Abstract
A live-attenuated dengue-2 virus strain S16803 vaccine candidate that is immunogenic and safe in humans was derived by 50 passages in primary dog kidney (PDK) cells. To identify mutations associated with attenuation of the dengue-2 PDK50 vaccine strain, we determined the nucleotide changes that arose during PDK passage of the dengue-2 virus. Thirteen mutations distinguished the PDK50 virus from low-passage parent resulting in amino acid substitutions in the premembrane (E89G), envelope (E202K, N203D), nonstructural proteins NS1 (A43T), NS2A (L181F), NS2B (I26V), and NS4B (I/T108T, L112F). In addition, the PDK50 virus contained a C to T change of nucleotide 57 in the 5′ non-coding region and four silent mutations of nucleotides 591, 987, 6471, and 8907. An infectious PDK50 cDNA clone virus was produced and characterized for growth kinetics in monkey (LLC-MK2, Vero) and mosquito (C6/36) cells. Identification of mutations in the vaccine strain and availability of an infectious clone will permit systematic analysis of the importance of individual or collective mutations on attenuation of dengue virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dengue viruses belong to the Flavivirus genus in the Flaviviridae family and are grouped as four separate serotypes, dengue 1, 2, 3, and 4. Dengue infection is endemic to tropical and subtropical regions inhabited by the Aedes mosquito vector, placing an estimated 2.5 billion people at risk [1]. In addition to subclinical infections, dengue viruses cause illnesses that range from a mild, flu-like syndrome with rash to severe dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS). Severe disease occurs most often in children and young adults and is associated with 1–5% mortality depending on the available treatment [2]. The majority of patients with DHF/DSS have experienced prior infection with a different serotype. One proposed mechanism for severe disease is enhanced infection because of formation of complexes between preexisting cross-reactive non-neutralizing antibodies, secondary infecting virus and Fc receptors on monocytes or macrophages [3]. Other factors proposed to contribute to the disease process include infecting strain, virus burden, and cross-reactive T lymphocytes [4–6].
The flavivirus genome consists of positive-sense single-stranded RNA approximately 11 kilobases long that is capped at the 5′ end and lacks a 3′ polyadenylated tail. The RNA encodes a single long open-reading frame flanked by 5′ and 3′ noncoding regions (NCRs) of variable size depending on the flavivirus. Three structural proteins, capsid (C), premembrane (prM), and envelope (E), and seven nonstructural (NS) proteins are co- and post-translationally cleaved from the polyprotein precursor by viral and cellular proteases, and occur in the polyprotein in the order C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 [7].
A licensed vaccine against dengue is not yet available. Because of the possibility of enhanced infection, simultaneous protection against all four dengue serotypes is thought to be needed. Investigators at the Walter Reed Army Institute of Research (WRAIR) developed a series of live-attenuated dengue 1–4 virus vaccine candidates by serially passaging each parental strain in primary dog kidney (PDK) cells [8]. The dengue-2 virus S16803 PDK50 vaccine candidate was passaged 50 times in PDK cells and was then amplified by 3 passages in fetal rhesus lung (FRhL) cells. Phenotypic characteristics of the PDK50 virus included temperature sensitivity, small plaque size, loss of neurovirulence for suckling mice and decreased incidence of viremia in monkeys [9].
Pilot studies of dengue-2 vaccine candidates PDK30, PDK40, and PDK50 in small groups of volunteers were conducted to determine the optimal passage level for vaccine candidate development [10]. Symptoms and immunological responses of ten volunteers for PDK30 and three volunteers for PDK40 and PDK50 were determined and showed progressive attenuation of the virus with increased passage of virus in PDK cells [10]. The PDK30 and 40 strains were considered to be under-attenuated, whereas the PDK50 strain was found to be safe and immunogenic in humans and was selected for further testing as a monovalent dengue-2 vaccine and in tetravalent combinations [11].
Here, we identify the mutations that evolved in the WRAIR dengue-2 PDK50 vaccine strain. To help determine which changes are associated with progressive attenuation, we also determined the nucleotide sequences for intermediate passage–level viruses PDK10, PDK30 and PDK40. We describe production of an infectious clone of the PDK50 strain and characterization of virus produced from the clone compared with PDK50 vaccine and low-passage parent viruses. Knowledge of mutations associated with attenuation of the dengue PDK50 strain and availability of a molecular clone to systematically analyze mutations may be important for designing improved flavivirus vaccines.
Materials and methods
Viruses
Dengue-2 virus strain S16803 was isolated in 1974 from a patient in Thailand, grown in mosquitoes and passaged four times in primary Green Monkey cells to become the low-passage parent virus [8]. Low-passage parent virus was passed 10, 30, 40, or 50 times in PDK cells and amplified three times in FRhL cells to make vaccine candidates [8]. All viruses were provided in lyophilized form by Dr. Ken Eckels, WRAIR.
Cells
African Green Monkey kidney epithelial (Vero) and Rhesus Monkey kidney epithelial (LLC-MK2) cells were grown at 37°C in a humidified incubator under 5% CO2 in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum and 50 μg of gentamicin sulfate per ml of culture medium. Aedes albopictus (C6/36) mosquito cells were grown at 28°C in the same medium supplemented with 1× nonessential amino acids, 0.1 mM sodium pyruvate, and 25 mM HEPES, pH 7.5.
Virus RNA and RT–PCR
Virus was suspended in 1.0 ml sterile water and RNA was prepared using a QIAamp viral RNA kit (Qiagen) and stored in aliquots at −80°C. For reverse transcription (RT), 10% of the RNA was combined with 50 pmol 3′-end primer 10723R (Table 1) and denatured for 3 min at 72°C, then chilled on ice and added to a 100 μl reaction mixture that contained 1× RT buffer, 10 mM dithiothreitol, 80 U RNasin RNase inhibitor (all from Invitrogen) and 0.5 mM each deoxynucleotide triphosphate (dNTP, Promega). This mixture was divided equally and 50 U superscript II reverse transcriptase (Invitrogen) was added to one half and the other half served as a negative control that lacked enzyme. Both reactions were incubated for 1 h at 40°C and terminated by incubation for 5 min at 95°C.
To amplify DNA by the polymerase chain reaction (PCR), 1 μl cDNA was added to a 49 μl reaction mixture that contained 1× PCR buffer, 0.2 mM each dNTP, 25 pmol PCR primers (Table 1), and either 2.5 U of high-fidelity (Pfu) DNA polymerase (Stratagene) to produce the four DNA fragments used for assembling the infectious clone or 2.5 U Expand DNA polymerase (Boehringer Mannheim) to produce six overlapping DNA fragments for sequence analysis. The PCR reaction mixtures were subjected to denaturation at 94°C for 5 min followed by 35 temperature cycles each being 94°C for 10 s, 45°C for 30 s, and 68°C (Expand) or 72°C (Pfu) for 6 min. Products were analyzed by 0.8% agarose gel electrophoresis.
DNA sequencing
Oligonucleotide primers were designed from the published sequence of dengue-2 virus New Guinea C (NGC) strain, GenBank accession number M29095. Prior to starting all sequence reactions, unincorporated PCR primers were removed by incubating the PCR product with 2 μl ExoSAP-IT reagent (USB) for 15 min at 37°C followed by heat inactivation of the reaction mixture for 15 min at 80°C. Sequence analyses of PCR products were performed on an ABI PRISM 3100 Genetic Analyzer using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). Each 20 μl sequence reaction contained 20 ng PCR product or 200 ng plasmid, 5–10 pmol primer and 7 μl sequence kit reagent. Sequence data were analyzed using Sequencher software (Gene Codes Corp).
To obtain 5′- and 3′-termini DNA for sequence analysis, the genome was circularized by ligation of the ends to each other, which served as a template for RT–PCR [12]. First, the 5′ cap structure was removed by treating viral RNA with 5 U tobacco acid pyrophosphatase in 1× buffer (Epicentre), 40 U RNasin and 1 mM ATP in a 30 μl reaction volume for 1 h at 37°C. Decapped RNA was extracted once with Aqua-Phenol (Ambion) and once with chloroform after which RNA was precipitated with 0.3 M sodium acetate/ethanol. The RNA was resuspended in water and ligated by incubation at room temperature for 3 h in a 20 μl reaction volume that contained 5 U T4 RNA ligase (New England Biolabs) in 1× ligase buffer, 10% DMSO, 1 mM ATP and 40 U RNasin RNase inhibitor. Ligated RNA templates were reverse transcribed to cDNA using the 5′-end reverse primer 246R and amplified by PCR using 3′-end forward primer 10158F and 246R 5′-end reverse primer (Table 1).
Construction of the dengue-2 PDK50 infectious clone
Overlapping dengue-2 PDK50 cDNA fragments were produced and used to replace the genomic sequences of dengue-2 NGC virus contained in the pRS424FLD2 infectious clone [13]. This was accomplished by sequential rounds of homologous recombination in the yeast strain Saccharomyces cerevisiae YPH857 between PDK50 DNA and corresponding sequences of the NGC DNA contained in the clone (Fig. 1). Following each round of homologous recombination, the chimeric yeast DNA was extracted and used to transform Escherichia coli STBL2 cells (Invitrogen) for amplification of the recombinant shuttle plasmid. A unique NotI enzyme restriction site was inserted directly downstream of the 3′ end of the PDK50 sequence in the completed clone to replace the non-unique SacI site in the plasmid.
Construction of a full-length dengue-2 PDK50 infectious clone. The dengue-2 PDK50 genome was reverse transcribed and amplified by PCR to produce overlapping DNA fragments used to replace dengue New Guinea C sequences in infectious clone pRS424FLD2 through sequential rounds of homologous recombination in yeast
Transformation of yeast and Escherichia coli
The yeast strain Saccharomyces cerevisiae YPH857 was a gift from Forrest Spencer (Johns Hopkins University, Baltimore, MD). Yeast were made competent by incubating log phase cells for 10 min in buffer containing 1 M Sorbitol, 10 mM bicine, pH 8.35, 40% (w/v) polyethyleneglycol-1000 [13]. For homologous recombination experiments, competent yeast were incubated with 1–2 μg DNA in a 500 μl reaction volume containing 40% (w/v) polyethylene glycol-1000 buffered with 200 mM bicine, pH 8.35, for 1 h at 30°C. Following incubation, cells were pelleted for 30 s at 3,000 rpm, washed once with buffer containing 150 mM sodium chloride and 10 mM bicine, pH 8.35, resuspended in the same buffer, and spread on standard minimal yeast media plates that lacked tryptophan to select for transformants [14]. Yeast DNA was prepared as described and transformed into chemically competent Escherichia coli STBL2 cells to amplify clone DNA [15].
RNA transcription
Plasmid containing the PDK50 clone DNA was linearized at the 3′ end of the dengue genome using a unique SacI site in hybrid NGC/S16803 PDK50 clones and a unique NotI site in the completed full-length PDK50 clone. The RNA transcripts were synthesized in 30 μl reaction mixtures containing 2 μg linearized DNA, 0.5 mM each rATP, rCTP, and rUTP, 0.1 mM rGTP, 0.5 mM cap analog m7G(5′)ppp(5′)G (New England Biolabs), 10 mM DTT, 40 U RNasin, 30 U SP6 RNA polymerase and 1× SP6 buffer (Promega). Reaction mixtures were incubated at 40°C for 1 h, and a 2 μl aliquot was examined by 0.8% agarose gel electrophoresis.
Electroporation
Infectivity of the RNA transcripts made from hybrid and completed PDK50 clones was confirmed by electroporating the transcripts into LLC-MK2 cells and analyzing for viral NS1 protein expression. Subconfluent cell monolayers were trypsinized and washed once with phosphate-buffered saline (PBS). Approximately 2 × 106 cells in 0.3 ml PBS were mixed with 10 μl RNA transcribed from the clone in a 0.4 cm gap electroporation cuvette (Biorad). The suspension was kept on ice for 10 min and then pulsed at 200 V and 950 μF, chilled briefly on ice, resuspended in 3.0 ml growth media, and seeded into 25 cm2 tissue culture flasks. The cell culture media was replaced with fresh media the next day and after 2 days, an aliquot of cells was removed from the 25-cm2 tissue culture flask by trypsinization and seeded onto a l-cm2 chamber slide (Nunc). The following day cells were rinsed twice with PBS to remove media, air-dried, fixed in ice cold acetone for 2 min, air-dried and analyzed for NS1 expression by indirect immunofluorescence using anti-NS1 monoclonal antibody 7E11 (produced at WRAIR) followed by FITC-labeled goat anti-mouse antibody. Once the full-length PDK50 clone was completed, RNA transcripts were prepared from the clone and transfected into LLC-MK2 cells by electroporation to make a high-titer virus stock of the PDK50 clone-derived virus.
Virus titration
Virus titers were determined by quantitative plaque assay of virus on LLC-MK2 cells in six-well plates. Serial dilutions of virus were adsorbed to cell monolayers for 1.5 h at room temperature after which the cells were overlaid with 3 ml of melted 1% SeaPlaque agarose (FMC Bioproducts) in Earle’s balanced salt solution without phenol red supplemented with 0.3% sodium bicarbonate, 10% FBS, 1 × each glutamine, MEM vitamins and amino acids (all from Invitrogen) and incubated at 37°C. After 8 days, plaques were stained with 2 ml of 1% SeaPlaque agarose containing 0.4 mg neutral red per ml. The plates were further incubated at 37°C overnight. Virus plaques were counted the following day.
Growth kinetics
Subconfluent monolayer cultures of LLC-MK2, Vero and C6/36 cells seeded in 25-cm2 flasks were infected with dengue-2 low-passage parent, PDK50 vaccine and PDK50 infectious clone-derived viruses at a multiplicity of infection of 0.01. Aliquots (0.5 ml) of culture fluid were removed daily for 10 days and stored at −80°C. The titer of dengue virus in each daily sample was determined by titration on LLC-MK2 cells.
Results
Sequence differences between parent and PDK-derived strains
Complete genomic nucleotide sequences were determined for dengue-2 S16803 low-passage vaccine parent, Genbank accession number GU289914.1, and its derivative PDK10, PDK30, PDK40 and PDK50 viruses. The analysis was performed on uncloned RT–PCR products and was therefore considered to yield a population average sequence. The PDK50 vaccine virus was found to differ from its low-passage parent by 13 mutations (Table 2). Eight mutations caused amino acid changes that were located in structural proteins prM (E89G) and E (E202K, N203D) and in nonstructural proteins NS1 (A43T), NS2A (L181F), NS2B (I26V) and NS4B (I/T108T and L112F). One mutation changed nucleotide 57 in the 5′ NCR from C to T and the remaining four mutations of nucleotides 591, 987, 6,471, and 8,907 were silent. The vaccine candidates that we sequenced are WRAIR pretransfection vaccine candidates and these sequences may not be identical to the unpublished sequence of the current WRAIR/GlaxoSmithKline vaccine candidate.
The evolution of mutations found in PDK50 was determined by analyzing the complete nucleotide sequences of intermediate passage–level viruses PDK10, PDK30 and PDK40 (Table 2), which were previously tested in small groups of human volunteers [10]. By PDK10, the average sequence had five nucleotide changes relative to parent virus, three of which caused amino acid changes in E (E202K), NS2A (L181F) and NS4B (I/T108T), whereas mutations of nucleotides 987 (E) and 6471 (NS3) were silent. By PDK30, two additional amino acid changes occurred in prM (E89G) and NS4B (L112F) proteins. Three more amino acid changes were found in E (N203D), NS1 (A43A/T) and NS2B (I26I/V) by PDK40 in addition to a C to T change at nucleotide 57 in the 5′ NCR, and two silent mutations at nucleotides 591 in prM and 8,907 in NS5. Between PDK40 and PDK50, mixtures in NS1 and NS2B were resolved to a single amino acid (NS1 A/T43T and NS2B I/V26V), and the G/A nucleotide mixture at position 8,907 in NS5 was resolved to A.
Sequence analysis of full-length PDK50 clone
The sequence of the complete dengue genome in the PDK50 clone sequence was determined directly from plasmid DNA and found to differ from the average sequence of the PDK50 virus at nucleotide positions 675, 7,558 and 9,334. These mutations are likely to represent a minor population of the PDK50 quasispecies virus or RT–PCR errors. Our goal was to produce a clone that contained the average sequence of the PDK50 genome. These three nucleotides were changed to the average PDK50 sequence using PCR products made from the PDK50 virus and additional rounds of homologous recombination in yeast followed by verification of sequence fidelity and clone infectivity.
Biological characterization of the infectious clone
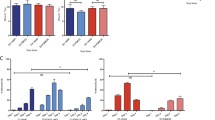
A high-titer stock of clone-derived PDK50 virus was generated by electroporating LLC-MK2 cells with RNA transcripts made from the final corrected clone. Low-passage parent, PDK50 vaccine and PDK50 infectious clone-derived viruses were compared for their growth kinetics in LLC-MK2, Vero and C6/36 cells (Fig. 2). The results showed that all three viruses grew similarly in all three cell types. Peak titers in Vero and C6/36 cells reached 106 pfu/ml. Peak titers in LLC-MK2 cells were lower, approximately 105 pfu/ml.
Growth kinetics of viruses. a LLC-MK2, b Vero, and c C6/36 cells grown in 25-cm2 flasks were infected with dengue-2 S16803 low-passage parent, PDK50 vaccine (PDK50V) and infectious clone-derived PDK50 (PDK50 IC) viruses at a multiplicity of infection of 0.01. Aliquots of culture fluid were removed daily for 10 days and stored at −80°C until use. The titer of DEN2 virus in each sample was determined by plaque assay on LLC-MK2 cells
Discussion
This study elucidated the nucleotide changes that occurred after passing dengue-2 virus strain S16803 in PDK cells following 10, 30, 40 and 50 passages. The study determined that the dengue-2 PDK50 virus differs from its low-passage parent by 13 mutations, 8 of which caused amino acid substitutions in structural proteins prM (E89G) and E (E202K, N203D), and nonstructural proteins NS1 (A43T), NS2A (L181F), NS2B (I26V) and NS4B (I/T108T, L112F). Four mutations of nucleotides 591 in prM, 987 in E, 6,471 in NS3 and 8,907 in NS5 were silent, and one changed a C to T in the 5′ NCR (Table 2). Most of these mutations are unique to the dengue-2 S16803 strain passaged in PDK cells but a few are similar in location to mutations found in other WRAIR PDK cell-adapted dengue strains [31]. Because the degree of attenuation of dengue-2 viruses progressively increased with PDK passage level, complete genomic sequences were derived for intermediate PDK passage-level strains PDK10, PDK30 and PDK40. Analysis of this data will aid future studies using the PDK50 infectious clone to isolate and evaluate mutations and their effects on virus biology, attenuation, and immune responses.
The mutation that changed the prM residue 89 from negatively charged glutamic acid to non-charged glycine occurred by PDK30. Residue 89 is position X of the four-residue consensus furin cleavage site R-X-K/R-R and is reported to be either aspartic or glutamic acid for dengue 1–4 [16]. Furin cleavage of prM occurs in the final stage of virus maturation and is required for subsequent rearrangement of the envelope protein and for virus infectivity [17]. The PDK30 mutation of prM 89 may contribute to attenuation by altering the rate of prM processing and ultimately virus maturation and infectivity. It has been shown that alterations of residues proximal to the furin cleavage site delayed export of chimeric dengue viruses out of infected cells and appeared to enhance the rate of furin processing of prM [16]. For a chimeric tick-borne encephalitis (prM-E)/dengue-4 virus, a valine to serine change in the first position downstream of the prM furin cleavage site significantly reduced mouse neurovirulence compared with the fatal infection caused by non-mutant chimeric virus; however, the effect of this mutation on furin cleavage of prM was not investigated [18]. The dengue-2 E89G substitution in prM is one of only two amino acid changes between PDK10 and PDK30. The other substitution between PDK10 and PDK30 occurs in NS4B (I/T108T).
Two E protein mutations, E202K and N203D, occurred by PDK10 and PDK40, respectively, and changed the encoded amino acid from acidic to basic at residue 202 and from polar non-charged to acidic at residue 203. The E protein is the major viral surface protein responsible for cell receptor binding and membrane fusion and it induces production of neutralizing antibodies. Three separate structural domains, central domain I, dimerization/fusion domain II and receptor binding/antigenic domain III, had been assigned based on the crystal structure of a soluble E protein fragment from tick-borne encephalitis and dengue-2 and -3 viruses [19–21]. A fourth domain, the C-terminal stem-anchor, was not part of crystallized proteins. On exposure to low pH in the endosome of infected cells, E protein dimers undergo a conformational change that rearranges the protein into trimers and exposes the domain II fusion peptide to initiate fusion of the virion with the endosomal membrane [22, 23]. This transformation involves transitions at the interfaces between domains I and II and domains I and III [22, 23]. Dengue-2 E protein residues 202 and 203 are located within the region that interconnects domains I and II. Mutations in the interconnecting regions between domains I and II and I and III have been found to occur in numerous flaviviruses that were adapted to grow in an alternate host and are thought to influence virus pathogenicity [24–30]. The other WRAIR PDK-passaged vaccine candidates also contain one or more mutations in regions that interconnect the E protein domains. The dengue-1 PDK20 strain has an E204K mutation at the domain I/II interface (personal observation), dengue-3 PDK30 has an I195N mutation at the domainI/II interface (unpublished observation, Bangti Zhao) and dengue-4 PDK20 has an N366N/S mutation at the domain I/III interface [31]. An understanding of how mutations at the E domain interfaces affect virulence and growth during vaccine manufacture is highly relevant to the development and safety testing of live-attenuated dengue vaccines.
The mutation that occurred in the 5′ NCR by PDK40 changed nucleotide 57 from C to T. Nucleotides in the 5′ NCR are predicted to be involved in complementary base pairing to form a stem structure and nucleotide position 57 of the dengue-2 genome is predicted to be involved in this base pairing, but is not predicted to participate in 3′ NCR and 5′ NCR complementary base pairing required for genome circularization [32]. A C to T change in dengue-2 PDK50 at this location might alter the RNA secondary structure and affect initiation of positive strand synthesis or RNA translation. The same nucleotide 57 C to T mutation occurred in a dengue-2 16881 PDK53 vaccine strain and when isolated using an infectious clone led to phenotypic changes in plaque size, replication in C6/36 cells and neurovirulence in newborn mice [33]. The C to T mutation was found to be genetically unstable in dengue-2 PDK53-vectored chimeric dengue-1, -3, and -4 viruses after growth adaptation to Vero cells [34]. For the dengue-2 S16803 PDK50 clone-derived virus, nucleotide 57 was unstable and reverted from T to parental C after one passage in FRhL cells (personal observation).
The five mutations in the PDK50 virus nonstructural genes that caused amino acid changes were in NS1 (A43T), NS2A (L181F), NS2B (I26V) and NS4B (I/T108T and L112F) proteins. Except for the enzymatic activities contained within NS3 and NS5, the roles of the other nonstructural proteins in virus replication and pathogenesis are not yet well defined. The conserved NS1 glycoprotein forms intercellular dimers and extracellular multimers and interacts with intracellular as well as cell surface membranes and other viral nonstructural proteins before being secreted into the extracellular medium [35, 36]. It is predicted to participate in RNA replication as it is localized within infected cells to presumed sites of RNA replication [37, 38]. Residue 43 in the NS1 glycoprotein is located near its N-terminus and is not part of the C terminal region required for dimerization [36, 37]. The A43T mutation changed the residue from hydrophobic to polar non-charged, which may affect NS1 interactions with other proteins or RNA. Other NS1 mutations have also been found to cause phenotypic changes in several flaviviruses [33, 38, 39].
The NS2A L181F mutation located near the C terminus of the protein occurred by PDK10 along with mutations in E (E202K) and NS4B (I/T108T) proteins. The NS2A protein is a small, multifunctional, membrane-associated protein shown to be involved in RNA replication, virus assembly, and modulation of the host antiviral response [40–42]. Residue 181 of dengue-2 S16803 virus is the middle leucine in the sequence VVSVSPLLLTSS. This sequence in NS2A is homologous to the sequence that forms the autophosphorylation site, KAVSPLLLTTT, of RNA-dependent protein kinase (PKR). The RNA-dependent PKR is regulated by double-stranded (ds) RNA and plays a significant role in the innate immune defense against virus infection. The N-terminal region contains a repeated domain that confers dsRNA-binding activity and the C-terminal region confers kinase activity. When activated by dsRNA, PKR undergoes conformational change that leads to phosphorylation of itself and the alpha-subunit of protein synthesis initiator 2 (eIF-2α). Phosphorylation of eIF-2α leads to inhibition of viral and cellular protein synthesis [43]. Numerous virus proteins have evolved mechanisms to inhibit PKR. Similar to the dengue-2 NS2A protein, the hepatitis C envelope (E2) protein contains the sequence RSEKSPLLLTTT that has homology with the PKR autophosphorylation site. The HCV E2 protein was shown to be inhibitory to PKR activity in cells and in vitro [44]. This raises the possibility that the sequence in the dengue-2 NS2A protein also has potential to serve as a pseudosubstrate for PKR and this protein should be investigated for interaction with and possible inhibition of PKR.
The PDK50 NS2B I26V mutation was found in PDK40 to be a mixture of the two residues and by PDK50, the mixture was resolved to be valine only. The I26V mutation is a conservative change with both isoleucine and valine being hydrophobic residues. The other two differences between PDK40 and PDK50 are at positions 2,548 in NS1 (A/T43T) and 8,907 (silent) in NS5. The NS2B protein is a cofactor required for viral NS3 protease cleavage of flavivirus nonstructural proteins at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, NS4A/NS4B, NS4B/NS5 junctions and at the C-terminus of the C protein [45, 46]. Three hydrophobic regions in NS2B flank a conserved 40-residue hydrophilic domain that participates in enzymatic processing [47, 48]. The I26V mutation is not located in these regions and its possible effect on attenuation is not known. Because only a small group of three volunteers were evaluated in the trial of the PDK40 strain, it is not certain that the resolution of mixtures in NS1 and NS2B in PDK40 to single amino acids in PDK50 was responsible for a real clinical difference.
The two NS4B mutations, I/T108T and L112F, are located in a central hydrophobic region in which other flavivirus vaccine strains develop mutations after readapting to grow in primate cells [49, 50]. For other flavivirus vaccine strains, mutations that arise in this region are found to be non-attenuating and even growth enhancing [49, 50]. A mutation occurred in this region of the NS4B protein in all four dengue WRAIR vaccine strains [31]. NS4B is a membrane-spanning hydrophobic protein with a highly conserved hydrophobicity profile but low sequence conservation among flaviviruses [51]. The NS4B protein was found to co-localize with the NS3 (protease and helicase) protein and with double-stranded RNA replication intermediates in cytoplasmic foci originating from the endoplasmic reticulum [52]. Using in vitro experiments, NS4B was shown to dissociate the NS3 protein from single-stranded RNA suggesting that it is involved in RNA replication through enhancing NS3 helicase activity [52]. The proposed membrane topology model localizes the dengue-2 virus NS4B I/T1089 (PDK10) and L112F (PDK30) mutations in the ER transmembrane domain [51]. Evidence was previously given that the NS4B protein of dengue, West Nile, and yellow fever viruses inhibits type I interferon signaling, and for dengue, the first 125 amino acids of NS4B were sufficient to cause the inhibition [53]. Knowledge of mutations relevant to enhancing virus growth in cell substrates or modulating innate antiviral immune responses is vital to the development of live-attenuated or inactivated dengue virus vaccines.
An infectious clone of dengue-2 PDK50 was produced and virus derived from the clone was shown to grow with the same kinetics as PDK50 vaccine and low-passage parental viruses in primate and mosquito cells. Results from the mutational analysis and availability of an infectious clone provide molecular tools for investigating the basis for dengue virus attenuation. The mutations will be isolated in future studies using the clone and systematically evaluated in assays that measure phenotypic differences. These are important steps that may help to develop assays to detect markers of attenuation and might be used to engineer improved molecular vaccines against dengue and other flavivirus infections.
References
World Health Organization Fact sheet No. 117, World Health Organization, Geneva, Switzerland (2002)
D.J. Gubler, Clin. Microbiol. Rev. 11, 480–496 (1998)
S.B. Halstead, Rev. Infect. Dis. 11(Suppl 4), S830–839 (1989)
D.W. Vaughn, S. Green, S. Kalayanarooj, B.L. Innis, S. Nimmannitya, S. Suntayakorn, T.P. Endy, B. Raengsakulrach, A.L. Rothman, F.A. Ennis, A. Nisalak, J. Infect. Dis. 181, 2–9 (2000)
D. Libraty, T.P. Endy, H.H. Houng, S. Green, S. Kayanarooj, S. Suntayakorn, W. Chansiriwongs, D.W. Vaughn, A. Nisalak, F.A. Ennis, A.L. Rothman, J. Infect. Dis. 185, 1213–1221 (2002)
J. Mongkolsapaya, T. Duangchinda, W. Dejnirattisai, S. Vasanawathana, P. Avirutnan, A. Jairungsri, N. Khemnu, N. Tangthawornchaikul, P. Chotiyarnwong, K. Sae-Jang, M. Koch, Y. Jones, A. McMichael, X. Xu, P. Malasit, G. Screaton, J. Immunol. 176, 3821–3829 (2006)
G. Wengler, D.W. Bradley, M.S. Collett, F.X. Heinz, R.W. Schlesinger, J.H. Strauss, Flaviviridae, in Virus Taxonomy: the Classification and Nomenclature of Viruses. Sixth Report of the International Committee on Taxonomy of Viruses, ed. by F.A. Murphy, C.M. Fauquet, D.H.L. Bishop, S.A. Ghabrial, A.W. Jarvis, G.P. Martelli, M.A. Mayo, M.D. Summers (Springer-Verlag, Vienna, 1995), pp. 415–427
K.H. Eckels, D.R. Dubois, R. Putnak, D.W. Vaughn, B.L. Innis, E.A. Henchal, C.H. Hoke Jr, Am. J. Trop. Med. Hyg. 69(6 Suppl), 12–16 (2003)
S.B. Halstead, N.J. Marchette, Am. J. Trop. Med. Hyg. 69(6 Suppl), 5–11 (2003)
N. Kanesa-Thasan, R. Edelman, C.O. Tacket, S.S. Wasserman, D.W. Vaughn, T.S. Coster, G.J. Kim-Ahn, D.R. Dubois, J.R. Putnak, A. King, P.L. Summers, B.L. Innis, K.H. Eckels, C.H. Hoke Jr, Am. J. Trop. Med. Hyg. 69(6 Suppl), 17–23 (2003)
W. Sun, R. Edelman, N. Kanesa-Thasan, K.H. Eckels, J.R. Putnak, A.D. King, H.S. Houng, D. Tang, J.M. Scherer, C.H. Hoke Jr, B.L. Innis, Am. J. Trop. Med. Hyg. 69(6 Suppl), 24–31 (2003)
C.W. Mandl, F.X. Heinz, E. Puchhammer-Stockl, C. Kunz, Biotechniques 10, 484–486 (1991)
S. Polo, G. Ketner, R. Levis, B. Falgout, J. Virol. 71, 5366–5374 (1997)
R.P. Valle, R.B. Wickner, J. Virol. 67, 2764–2771 (1993)
L. Zeng, B. Falgout, L. Markoff, J. Virol. 72, 7510–7522 (1998)
E.P. Kelly, B. Puri, W. Sun, B. Falgout, Vaccine 288, 3030–3037 (2010)
P. Keelapang, R. Sriburi, S. Supasa, N. Panyadee, A. Songjaeng, A. Jairungsri, C. Puttikhunt, W. Kasinrerk, P. Malasit, N. Sittisombut, J. Virol. 78, 2367–2381 (2004)
F. Guirakhoo, F.X. Heinz, C.W. Mandl, H. Holzmann, C. Kunz, J. Gen. Virol. 72, 1323–1329 (1991)
A.G. Pletnev, M. Bray, C.J. Lai, J. Virol. 67, 4956–4963 (1993)
F.A. Rey, F.X. Heinz, C. Mandl, C. Kunz, S.C. Harrison, Nature 375, 291–298 (1995)
Y. Modis, S. Ogata, D. Clements, S.C. Harrison, Proc. Natl. Acad. Sci. USA 100, 6986–6991 (2003)
Y. Modis, S. Ogata, D. Clements, S.C. Harrison, J. Virol. 79, 1223–1231 (2005)
S. Bressanelli, K. Stiasny, S.L. Allison, E.A. Stura, S. Duquerroy, J. Lescar, F.X. Heinz, F.A. Rey, EMBO J. 23, 728–738 (2004)
Y. Zhang, W. Zhang, S. Ogata, D. Clements, J.H. Strauss, T.S. Baker, R.J. Kuhn, M.G. Rossmann, Structure 12, 1607–1618 (2004)
E. Lee, R.C. Weir, L. Dalgarno, Virology 232, 281–290 (1997)
K.V. Pugachev, F. Guirakhoo, S.W. Ocran, F. Mitchell, M. Parsons, C. Penal, S. Girakhoo, S.O. Pougatcheva, J. Arroyo, D.W. Trent, T.P. Monath, J. Virol. 78, 1032–1038 (2004)
W.F. Tang, Y. Eshita, M. Tadano, K. Morita, Y. Makino, Microbiol. Immunol. 49, 285–294 (2005)
C.N. Duarte dos Santos, M.P. Frenkiel, M.P. Courageot, C.F. Rocha, M.C. Vazeille-Falcoz, M.W. Wien, F.A. Rey, V. Deubel, P. Despres, Virology 274, 292–308 (2000)
C.Y. Huang, S. Butrapet, D.J. Pierro, G.J. Chang, A.R. Hunt, N. Bhamarapravati, D.J. Gubler, R.M. Kinney, J. Virol. 74, 3020–3028 (2002)
T.P. Monath TP, J. Arroyo, I. Levenbook, Z.X. Zhang, J. Catalan, K. Draper, F.J. Guirakhoo, J. Virol. 76, 1932–1943 (2002)
F. Guirakhoo, Z. Zhang, G. Myers, B.W. Johnson, K. Pugachev, R. Nichols, N. Brown, I. Levenbook, K. Draper, S. Cyrek, J. Lang, C. Fournier, B. Barrere, S. Delagrave, T.P. Monath, J. Virol. 78, 9998–10008 (2004)
D.E. Alvarez, M.F. Lodeiro, S.J. Luduena, L.I. Pietrasanta, A.V. Gamarnik, J. Virol. 79, 6631–6643 (2005)
S. Butrapet, C.Y.H. Huang, D.J. Pierro, N. Bhamarapravati, D.J. Gubler, R.M. Kinney, J. Virol. 74, 3011–3019 (2000)
S. Butrapet, R.M. Kinney, C.Y.H. Huang, J. Virol, Methods 131, 1–9 (2006)
A.J. Crooks, J.M. Lee, L.M. Easterbrook, A.V. Timofeev, J.R. Stephenson, J. Gen. Virol. 75, 3453–3460 (1994)
M.J. Pryor, P.J. Wright, Virology 194, 769–780 (1993)
J.M. Mackenzie, M.K. Jones, P.R. Young, Virology 220, 232–240 (1996)
I.R. Muylaert, R. Galler, C.M. Rice, J. Virol. 71, 291–298 (1997)
I.E. Muylaert, T.J. Chambers, R. Galler, C.M. Rice, Virology 222, 159–168 (1996)
E.G. Westaway, J.M. Mackenzie, A.A. Khromykh, Adv. Virus Res. 59, 99–140 (2003)
B.M. Kummerer, C.M. Rice, J. Virol. 76, 4773–4784 (2002)
W.J. Liu, X.J. Wang, D.C. Clark, M. Lobigs, R.A. Hall, A.A. Khromykh, J. Virol. 80, 2396–2404 (2006)
M.J. Clemens, Int. J. Biochem. Cell. Biol. 29, 945–949 (1997)
D.R. Taylor, S.T. Shi, P.R. Romano, G.N. Barber, M.M.C. Lai, Science 285, 107–110 (1999)
B. Falgout, M. Pethel, Y.M. Zhang, C.J. Lai, J. Virol. 65, 2467–2475 (1991)
T.J. Chambers, A. Grakoui, C.M. Rice, J. Virol. 65, 6042–6050 (1991)
P. Niyomrattanakit, P. Winoyanuwattikun, S. Chanprapaph, C. Angsuthanasombat, S. Panyim, G. Katzenmeier, J. Virol. 78, 13708–13716 (2004)
P. Erbel, Schiering, A. D’Arcy, M. Renatus, M. Kroemer, S.P. Lim, Z. Yin, T.H. Keller, S.G. Vasudevan, U. Hommel, Nat. Struct. Mol. Biol. 13, 372–373 (2006)
J.E. Blaney Jr., G.G. Manipon, C.Y. Firestone, D.H. Johnson, C.T. Hanson, B.R. Murphy, S.S. Whitehead, Vaccine 21, 4317–4327 (2003)
J.E. Blaney Jr., C.T. Hanson, C.Y. Firestone, K.A. Hanley, B.R. Murphy, S.S. Whitehead, Am. J. Trop. Med. Hyg. 71, 811–821 (2004)
S. Miller, S. Sparacio, R. Bartenschlager, J. Biol. Chem. 281, 8854–8863 (2006)
I. Umareddy, A. Chao, A. Sampath, F. Gu, S.G. Vasudevan, J. Gen, Virol 87, 2605–2614 (2006)
J.L. Munoz-Jordan, M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W.I. Lipdin, A. Garcia-Saster, J. Virol. 79, 8004–8013 (2005)
Acknowledgments
The authors would like to thank Dr. Ken Eckels, Walter Reed Army Institute of Research, for providing the lyophilized low-passage parent and PDK10, PDK30, PDK40 and PDK50 vaccine candidate viruses. The studies were supported by the United States Army Medical Research and Materiel Command.
Disclaimer
The views expressed herein are those of the authors and do not necessarily represent those of the Department of the Army, Department of Defense or the Food and Drug Administration.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kelly, E.P., Polo, S., Sun, W. et al. Evolution of attenuating mutations in dengue-2 strain S16803 PDK50 vaccine and comparison of growth kinetics with parent virus. Virus Genes 43, 18–26 (2011). https://doi.org/10.1007/s11262-011-0602-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0602-z