was similar to that of previously described bipartite begomoviruses predicted to encode two ORFs (Table 2). Despite the fact that the BYVDV DNA-A component sequence is quite divergent relative to those of other known begomoviruses, the movement protein gene of the virus was very similar (94% identical) to that of ToLCNDV [IN:Karnal:OY81A:04] (GU112083) and its nuclear shuttle protein gene was similar (97.5% identical) to that of ToLCNDV [AM286435, (Table 4)]. These high degrees of similarity were also observed for the full DNA-B sequence compared with other bipartite begomoviruses. Curiously, the BYVDV DNA-B was most similar (87.8%) to that of a ToLCNDV (AY158080) isolated from Potato in New Delhi (Fig. 3b). Since the sample from which BYVDV was obtained had no associated betasatellite and its genomic arrangement was identical to that of bipartite begomoviruses with iteron sequence (GGTGT) identical to its DNA-A component, it is very likely that BYVDV is a bipartite begomovirus. BYVDV therefore represents the first known YVMD associated bipartite begomovirus.
Detection of recombination
The varying patterns of pairwise sequence similarity seen for different genomic regions of BYVDV suggest that this virus is a recombinant. To test for the evidence of recombination within the BYVDV genome, the sequences of DNA-A and DNA-B components from different begomoviruses infecting malvaceous, solanaceous, cucurbits, and pulse species were retrieved from GenBank, aligned together with BYVDV, and used to construct a neighbor-network (using the program Splits-Tree version 4.11.3). Such networks are capable of graphically displaying patterns of non-tree-like evolution such as those expected in the presence of recombination. The neighbor-network of both the DNA-A and DNA-B components clearly substantiated evidence of phylogenetic conflicts within the analyzed sequences, and indicated that nearly every sequence within the network had potentially been the recipient of horizontally acquired sequences at some time during their evolutionary past.
In order to further test for evidence of recombination and characterize potential individual recombination events within the BYVDV genome, we analyzed the sequences with the computer program, RDP. These analyses provided strong evidence for the presence of past recombination events in both the DNA-A and DNA-B components of the BYVDV sequence (Table 5). Specifically, they indicated that the BYVDV DNA-A very clearly contains sequences derived through recombination from Indian ToLCNDV-like parent (p < 5.1 × 10−22 for all six recombination detection methods applied) and an Indian BYVMV-like parent (p < 3.6 × 10−24 for all six recombination detection methods applied). Although less obviously recombinant, the analysis also revealed that the BYVDV DNA-B was very likely derived through recombination among the DNA-B components of different ToLCNDV DNA-Bs (Table 5).
Discussion
In the tropics and subtropics, begomoviruses are emerging as a major threat to the sustainable cultivation of many economically important crop species [31]. These viruses have very high mutation [32] and recombination [20] rates, and display a remarkable degree of evolutionary adaptability. Whereas new strains and variants are perpetually generated through recombination within frequently occurring mixed infections, associations with pathogenicity and host-range modulating satellite molecules, human mediated movements of viruses into new ecosystems and movements of viruses into new species by polyphagous invasive whitefly biotypes creating a complex adaptive landscape with a rich variety of niches for begomoviruses to fill [32, 33]. YVMD of bhendi is spreading rapidly throughout India, affecting plants at all growth stages, and resulting in plants yielding unmarketable fruits and clearly a most serious of the newly emerging/reemerging begomovirus-associated diseases in India [7, 34, 35].
Despite its importance, the etiology of YVMD across much of India is unknown and has been shown to be caused by betasatellite associated monopartite begomovirus species. At least part of the reason for this is, most of the begomoviruses that have been characterized so for from malvaceous crops are monopartite and are associated with a betasatellites and alphasatellites [2, 3, 5, 36, 37]. The exceptions to this have been the malvaceous species infecting begomoviruses of the New World where it appears that betastellites do not occur [38, 39]. It is demonstrated here for the first time that at least one of the virus species contributing to YVMD in bhendi is an efficiently whitefly transmitted bipartite begomovirus that appears most closely related among the currently classified Indian begomoviruses to ToLCNDV.
The amplification of identical DNA-A and DNA-B components and failure to amplify and detect betasatellite in both field samples as well as whitefly inoculated glasshouse samples suggests that the virus is bipartite. In addition to this, the dot blot detection for the presence of betasatellite also strengthens the evidence for the association of DNA-B with the virus under present study ruling out any probable chance contamination of the sample with DNA-B. Further, if it was a contamination it would not have picked up by the whitefly, which is a vector for spread of the virus in nature. The analyses of sequences showed that the iteron of both DNA-A and B are identical (GGTGT). The recent report on recognition of iteron by Rep suggests that, begomoviruses may tolerate some variation in iteron sequences without deleterious effects on Rep recognition [40]. However, in the present study, since the iteron of both DNA-A and B components are identical and DNA-B was recovered from whitefly inoculated sample without betasatellite contamination, if the above hypothesis holds good, then in all probability the Rep should bind to the iteron of B component with respect to BYVDV.
The DNA-A and DNA-B component of the virus has less than 89% identity to any currently classified begomovirus for which we have tentatively named as Bhendi yellow vein Delhi virus [BYVDV-IN (India: Delhi: okra)], and classified as a new species [30]. At least part of the reason that the BYVDV isolate characterized here does not closely resemble any other begomovirus species is that, the BYVDV DNA-A has likely arisen through at least two separate recombination events between BYVMV and ToLCNDV parental viruses. Although at least some of the detected recombination breakpoints within the BYVDV DNA-A and DNA-B are the near sites in the IR and the 3′ half of the C1 ORF which are known recombination hotspots [12, 41]. One of the sites within the V1 ORF falls in a known recombination cold-spot. Although the possibility remains that the recombination event that generated the BYVDV DNA-A essentially by introducing a ToLCNDV-like fragment of C1, IR, and V2 ORF into a BYVMV. The genomic background might have provided BYVDV with an adaptive advantage over its parents and contributed to its emergence, this will need to be assessed further in laboratory studies.
In conclusion, these results show for the first time that a previously unidentified recombinant bipartite begomovirus is part of the complex of viruses that cause YVMD in bhendi. The sequence of this virus can now be used to design diagnostic techniques to identify its presence within YVMD infected plants and should help in further epidemiological studies of this disease. A rich diversity of viruses infecting Indian bhendi is of concern, since this situation undoubtedly increases incidences of mixed infections and increases the possibility of yet more novel recombinant viruses arising within this species. Together with the already existing diversity of begomoviruses infecting bhendi, such novel recombinants will probably further undermine efforts to control the virus using transgenic or inbred resistance strategies.
References
S.G. Lazarowitz, Crit. Rev. Plant Sci. 11, 327–349 (1992)
R.W. Briddon, S. Mansoor, I.D. Bedford, M.S. Pinner, K. Saunders, J. Stanley, Y. Zafar, K. Malik, P.G. Markham, Virology 285, 234–243 (2001)
R.W. Briddon, S.E. Bull, I. Amin, A.M. Idris, S. Mansoor, I.D. Bedford, P. Dhawan, N. Rishi, S.S. Siwatch, A.M. Abdel-Salam, J.K. Brown, Y. Zafar, P.G. Markham, Virology 312, 106–121 (2003)
R.W. Briddon, C.M. Fauquet, J.K. Brown, E. Moriones, J. Stanley, M. Zerbini, X. Zhou, Arch. Virol. 153, 763–781 (2008)
K. Saunders, I.D. Bedford, R.W. Briddon, P.G. Markham, S.M. Wong, J. Stanley, Proc. Natl. Acad. Sci. U S A 97, 6890–6895 (2000)
R. Usha, in Characterization, diagnosis & management of plant viruses, ed. by G.P. Rao, P.L. Kumar, R.L. Holguín-Peña (Studium Press, Houston, 1980), pp. 387–392
S.P. Capoor, P.M. Varma, Indian J. Agric. Sci. 20, 217–230 (1950)
J. Jose, R. Usha, Virology 305, 310–317 (2003)
S.E. Bull, R.W. Briddon, W.S. Sserubombwe, K. Ngugi, P.G. Markham, J. Stanley, J. Gen. Virol. 87, 3053–3065 (2006)
R.V. Chowdareddy, J. Colvin, V. Muniyappa, S.E. Seal, Arch. Virol. 150, 845–867 (2005)
H. Delatte, D.P. Martin, F. Naze, R.W. Golbach, B. Reynaud, M. Peterschmitt, J.M. Lett, J. Gen. Virol. 86, 1533–1542 (2005)
P. Lefeuvre, D.P. Martin, M. Hoareau, F. Naze, H. Delatte, M. Thierry, A. Varsani, N. Becker, B. Reynaud, J.M. Lett, J. Gen. Virol. 88, 3458–3468 (2007)
S. Mansoor, R.W. Briddon, S.E. Bull, I.D. Bedford, A. Bashir, M. Hussain, M. Saeed, M.Y. Zafar, K.A. Malik, C. Fauquet, P.G. Markham, Arch. Virol. 148, 1969–1986 (2003)
F.J. Morales, Adv. Virus Res. 67, 127–162 (2006)
J. Ndunguru, J.P. Legg, T.A.S. Aveling, G. Thompson, C.M. Fauquet, Virol. J. 2, 21 (2005)
A.I. Sanz, A. Fraile, J.M. Gallego, J.M. Malpica, F. Garcia Arenal, J. Mol. Evol. 49, 672–681 (1999)
Y.C. Zhou, M. Nonssourou, Kon, M.R. Rajas, H. Jiang, L.F. Chen, K. Gamby, R. Foster, R.L. Gilbertson, Arch. Virol. 153, 693–706 (2008)
L.C. Berrie, E.P. Rybicki, M.E.C. Rey, J. Gen. Virol. 82, 53–58 (2001)
H. Jeske, M. Lutgemeier, W. Preiss, EMBO J. 20, 6158–6167 (2001)
M. Padidam, S.Y. Sawyer, C.M. Fauquet, Virology 265, 218–225 (1999)
X.P. Zhou, Y.L. Liu, L. Calvert, C. Munoz, G.W. Otim-Nape, D.J. Robinson, B.D. Harrison, J. Gen. Virol. 78, 2101–2111 (1997)
W.H. Schnippenkoetter, D.P. Martin, J.A. Willment, E.P. Rybicki, J. Gen. Virol. 82, 3081–3090 (2001)
J.J. Doyle, J.L. Doyle, Focus 12, 13–15 (1990)
R.W. Briddon, S.E. Bull, S. Mansoor, I. Amin, P.G. Markham, Mol. Biotechnol. 20, 315–318 (2002)
R.E. Boulton, G.J. Jellis, D.C. Balcoumbe, A.M. Squire, in proceedings of the british crop protection conference, British crop protection council, Thorton Heath, surrey, 1984, pp. 177–180
T.A. Hall, Nucl. Acids Symp. Ser. 41, 95–98 (1999)
K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei, S. Kumar, Mol. Biol. Evol. (2011). doi:10.1093/molbev/msr121
D.H. Huson, D. Bryant, Mol. Biol. Evol. 23, 254–267 (2006)
D.P. Martin, P. Lemey, M. Lott, M. Vincent, D. Posada, P. Lefeuvre, Bioinformatics 26(19), 2462–2463 (2010)
C.M. Fauquet, R.W. Briddon, J.K. Brown, E. Moriones, J. Stanley, M. Zerbini, X. Zhou, Arch. Virol. 153, 783–821 (2008)
J.E. Polston, P.L. Anderson, Plant Dis. 81, 1358–1369 (1997)
A. Varma, V.G. Malathi, Ann Appl Biol. 142, 145–164 (2003)
S. Paull, R. Ghosh, S. Chaudhuri, S.K. Ghosh, A. Roy, J. Plant Pathol. 91(3), 637–647 (2009)
V. Muniyappa, in Vectors of Plant Pathogens, ed. by K.F. Harris, K. Maramorosch (Academic Press, New York, 1980), pp. 39–85
S.J. Singh, Indian J. Mycol. Plant Pathol. 10, 35–39 (1980)
R.W. Briddon, J. Stanley, Virology 344, 198–210 (2006)
V. Venkataravanappa, C.N.L. Reddy, P. Swaranalatha, S. Jalali, R.W. Briddon, M.K. Reddy, Virol. J. 8, 855 (2011)
A.P. Graham, D.P. Martin, M.E. Roye, Virus Genes 40, 256–266 (2010)
C.H. Zepeda, M. Ali, Idris, G. Carnevali, J.K. Brown, O.A.M. Valenzuela, Virus Genes 35, 369–377 (2007)
M. Shafiq, S. Asad, Y. Zafar, R.W. Briddon, S. Mansoor, Virol. J. 7, 367 (2010)
P. Kumari, A.K. Singh, B. Chattopadhyay, S. Chakraborty, Virus Res. 152, 19–29 (2010)
Acknowledgments
The research was supported by ICAR NETWORK project on development of diagnostics to emerging plant viruses, Indian Council of Agricultural Research, Government of India, New Delhi. We thank DP Martin, Computational Biology Group, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, South Africa for going through the manuscript and suggesting the corrections.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.

11262_2012_732_MOESM1_ESM.jpg
S1 Detection of BYVDV in the total DNA isolated from naturally infected bhendi samples by dot blot hybridization using nonradioactive digoxginin labelled Probe of betasatellite of Bhendi yellow vein Dehli virus (the dot circle depicts the +ve betasatellite clone). (JPEG 4 kb)
Rights and permissions
About this article
Cite this article
Venkataravanappa, V., Lakshminarayana Reddy, C.N., Jalali, S. et al. Molecular characterization of distinct bipartite begomovirus infecting bhendi (Abelmoschus esculentus L.) in India. Virus Genes 44, 522–535 (2012). https://doi.org/10.1007/s11262-012-0732-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0732-y