Abstract
Okra mosaic virus (OkMV) is a tymovirus infecting members of the family Malvaceae. Early infections in okra (Abelmoschus esculentus) lead to yield losses of 12–19.5%. Besides intensive biological characterizations of OkMV only minor molecular data were available. Therefore, we determined the complete nucleotide sequence of a Nigerian isolate of OkMV. The complete genomic RNA (gRNA) comprises 6,223 nt and its genome organization showed three major ORFs coding for a putative movement protein (MP) of Mr 73.1 kDa, a large replication-associated protein (RP) of Mr 202.4 kDa and a coat protein (CP) of Mr 19.6 kDa. Prediction of secondary RNA structures showed three hairpin structures with internal loops in the 5′-untranslated region (UTR) and a 3′-terminal tRNA-like structure (TLS) which comprises the anticodon for valine, typical for a member of the genus Tymovirus. Phylogenetic comparisons based on the RP, MP and CP amino acid sequences showed the close relationship of OkMV not only to other completely sequenced tymoviruses like Kennedya yellow mosaic virus (KYMV), Turnip yellow mosaic virus (TYMV) and Erysimum latent virus (ErLV), but also to Calopogonium yellow vein virus (CalYVV), Clitoria yellow vein virus (CYVV) and Desmodium yellow mottle virus (DYMoV). This is the first report of a complete OkMV genome sequence from one of the various OkMV isolates originating from West Africa described so far. Additionally, the experimental host range of OkMV including several Nicotiana species was determined.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family Tymoviridae comprises the three genera: Tymovirus, Marafivirus and Maculavirus whose members share several characteristics like structural organization and physiochemical properties of virus particles [1]. The demarcating criteria among the three genera are the genome organization and the number and position of open reading frames (ORF) [2, 3]. Members of the genus Tymovirus comprise three ORFs coding for replication-associated proteins (RNA-dependent RNA polymerase (RdRp), methyltransferase and helicase) which are processed by a virus-encoded papain-like protease from one large protein [4], a putative movement protein (MP) with suppression of RNA silencing function and the coat protein (CP).
Okra mosaic virus (OkMV) was first reported in 1969 to cause chlorosis, mosaic and vein banding on the leaves of Abelmoschus esculentus (okra plant) in the Ivory Coast [5]. Besides the two Ivory Coast strains (OkMV-IC and OkMV-HR), other OkMV strains were described from Nigeria (OkMV-N, OkMV-NIN, OkMV-Nig and OkMV-Indigofera) which seem to be closely related but distinct strains of the same virus as they show differences in symptom expression and host range [6–11]. Typical symptoms on OkMV infected A. esculentus are dark and light green mosaic patterns or regular vein chlorosis of leaves. OkMV host plant species were found in families Amaranthaceae, Chenopodiaceae, Convolvulaceae, Cucurbitaceae, Euphorbiaceae, Leguminosae, Malvaceae and Solanaceae [5–7, 11, 12]. OkMV-IC is transmitted mechanically and by the flee-beetle Podagrica decolorata but not through seed or dodder [12]. For OkMV-N two additional beetle vectors were described, namely P. sjostedti and P. uniforma [13]. In okra plantings, the OkMV disease incidence ranges from 30% to 89% with 12–19.5% yield losses [14, 15]. Yield losses are mainly caused by reduction in number of fruits per plant (−34.7%) and reduced fruit length (−11.3%) in diseased okra [15].
The isometric OkMV particles are 28 nm in diameter with icosahedral symmetry. On the basis of base composition of the single-stranded RNA, the presence of empty protein shells, the high thermal inactivation point and the serological relationships to Turnip yellow mosaic virus (TYMV), OkMV was proposed, in 1973, to be a member of the tymovirus group [5].
Despite intense biological characterizations of different OkMV isolates, only minor information is available on the OkMV nucleotide sequence. In 1997, A.M. Mackenzie and colleagues (Australian National University, Canberra) described a small part of the OkMV genome including the CP nucleotide sequence and the 3′ untranslated region (UTR) (GenBank No. AF035202). Biological properties and the phylogenetic comparisons of partial genome sequences gave the first information about assigning OkMV to a tymoviral genus and only the complete sequence of the virus genome guarantees a final authentic taxonomic status. In this article, we report the first complete nucleotide sequence (nt) of the OkMV genomic RNA. Our sequence analysis and phylogenetic comparisons clearly identify OkMV as a member of the genus Tymovirus with close relationship not only to Kennedya yellow mosaic virus (KYMV) and TYMV on complete genome level, but also to Calopogonium yellow vein virus (CalYVV), Clitoria yellow vein virus (CYVV) and Desmodium yellow mottle virus (DYMV) when CP sequences were compared.
Materials and methods
Virus source and experimental host range
Freeze-dried leaf material of OkMV (DSMZ PV-0264) infected Vigna unguiculata cv Ife Brown was transmitted by mechanical inoculation to Nicotiana hesperis and N. benthamiana and the infected plants were kept under greenhouse conditions at 24°C heating temperature with 15 h day time and auxiliary lighting (5,000 lux). For mechanical inoculations, leaves were rubbed with plant sap from infected plants (1:10, wt/vol) or freeze-dried material (1:5, wt/vol) in 0.03 M HEPES (pH 7) inoculation buffer supplemented with celite. The OkMV isolate is available from DSMZ (order PV-0264).
To determine the experimental host range, 12 plant species belonging to the families of Solanaceae (N. benthamiana, N. occidentalis, N. clutinosa, N. clevelandii, N. glutinosa, N. rustica, N. tabacum ‘Xanthi’, Solanum melongena), Chenopodiaceae (Chenopodium capitatum, C. quinoa), Malvaceae (A. esculentus) and Fabaceae (Vigna oblongifolia) at the 4–6 leaf stage (Table 2) were mechanically inoculated as described above. Except for V. oblongifolia, where four test plants were inoculated, a minimum of 10 plants per plant species were inoculated in the host range experiment. Predominantly, Nicotiana species, the typical laboratory host plants for plant viruses, were used. Control plants were treated with inoculation buffer only. Total RNA was extracted from newly developed leaves 3 weeks after mechanical inoculation and systemic OkMV infections were detected by RT-PCR using 0.5 U of Avian myeloblastosis virus Reverse Transcriptase (Finnzymes) and 0.5 U of Taq DNA Polymerase (Promega) in 50 μl reactions. Standard one-step RT-PCRs were performed due to Promegas recommendations with 60 min at 42°C for cDNA synthesis and 35 PCR cycles (94°C/30 s; 58°C/30 s; 72°C/30 s). The following primers were used to detect systemic OkMV infections by RT-PCR: OkMVDs (5′-ACACTCCCCTGCCAAAGACC-3′) and OkMVEas (5′-TCGTTGACCTTGTGTTGGGC-3′). Test plants were inspected for symptom development until 7 weeks postinoculation (pi).
RNA extraction and purification of OkMV particles
The extraction of total plant RNA was done as described earlier [16] except for an additional DNAse (5 U) and proteinase K (12 U) treatment of the overnight-dissolved precipitate followed by phenol/chloroform purification prior the final ethanol/NaOAc precipitation step. Another RNA extraction protocol [17] was used to extract total RNA from highly viscous plant sap of A. esculentus. Ten gram of leaf tissue was used to extract dsRNA following established procedures [18, 19]. Extracted dsRNA was fractionated by electrophoresis in 1% agarose gel and eluted in 20 μl Tris–EDTA (10 mM/0.1 mM; pH 7.5).
Purification of OkMV was essentially done as described earlier [20] including centrifugation in 10–40% sucrose gradients. RNA was extracted from the final proteinaceous ultracentrifugated pellet by proteinase K treatment, phenol/chloroform extraction and subsequent RNA precipitation. The OkMV particles recovered from the final ultracentrifugated pellet were negatively stained with 1% uranyl-acetate and visualized by electron microscopy.
OkMV sequence determination
On the basis of a 3′-end 681 nt sequence fragment of the OkMV genome available (GenBank No. AF035202) specific F1s and F1as primers were designed and used for cDNA synthesis using avian myeloblastosis virus reverse transcriptase (Promega Corp.) to amplify the entire OkMV CP. PCR reactions were performed using Phusion DNA polymerase (Finnzymes) in HF-buffer, nevertheless GC-buffer provided by Finnzymes was used for the amplification of fragments F9 and F10 according to the manufacturer’s recommendations. All primers used to determine the complete OkMV nucleotide sequence are listed in Table 1. Different methods were applied to determine the complete genome sequence of OkMV. Fragments F6, F9 and F10 were amplified using sequence specific sense and anti-sense primers selected from bordering fragments. For the amplification of fragments F2, F3 and F8, gene specific primers were designed from newly determined sequences and the degenerated primers F2s, F3s and F8as were selected from conserved tymoviral nucleotide stretches. The degenerated primers were designed based on conserved motifs determined by multiple nucleotide sequence alignments of complete tymoviral genomes of Eggplant mosaic virus (EMV), Erysimum latent virus (ErLV), Kennedya yellow mosaic virus (KYMV), Ononis yellow mosaic virus (OYMV), Physalis mottle virus (PhyMV) and TYMV. In a similar approach, fragment F7 was amplified by using a primer including eight highly conserved nucleotides (as determined from alignments) together with a random primer. Amplification of fragment F4 was done by accomplishing a standard gene walking approach [22] implementing a sequence specific and a random primer. A sequence specific and a consensus-degenerate hybrid oligonucleotide primer (CODEHOP), which was designed by the CODEHOP PCR design program [23], was used to amplify fragment F5.
The OkMV 5′- and 3′-terminal sequences were determined by applying the SMART method [21]. For this, OkMV dsRNA was used as template for cDNA synthesis using Superscript II (Invitrogen Corp.) and sequence specific sense (3′-end) and anti-sense (5′-end) primers.
Extracted total RNA, dsRNA or RNA purified from virions was heated at 95°C for 5 min and immediately cooled on ice prior to cDNA synthesis. Amplified PCR fragments were cloned into pGEM-T easy vector (Promega Corp.) and subsequently sequenced. The complete OkMV sequence of the Nigerian isolate is available at GenBank (EF554577).
Sequence analysis and phylogenetic comparisons
Sequence alignments were performed employing Vector NTI 10.0.1 ContigExpress and AlignX software (Invitrogen Corp.) and the NCBI BLAST program [24]. Phylogenetic trees based on multiple sequence alignments created by CLUSTAL X V.1.8 [25] were visualized using TreeView V.1.6.6 [26]. RNA secondary structure prediction was performed by using pknotsRG [27] and the Pseudoviewer 3 software [28]. Conserved domains were identified according to the method described earlier for methyltransferases [29] proteases [30], helicases and RdRps [31] and also by using the protein motif scan software MyHits [32].
Results
OkMV sequence determination
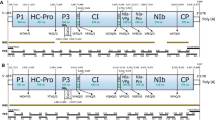
The entire OkMV genome was determined by RT-PCR reactions using total RNA, dsRNA or RNA extracted from purified virus particles. The overlapping 12 PCR fragments covering the entire OkMV genome are shown in Fig. 1a. Sequence determination started with a stretch of known OkMV nucleotide sequence covering the CP region at the 3′-terminal end of the genome. The specific primers F1s and F1as, based on the known sequence, were used to amplify the complete OkMV CP nucleotide sequence (Fig. 1a, F1). Subsequently, different methods were used to determine the complete OkMV genome sequence (Table 1). A standard gene walking approach using a specific and a random annealing primer was used to amplify fragment F4. Except for the terminal ends, all other partial genomic sequences were determined by using sequence specific or random annealing primers in combination with primers chosen from multiple nucleotide or amino acid alignments (conserved nucleotide stretches and CODEHOPs) of other tymoviruses. The conserved 8 nucleotides 5′-TTCATGAA-3′ found in tymovirus genome sequences about 480 nt downstream of the 5′-end were used to design primer F7s. Another highly conserved stretch of the 11 nucleotides 5′-TGGGACAA(C/T)CT-3′, found about 1,130 nt downstream of the 5′-end, was included in the degenerated anti-sense primer F8as. Taken together, all primer sequences selected from tymoviral multiple alignments or by the CODEHOP strategy were also conserved in the OkMV genome sequence (as shown by overlapping PCR fragments) and proved to be suitable to determine the complete OkMV genome sequence.
(a) Genome organization of OkMV and position of overlapping cDNA clones F1–F12 used for sequence determination. Three major ORFs are indicated by grey boxes with nucleotide positions of the start and stop codons, respectively. Mr of putative protein products is shown. (b) Electron micrograph showing full and empty (stained in the middle) OkMV particles purified from N. hesperis after uranyl acetate staining. Scale bar indicates 50 nm
Specific sense (3′-end) or anti-sense primers (5′-end) were designed from the known sequences of fragment F1 and F7, respectively; those primers together with the standard SMART primer were used for cDNA synthesis. F11 and F12 fragments could be amplified by using internal specific primers together with the internal SMART primer. The first 8 nt of the OkMV gRNA are 5′-GTAATCTT-3′, having the first four residues identical to that of TYMV, OYMV, KYMV and EMV. The last 8 nt of the OkMV 3′-UTR are 5′-TCGAGACC-3′ and the terminal 3 nt are identical with that of TYMV and OYMV.
After comparing the newly known amino acid (aa) sequence of putative OkMV RdRp with that of other tymovirus RdRp’s, we found an internal deletion of about 360 nt in fragment F4. Therefore, sequence specific primers were selected to amplify fragment F6 which covered the missing sequence in F4. The reason for the deletion in F4 is unknown, but is most probably artificially and PCR-generated.
Fragments F1–F8 were amplified by RT-PCR using total RNA or dsRNA extracted from OkMV infected plants. Since attempts to amplify the 1,800 nt region from F5 and F8 failed, RNA from purified OkMV particles was used to amplify fragments F9 and F10. Electron-microscope captured purified virions (Fig. 1b) showed a high number of apparently empty virion shells. The RNA from virions was sufficient for RT-PCR amplification of fragments F9 and F10 and the missing nucleotide stretch of about 1,800 nt spanning F5 and F8.
Genome organization and tymoviral characteristics of OkMV
The genome organization of OkMV was determined by alignment of overlapping cDNA fragments as shown in Fig. 1a. The complete genomic RNA (gRNA) consists of 6,223 nt and is available in the GenBank (Accession number EF554577). The genome sequence reveals a typical tymovirus base ratio with a high percentage of cytosine residues (18.7 A, 44.8 C, 14.9 G and 21.6 U). On the gRNA three major ORFs were found, which were similar in size and arrangement as shown for other tymoviruses. The first ORF (residues 85–2,100) on the gRNA is 2,016 nt in length and was predicted to encode a putative protein product of Mr 73.1 kDa. Comparative Blast analysis revealed high aa sequence homology with other tymoviral MPs. All conserved motifs described for tymovirus MPs [33] were found in the OkMV MP, except a single change where the conserved aa residue Arg124 is replaced by Gly124. The largest ORF (residues 92–5,542) is 5,451 nt in length and encodes the putative OkMV replication associated protein (RP). This ORF almost overlaps the MP ORF and starts, as shown for all other tymoviruses, 7 nt downstream. The putative RP of Mr 202.4 kDa contains conserved domains of methyltransferases (aa 70–199), viral peptidases (aa 759–867), helicases (aa 950–1,185) and RdRps (aa 1,323–1,769) and a proline-rich region (aa 429–717). For TYMV two aa residues (Cys783 and His869) are essential for the proteinase active site [29] which in OkMV could be mapped in the RP at Cys765 and His851, respectively. Another aa residue (Gly803) which is conserved in all tymovirus proteinase domains, was also found in the OkMV RP. The third and smallest ORF (residues 5,569–6,135) is 567 nt in length and encodes the OkMV CP with a Mr 19.6 kDa. The tymovirus CP is predicted to be translated from a subgenomic RNA (sgRNA) having the highly conserved 16 nt tymobox as sgRNA promoter [34, 35]. An analogous sequence upstream of the sgRNA transcription initiation site is found for OkMV at nt position 5,541–5,556 in the non-coding region (26 nt) separating RP and CP ORFs sharing 15/16 identical nucleotides of the tymobox. In contrast to other tymoviruses, the OkMV tymobox is completely located in the untranslated region upstream of the CP coding ORF and is not overlapped by the RP ORF. All conserved aa patterns described for tymovirus CPs [33] were found in the OkMV CP also.
The OkMV 5′-UTR is 84 nt in length and typical tymovirus hairpin structures were found in the first 130 nt of the OkMV genome (Fig. 2a). Three imperfect hairpins (HP) were predicted in the OkMV non-coding and coding 5′-terminal region with protonable internal loops implementing typical C–C mismatches [36] which in TYMV are shown to be important for viral packaging [37]. In contrast to other tymoviruses, the MP and RP AUG start codons were found within HP2 and HP3. The 3′-UTR is 88 nt in length and part of this sequence can be folded into an aminoacetylatable tRNA-like structure (TLS) similar to other tymoviruses [38]. For TYMV it was shown that the TLS acts as a translational enhancer [39]. The OkMV TLS is located upstream of a pseudoknot with a typical T-stem hairpin having a possible D-loop-T-loop interaction [40]. The 5′-region of the OkMV TLS contains the anticodon for valine (Fig. 2b).
Predicted secondary structures for the 5′- and 3′-termini of OkMV gRNA. (a) Three hairpins (HP) with internal loops were predicted in the 130 5′-terminal nucleotides including the MP and RP AUG start codons (arrows). (b) tRNA-like structure at the 3′-terminal end of OkMV gRNA with the CAC anticodon for valine (arrow), a pseudoknot and T- and D-like loops with possible tertiary interaction (dotted line)
Phylogenetic comparisons
The complete nucleotide sequence and the aa sequences of OkMV MP, RP and CP were aligned with corresponding regions of other tymoviruses. Overall genome nucleotide identity of OkMV with other tymoviruses was in the range of 54–63% showing highest identity to KYMV (63%) and TYMV (60%) and least identity to ErLV (54%).
Comparison of our OkMV sequence with the OkMV CP aa sequence reported earlier (GenBank No. AF035202) revealed an aa identity of 89% (87% nucleotide identity) slightly below the 90% demarcation criteria for tymoviruses listed in the VIIIth ICTV Report on Taxonomy of Viruses. Based on the CP aa sequence, closest phylogenetic relationship was found with KYMV (65%; Fig. 3), similar to what was observed for the complete nucleotide sequence of OkMV. Much less conservation was generally found in tymovirus MPs. Nevertheless, in the phylogenetic tree based on MP alignments from several tymoviruses, OkMV clearly clusters with KYMV (34% identity) and TYMV (32% identity) (Fig. 3). The same observation was made when comparing 12 available tymovirus RP aa sequences with OkMV (Fig. 3), where highest sequence identities were found with KYMV (60%) and TYMV (54%). OkMV, KYMV, TYMV and ErLV formed a subcluster in the phylogenetic tree based on RP or MP aa sequences, though this picture is different when CP aa sequences are taken as the basis for the phylogenetic tree. Except for KYMV (65%), OkMV shows higher identity in CP region to CalYVV (62%), CYVV (61%) and DYMoV (60%), for which neither MP nor RP sequences are available as yet, than to TYMV (45%) or ErLV (34%).
Phylogenetic trees based on multiple aa sequence alignments, (CP) Tymovirus coat proteins, (MP) movement proteins and (RP) replication-associated proteins. Numbers at nodes indicate bootstrap values for 1,000 replicates. Bootstrap values below 50% are not shown. Genbank accession numbers of proteins are given in brackets. Scale bar represents 0.1 aa substitutions per site related to branch length. Percent marks next to the alignments represent pairwise aa sequence identities compared to OkMV
Experimental host range
The results of the experimental host range studies are summarized in Table 2. Systemic OkMV infections were detected in Nicotiana occidentalis, N. clevelandii, N. benthamiana, N. hesperis, A. esculentus and Vigna oblongifolia. Differences in symptom expression were observed between the evaluated plant species, though all the test plants showing symptoms were positively identified for OkMV infection using RT-PCR. On only one infected N. occidentalis plant, strong yellowing of intercostal leaf areas on systemically infected leaves was observed 3 weeks pi (Fig. 4c) with recovery and absence of symptoms 7 weeks pi. Similar symptoms with pale yellowing and chlorotic vein banding patterns were detected on infected N. benthamiana plants but fainting to complete absence resulted on most of the plants 5–6 weeks pi. OkMV-infected N. clevelandii plants showed systemic dark and light green mosaic patterns as early as 2 weeks pi, but symptoms were not detected on newly developed leaves 5–6 weeks pi. N. hesperis-infected plants showed yellowing and chlorotic patterns on leaves and leaves were smaller in size compared to those of healthy control plants. These symptoms appeared 1–2 weeks pi and were also found on newly developed leaves. Interestingly, yellowing symptoms were observed on stems of infected N. hesperis plants. OkMV-infected young leaves of V. oblongifolia (Fig. 4d) showed yellow mosaic pattern and leaf enation 2 weeks pi and those symptoms persisted throughout the experiment. Typical symptoms of an OkMV infection were detected on okra plants (A. esculentus) as early as 1–2 weeks pi. Yellowing/chlorosis was observed first on leaf edges, and the leaf midrib followed by subsequent spread out to the entire leaf area (Fig. 4a). Light green mosaic and strong vein banding patterns were the prominent symptoms on these leaves. The next appearing 1–2 leaves showed a nearly complete yellowing intercepted by strong dark green mosaic patterns (Fig. 4b). Only the first 3–5 leaves were symptomatic on A. esculentus and no symptoms were detected on other newly emerging leaves. In all experiments, no symptoms were detected on test plants which were treated with inoculation buffer only. This is the first report that V. oblongifolia, N. occidentalis and N. hesperis are OkMV host plants.
Symptoms observed on OkMV-infected test plants. Typical symptoms development like (a) vein chlorosis and (b) strong mosaic pattern, on the first three leaves of infected A. esculentus. (c) Healthy control (left) and an infected leaf showing intercostal leaf yellowing (right) on N. occidentalis. (d) Healthy control (left) and infected leaf showing yellow chlorosis (right) on V. oblongifolia
Discussion
The complete nucleotide sequence of OkMV was determined and sequence analysis clearly confirmed that OkMV resembles several features of a typical member of the genus Tymovirus having three major ORFs on the gRNA, 3′-terminal hairpins, a tymobox and 5′-terminal TLS. This is the first complete genome sequence of a tymovirus infecting Malvaceae.
Phylogenetic comparisons of CP sequences showed closest relationship to KYMV and CYVV, another tymovirus infecting plant species of the family Malvaceae. It is only speculative, but subclustering of OkMV with ErLV and TYMV in MP and RP phylogenetic trees might change when more sequence information of tymoviruses become available both of which cluster with OkMV in the CP phylogenetic tree. Interestingly, the Nigerian OkMV-isolate presented here showed a remarkable low CP sequence identity to the isolate available at the GenBank. Even if the CP aa identity is slightly below the 90% demarcation criteria for tymoviruses, we still consider these viruses as isolates of OkMV. Recently, it was shown for British TYMV isolates that CP nucleotide sequence homologies can be as low as 88% and even lower (74%) when compared with Australian TYMV isolates [41].
The difference in the 3′-terminal nucleotide between the earlier published (5′-CCA-3′) and our presented OkMV sequence (5′-CC-3′) is in agreement with reports that 80% of encapsidated TYMV RNAs have a CC 3′-termini lacking a terminal adenosine residue [42, 43]. Shortly after infection, a host CCA nucleotidyltransferase is supposed to add the terminal adenosine residue which is important for expression of full tRNA-like properties and minus-strand synthesis [44] and this should be valid for OkMV, too.
Purified OkMV RNA combined with a PCR-buffer optimized for the amplification of GC-rich templates resolved our problems to amplify fragments F9 and F10. Difficulties to amplify C-rich templates are well known and were also reported for the tymovirus PhyMV [45] where C-rich nucleotide stretches in PhyMV genomic regions comparable to the ones covered by F9 and F10 hindered efficient amplifications. The described degenerated primers based on conserved tymovirus nucleotide motifs and the CODEHOP strategy can be useful for the determination of complete genome sequences of other tymoviruses.
There are several reports about OkMV isolates with varying host plant spectra [46]. The described isolates exclusively originate from Nigeria (OkMV-N, OkMV-Nig and OkMV-Indigofera from Ibadan [6, 10, 11, 47] or OkMV-NIN from Nsukka [7, 8]) or from the Ivory Coast (OkMV-IC from Bouaké [5, 10, 11] or OkMV-HR from Abidjan [11]). The symptoms observed in our host range experiments are in agreement with those of earlier reports of Nigerian OkMV isolates [6, 9, 47] except that we found a single OkMV-infected N. occidentalis plant and neither systemic infections nor clear local lessions in C. quinoa.
Okra mosaic virus-induced symptoms on A. esculentus were only detectable on the first 3–5 leaves and not on further leaves. This is in agreement with earlier reports that OkMV is not uniformly distributed in A. esculentus [47] and younger leaves were asymptomatic [5, 7]. Nevertheless, asymptomatic leaves still contained OkMV as was shown by grafting experiments. Similar results were reported for another tymovirus, namely chayote mosaic virus (ChMV), where asymptomatic systemic infections could be found in several plant species [48]. Our host range experiment showed that beside A. esculentus, N. clevelandii, N. occidentalis and to some extent N. benthamiana plants recovered from OkMV-induced symptoms 5–7 weeks pi. This is in contrast to earlier reports [5] where OkMV-infected N. clevelandii showed persistence of symptoms and not a recovery phenotype. We did not do any intense testing of symptomless leaves, but OkMV might still be present in these leaves and the virus titre might be reduced most probably by a plant immune response like RNA-silencing. We cannot exclude the possibility that environmental conditions are the reason for the asymptomatic leaves as it was shown for petunia vein banding virus (PetVBV) that symptoms on N. benthamiana and Nicandra physalodes were only observed during autumn and winter [49]. We did not detect any clear OkMV-induced chlorotic local [47] or systemic [7, 11] lesions on C. quinoa as was reported earlier for OkMV-N, and also any persistence of symptoms in this plant species as was reported for OkMV-IC [5, 11]. Therefore, C. quinoa and possibly other Chenopodiaceae species are still regarded as discriminating hosts between the Nigerian isolate OkMV-N and the Ivory Coast isolate OkMV-IC.
Among the newly described OkMV host plants V. oblongifolia, N. occidentalis and N. hesperis, the latter one is an optimal OkMV laboratory propagation host as symptoms persist over a long period and high amounts of virus particles can be extracted.
The complete genome of OkMV presented here broadens the range of available tymovirus sequence information and confirms all typical tymovirus characteristics reported earlier. As the host spectrum and the geographical origin of the OkMV isolate stored earlier in the GenBank (No. AF035202) are unknown, it would be interesting to determine the complete genome of a second geographical distinct OkMV isolate, e.g. the Ivory Coast isolates OkMV-IC or OkMV-HR. These isolates differ from the Nigerian isolate described here, at least in their ability to show persistent symptoms on C. quinoa and several other Chenopodium species [5, 11, 47], and differences might be related to genome sequence variations. But different symptomatology is most probably not only based on CP sequence variation, but also on differences in the MP and RP coding region as was proposed for seven Australian TYMV isolates with deviating symptoms in Cardamine robusta and in Chinese cabbage plants [50]. Therefore, future research implementing complete genome sequence information of other OkMV isolates might shed light onto the basis of these differences.
References
G.P. Martelli, S. Sabanadzovic, N.A.G. Sabanadzovic, M.C. Edwards, T.W. Dreher, Arch. Virol. 147, 1837 (2002)
T.W. Dreher, M.C. Edwards, A.J. Gibbs, A.-L. Haenni, R.W. Hammond, I. Jupin, R. Koenig, S. Sabanadzovic, N. Abou Ghanem-Sabanadzovic, G.P. Martelli, in Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses ed. by C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball (Elsevier Academic Press, San Diego, 2005), pp. 1067–1076
G.P. Martelli, S. Sabanadzovic, N. Abouh-Ghanem Sabanadzovic, P. Saldarelli, Arch Virol 147, 1847 (2005)
K.L. Bransom, J.J. Weiland, T.W. Dreher, Virology 184, 351 (1991)
L. Givord, L. Hirth, Ann. Appl. Biol. 74, 359 (1973)
A.O. Lana, R.F. Bozarth, Phytopath. Z. 83, 77 (1975)
E.C.K. Igwegbe, Plant Dis. 67, 320 (1983)
E.C.K. Igwegbe, A.D. Hewings, V.D. Damsteegt, W.M. Dowler, Phytopathology 77, 987 (1987)
G. Thottappilly, Acta Virol. 40, 233 (1996)
R.F. Bozarth, A.O. Lana, R. Koenig, J. Reese, Phytopathology 67, 735 (1977)
L. Givord, Ann. Phytopathol. 9, 53 (1977)
L. Givord, L. Den Boer, Ann. Appl. Biol. 94, 235 (1980)
A.O. Lana, T.A. Taylor, Ann. Appl. Biol. 82, 361 (1976)
G.I. Atiri, M.F. Ivbijaro, A.D. Oladele, Trop. Agri. 68, 178 (1991)
J. Ndunguru, A.C. Rajabu, Scientia Hort. 99, 225 (2004)
T.C. Verwoerd, B.M. Dekker, A. Hoekema, Nucleic Acids Res. 25, 2362 (1980)
I. Bekesiova, J.P. Nap, L. Mlynarova, Plant Mol. Biol. Rep. 17, 269 (1999)
A.J. Dodds, J.T. Morris, L.R. Jordan, Ann. Rev. Phytopathol. 22, 151 (1984)
A.J. Dodds, in DsRNA in diagnosis, ed. by R.E.F. Matthews (CRC Press, Florida, 1993), pp. 274–289
A. Gibbs, A.M. Mackenzie, Meth. Mol. Biol. 81, 219 (1998)
M. Matz, D. Shagin, E. Bogdanova, O. Britanova, S. Lukyanov, L. Diatschenko, A. Chenchik, Nucleic Acids Res. 27, 1558 (1999)
J.D. Parker, P.S. Rabinovitch, G.C. Burmer, Nucleic Acids Res. 19, 3055 (1991)
T.M. Rose, Virol. J. 2, 20 (2005)
S.F. Altschul, T.L. Madden, A.A. Schäffer, Nucleic Acids Res. 25, 3389 (1997)
J.D. Thompson, T.J. Gibson, F. Plewniak, F. Jeanmougin, D.G. Higgins, Nucleic Acids Res. 25, 4876 (1997)
R.D.M. Page, Comput. Appl. Biosci. 12, 357 (1996)
J. Reeder, R. Giegerich, BMC Bioinf. 5, 104 (2004)
Y. Byun, K. Han, Nucleic Acids Res. 34 (web server issue), W416 (2006)
M.N. Rozanov, E.V. Koonin, A.E. Gorbalenya, J. Gen. Virol. 73, 2129 (1992)
M.N. Rozanov, G. Drugeon, A.L. Haenni, Arch. Virol. 140, 273 (1995)
E.V. Koonin, V.V. Dolja, Crit. Rev. Biochem. Mol. Biol. 28, 375 (1993)
M. Pagni, V. Ioannidis, L. Cerutti, M. Zahn-Zabal, C.V. Jongeneel, L. Falquet, Nucleic Acids Res. 32(web server issue), W332 (2004)
R. Koenig, C.W. Pleij, D.E. Lesemann, S. Loss, H.J. Vetten, Arch. Virol. 150, 2325 (2005)
S. Ding, J. Howe, P. Keese, A. Mackenzie, D. Meek, M. Osorio-Keese, M. Skotnicki, P. Srifah, M. Torronen, A. Gibbs, Nucleic Acids Res. 18, 1181 (1990)
J. Schirawski, A. Voyatzakis, B. Zaccomer, F. Bernardi, A.L. Haenni, J. Virol. 74, 11073 (2000)
K. Hellendoorn, P.J.A. Michiels, R. Buitenhuis, C.W.A. Pleij, Nucleic Acids Res. 24, 4910 (1996)
H.H. Bink, J. Schirawski, A.L. Haenni, C.W. Pleij, J. Virol. 77, 7452 (2003)
T.W. Dreher, J.B. Goodwin, Nucleic Acids Res. 26, 4356 (1998)
D. Matsuda, T.W. Dreher, Virology 321, 36 (2004)
M.H. de Smit, A.P. Gultyaev, M. Hilge, H.H. Bink, S. Barends, B. Kraal, C.W. Pleij, Nucleic Acids Res. 30, 4232–4240 (2002)
E.J. Mitchell, J.M. Bond, Arch. Virol. 150, 2347 (2005)
J.P. Briand, G. Jonard, H. Guilley, K. Richards, L. Hirth, Eur. J. Biochem. 72, 453 (1977)
M. Silberklang, A. Prochiantz, A.L. Haenni, U.L. Rajbhandary, Eur. J. Biochem. 72, 465 (1977)
T.W. Dreher, Mol. Plant Pathol. 5, 367 (2004)
C.T. Ranjith-Kumar, K. Gopinath, A.N.K. Jacob, V. Srividhya, P. Elango, H.S. Savithri, Arch. Virol. 143, 148 (1998)
P.L. Guy, J.L. Dale, M.A. Adena, A.J. Gibbs, Plant Pathol. 33, 337 (1984)
A.O. Lana, R.M. Gilmer, H.D. Cheda, D.O. Fatokun, Plant Dis. Rep. 58, 616 (1974)
J.J. Bernal, I. Jiménez, M. Moreno, M. Hord, C. Rivera, R. Koenig, E. Rodríguez-Cerezo, Phytopathology 90, 1098 (2000)
M.A.V. Alexandre, L.M.L. Duarte, E.B. Rivas, C.M. Chagas, M.M. Barradas, R. Koenig, Plant Dis. 84, 739 (2000)
C.M. Hayden, A.M. Mackenzie, A.J. Gibbs, Arch. Virol. 143, 191 (1998)
Acknowledgements
We thank Dirk Bellstedt, Kees Pleij and Theo Dreher for scientific discussion and Agnes Pietruszka and Marianne Koerbler for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this article have been submitted to the Genbank nucleotide sequence database and have been assigned the accession number EF554577.
Rights and permissions
About this article
Cite this article
Stephan, D., Siddiqua, M., Ta Hoang, A. et al. Complete nucleotide sequence and experimental host range of Okra mosaic virus . Virus Genes 36, 231–240 (2008). https://doi.org/10.1007/s11262-007-0181-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-007-0181-1