Abstract
The human papillomavirus type 16 (HPV-16) L1 capsid protein is very poorly expressed from cDNA expression plasmids transiently transfected into mammalian cells. The results described herein demonstrate that inhibition of HPV-16 L1 expression from L1 cDNAs correlates with the presence of splicing regulatory sequences in the L1 coding region. This inhibitory effect correlates with the binding of hnRNP A1 to the RNA elements. Similar to unutilised splice sites that may retain mRNAs in the nucleus, regulatory splicing RNA elements may also inhibit gene expression in the absence of splicing. The results presented here explain the inefficient expression of HPV-16 L1 protein from the wild type L1 cDNA expression plasmids in mammalian cells. These results may be of general interest since alteration of RNA sequences to prevent unwanted RNA–protein interactions may increase expression of many different genes in transient transfections or after plasmid uptake in DNA vaccination approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human papillomavirus type 16 (HPV-16) genome contains an early and a late region [1]. The late region encodes the two capsid proteins L1 and L2 that are expressed exclusively in terminally differentiated keratinocytes, late in infection [2]. Expression of papillomavirus L1 and L2 from mammalian expression plasmids transfected into various mammalian cells has been very inefficient [3]. In case of HPV-16 L1, expression of L1 was achieved after insertion in cis of the HIV-1 Rev-responsive element and expression in trans of the HIV-1 Rev export protein [3]. Similarly, the presence of the simian retrovirus type 1 constitutive transport element on the L1 mRNA could induce HPV-16 L1 production [3]. These results suggested that the processing or transport of the L1 mRNA was affected by inhibitory RNA sequences within the L1 protein-coding region [4]. Indeed, hybrids between the efficiently expressed EIAV gag gene and HPV-16 L1 mapped inhibitory RNA elements to the 5′-end of L1 [5]. The presence of distinct RNA elements was proven when mutations that were introduced in the first 514 nucleotides of L1 that changed the RNA but not the protein sequence of L1 resulted in high expression of L1 mRNA and protein from CMV driven expression plasmids transfected into human cells [5]. In addition, mice immunised with a plasmid expressing the mutant L1 sequence developed neutralising antibodies and cell-mediated immune responses against HPV-16 L1, whereas mice immunised with a plasmid encoding the wild type HPV-16 L1 did not [6]. A functional analysis of the HPV-16 mutant L1 sequence revealed that splicing silencers in the L1 RNA had been destroyed by the L1 mutations [7], while splicing enhancers were intact [8].
Materials and methods
Plasmid constructions
CMV-driven expression plasmids
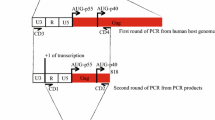
To generate plasmids pC16WT1, pC16WT2 and pC16WT3, the L1-encoding BamHI-XhoI sequences in previously described plasmids pCL1H:I, pCL1H:II and pCL1H:III [5] were replaced with a PCR fragment encoding CAT that was generated with primers CATBAM and CATXHO (Table 1). pC16WTL1 and pC16mutL1 were generated in the same way except that CAT was inserted into pC16L1 and Pc16L1MUT123 [5]. To generate plasmids pCC184-366L1 and pCC184-366L1M, HPV-16 wt or mutant L1 sequences were first PCR-amplified using the following two primer pairs: 16L1(184)S/SalI and 16L1(366)AS or 16L1M(184)S/SalI and 16L1M(366)AS. The PCR fragments were then separately ligated to a PCR fragment encoding CAT, generated with primers CATBAM and CATXHO. The ligated fragment was subsequently introduced into pCL086 [5] with SalI and BamHI, which resulted in the two in-frame L1-CAT hybrids. The constructed plasmids were all subjected to sequencing. pPLW1, pPLW2 and pPLW3 were generated by digesting the plasmids pCL1H:I, pCL1H:II and pCL1H:III [5] with BssHII and BamHI, and transferring the digested fragments into pBEL-pAEPL which was also digested with the same emzymes, respectively. To construct plasmids pC1-129WT, pC1-129mut, pC1-36WT and pC94-129WT, plasmids pT1-129WT, pT1-129mut, pT1-36WT and pT94-129WT were digested with SalI and MluI, and HPV fragments were cloned into the CAT vector described above.
T7 promoter driven plasmids
To generate plasmid pT178-366, PCR amplification was performed from pBELDPU with primers 16L1(178SalI)S and 16L1(366)AS [7]. The PCR fragment was transferred into pT7 by SalI and BamHI. To generate plasmids pT1-36WT, pT94-129WT and pT94-129mut, the following pairs of oligos were annealed: 1–36(S) and 1–36(AS), 94–129(S), and 94–129(AS) and 16L194129MUT(S) and 16L194129MUT. The fragments were separately inserted into SalI and MluI digested pT7 plasmid. pT178-226, pT1-129WT and pT1-129mut have been described previously [7, 8].
Transfections
Transfections were performed in HeLa cells according to the Fugene 6 method (Roche Molecular Biochemicals). Briefly, 1 μg of DNA was mixed with 3 μl of Fugene 6, and subsequently added in 200 μl aliquots consisting of DNA, Fugene 6 and DMEM medium to 60-mm plates containing sub-confluent HeLa cells. The transfected cells were harvested at 24 h post-transfection. The data variation in each transfection experiment was less than 20%.
RNA extraction and Northern blotting
Total and cytoplasmic RNA was prepared according to the RNeasy Mini protocol (Qiagen). Northern blot analysis was performed by the separation of 10 μg of total or cytoplasmic RNA on a 1% agarose gels containing 2.2 M formaldehyde, followed by transfer to a nitrocellulose filter and hybridisation, as described previously [5]. The L1 and CAT probes indicated in Figs. 1 and 2 were generated by digestion of pC16L1 [5] or pL1-129WTCAT [5] with BamHI and XhoI followed by gel purification and labelling of the DNA fragments. Random priming of the DNA probe was performed using a Decaprime kit (Ambion) according to the manufacturer’s instructions. All Northern blots were quantified in a Bio-Rad phosphorimager (GS-250).
Schematic representation of the HPV-16 genome. Boxes indicate the protein coding regions. Numbers refer to nucleotide positions in the HPV-16R sequence [9]. The major p97 promoter and the differentiation dependent promoter p670 are indicated. Splice sites and polyadenylation signals are shown. pAE, early polyA signal; pAL, late polyA signal; CMV, human cytomegalovirus immediate-early promoter; SD, 5′ss; SA, 3′ss. ATG is a start codon of L1coding region. The star indicates the splicing silencer in nucleotide position 178–226 of L1
(a) The structure of the pBEL-pAEPL [10] expression plasmid is shown. The early polyadenylation signal pAE has been deleted to induce detectable levels of late mRNAs [7]. The late UTR which contains RNA instability elements [11] was originally deleted to increase the chances of obtaining detectable levels of late mRNAs [7]. The L1 probe is indicated. The wild type and mutant L1 sequences inserted into the polylinker are shown. Plasmid names are on the left. (b and c) Northern blots of total RNA extracted from HeLa cells transfected with the indicated plasmids hybridised to the L1 probe (Fig. 1). Spliced mRNA as a percentage of total late RNA in each lane is indicated at the bottom of the gel. The predicted late mRNAs are displayed to the right. The data variation in each transfection experiment was less than 20%
CAT-ELISA
To monitor CAT protein levels, transfected HeLa cells were harvested as described previously. The levels of chloramphenicol acetyltransferase (CAT) protein were quantified using a CAT antigen capture enzyme-linked immunosorbent assay (ELISA; Roche Molecular Biochemicals). All CAT quantitations were normalised to the protein concentration of the cell extract, as determined by the Bradford method.
UV cross-linking and preparation of cellular extracts
In vitro synthesis of radiolabelled and unlabelled RNA was performed on linearised plasmid DNA. In vitro transcription was performed with T7-RNA polymerase in the presence of (32P)-UTP, as previously described [12]. The radiolabelled RNAs were purified by phenol–chloroform extraction and EtOH precipitation, and resuspended in water. UV cross-linking and synthesis of radiolabelled RNA was performed as previously described [12]. Around 105 cpm radiolabelled RNA was used in each UV cross-linking reaction. HeLa cell nuclear extracts were prepared according to the method of Dignam [13].
Results
Binding of the 35 kDa hnRNP A1 to HPV-16 L1 RNA correlates with inhibition of gene expression in the absence of splicing
We have previously reported that the HPV-16 L1 coding region contains sequences that strongly inhibit expression of L1 from L1 cDNAs [3–5] (Fig. 1). In contrast, a mutant L1 cDNA in which the wobbling base in the AU-rich HPV-16 genome was changed, in many cases to a G or a C to reduce the high AU-content, expressed high L1 mRNA and protein levels [5]. Care was taken not to introduce rare codons. We have also shown that the first 514 nucleotides of L1 contain splicing silencers that are inactivated in the genetically altered L1 mutant sequence [7]. To investigate the link between inhibition of L1 mRNA splicing and inhibition of L1 cDNA expression, we first compared the inhibitory effect on gene expression with the inhibitory effect on splicing by three hybrids between wt and mutant L1 sequences. The first 514 nts of L1 were divided into three regions (1, 2 and 3) (Fig. 2a). Hybrids between wt and mutant fragments 1, 2 and 3 were generated and inserted into pBEL-pAEL [7], which contains a polylinker between the L1 ATG and the BamHI site 514 nucleotides further down (Fig. 2a). Analysis of late mRNA production in cells transfected with the pBEL-pAEL-derived plasmids demonstrated that the strongest inhibitory effect on splicing was exerted by fragment 1 (Fig. 2b). Fragments 2 and 3 do not seem to inhibit splicing in this context. Fragment 1 also contains splicing enhancers that are not destroyed by the mutations introduced in L1, explaining why pPLW2 produces more spliced L1 mRNA than pPLW1 [8]. The splicing efficiency was much lower in pPL178-366 (Fig. 2c), since the first 177 nucleotides containing the enhancers had been deleted. Plasmid pPL1-520, which contained the full wild type L1 sequence, produced the lowest levels of spliced L1 mRNA of all plasmids (Fig. 2b), suggesting that multiple splicing silencers were located in the first 514 nucleotides of L1. In contrast, pPL1-520M, which contained HPV-16 L1 sequences derived from the mutant L1 gene [5] described above, produced primarily spliced mRNAs, as expected (Fig. 2b).
We wished to compare the inhibitory activity of the hybrids on splicing to the inhibitory effect on the expression of a reporter gene in the absence of splicing. The first 514 nucleotides of the five L1 sequences shown in Fig. 2a were therefore inserted in frame with the CAT reporter gene, resulting in the plasmids shown in Fig. 3a. Analysis of RNA levels produced by these plasmids in transfected HeLa cells revealed that the full wild type 514 nucleotides reduced mRNA levels to the highest extent, whereas the full mutant 514 nucleotides reduced mRNA levels to the lowest extent (Fig. 3b). One of the hybrids, pC16WT1 reduced mRNA levels to the highest extent, whereas both pC16WT2 and pC16WT3 had a smaller effect on mRNA levels (Fig. 3b). Similar results were obtained when CAT protein levels were monitored (Fig. 3c). We concluded that there was a good correlation between inhibition of CAT expression and inhibition of splicing (Figs. 2b and 3b). In addition, the 178–366 sequences that inhibited splicing very strongly in the absence of the first 177 nucleotides of L1 (Fig. 2c), also inhibited CAT mRNA and protein production when fused in frame with CAT (Fig. 4a and b). The mutant 184–366 L1 sequence did not reduce CAT mRNA and protein levels to the same extent (Fig. 4b and c), further demonstrating a correlation between inhibition of splicing and inhibition of expression of an unspliceable mRNA.
(a) Schematic representation of the HPV-16 L1-CAT in frame hybrids. Boxes indicate the protein coding regions. The CMV promoter is indicated. Plasmid names are on the left. Numbers refer to nucleotide positions in HPV-16 L1, position 1 being A in the AUG. The CAT probe is indicated. (b) Northern blots of total RNA extracted from HeLa cells transfected with the indicated plasmids hybridised to the CAT probe (Fig. 3a). The quantitation of the RNA levels in a phosphorimager is shown in the graph below. Northern blots were quantified in a Bio-Rad phosphorimager (GS-250). (c) CAT protein levels produced by the indicated plasmids in the transfected cells were monitored by a CAT-capture ELISA as described in Materials and methods. Arbitrary CAT units are shown
(a) Schematic representation of the HPV-16 L1-CAT in frame hybrids. Boxes indicate the protein coding regions. The CMV promoter is indicated. Plasmid names are on the left. Numbers refer to nucleotide positions in HPV-16 L1, position 1 being A in the ATG. An in frame ATG was inserted upstream of position 184 in L1. (b) Northern blots of total RNA extracted from HeLa cells transfected with the indicated plasmids hybridised to the CAT probe (Fig. 2a). (c) The quantitation of the CAT RNA and protein levels is shown in the graph. CAT protein levels produced by the indicated plasmids in the transfected cells were monitored by a CAT-capture ELISA. Arbitratry CAT protein units are shown. Northern blots were quantified in a Bio-Rad phosphorimager (GS-250)
We speculated that the 35 kDa hnRNP A1 protein that binds to the HPV-16 L1 splicing silencers mediated the inhibitory effect on CAT expression. To test this idea, we used UV cross-linking to identify proteins that interacted with the 178–366 sequence. HPV-16 L1 sequences under the control of the bacteriophage T7 promoter were constructed and used for in vitro synthesis of radiolabelled RNA (Fig. 5a). The results revealed that the 178–366 fragment that inhibited splicing (Fig. 2c), and CAT expression in the absence of splicing (Fig. 3b), interacted strongly with the same 35 kDa protein that binds to a previously described splicing silencer located between L1 nucleotide positions 178–226 (Fig. 5b). This protein has been identified previously as hnRNP A1.
(a) Schematic representation of the HPV-16 L1 mRNA and the HPV-16 L1-derived RNA probes used in UV cross-linking. The numbering of the sequences starts at the A in the L1 ATG. The T7 bacteriophage promoter is indicated. Plasmid names are on the left. (b–e) UV cross-linking of nuclear extract to the indicated radiolabelled RNAs. The p35 protein cross-linking specifically to HPV-16 wt L1 sequences that inhibit gene expression and inhibit splicing [10]. The arrow indicates p55, a 55 kDa protein that appears to bind to the majority of the L1 sequences
The 129WT sequence strongly inhibits CAT when fused to the CAT gene, while the mutant 1-129mut does not (Fig. 6a and b). The 35 kDa protein cross-linked to the first 129 nucleotides of HPV-16 L1 (129WT) that have been shown previously to inhibit splicing [8] (Fig. 5c], but not to a mutant variant of the first 129 nucleotides (129mut) (Fig. 5c), that does not inhibit splicing [8]. Mapping of the inhibitory sequence in the first 129 nucleotides of L1 by fusion to CAT (Fig. 6a) revealed that nucleotides 94–129 strongly inhibited CAT production (Fig. 6b), whereas nucleotides 1–36 did not (Fig. 6b). The latter probe interacted with an unidentified 55 kDa protein indicated with an arrow (Fig. 6b). UV cross-linking experiments to probes spanning wild type nucleotides 1–36 or 94–129, revealed that the 35 kDa protein cross-linked to 94-129WT (Fig. 5d), but not to 1-36WT (Fig. 5e). Neither did p35 cross-link to nucleotides 94–129 derived from the mutant L1 sequence (94–129mut) (Fig. 5d), as expected. We concluded that all sequences that inhibited CAT expression also cross-linked to p35 (hnRNP A1). Therefore, binding of hnRNP A1 to HPV-16 L1 cording region correlated with inhibition with inefficient L1 expression from HPV-16 L1 cDNAs.
(a) Schematic representation of the HPV-16 L1-CAT in frame hybrids. Boxes indicate the protein coding regions. The CMV promoter is indicated. Plasmid names are on the left. Numbers refer to nucleotide positions in HPV-16 L1, position 1 being A in the ATG. A in frame ATG was inserted upstream of position 184 in L1. (b) CAT protein levels produced by the indicated plasmids after transfection into HeLa cells. CAT protein levels were monitored by a CAT-capture ELISA as described in Materials and Methods. Arbitrary CAT protein units are shown
Discussion
Many viral genes are poorly expressed from mammalian expression plasmids. In some cases, this may be due to presence on the mRNAs of unutilised splice sites [14–16]. These splice sites are presumably interacting with the splicing machinery, but fail to splice the message. As a result, the mRNAs are retained in the nucleus and degraded. However, in other mRNAs, the presence of unutilised splice site clearly has no inhibitory effect [17]. It has also been shown that it is difficult to express a number of many retroviral genes even though the splice sites had been carefully deleted [17–20]. These mRNAs were invariably derived from alternatively spliced mRNAs and were often overlapping protein coding regions. It was proposed that these mRNAs contained regulatory RNA elements that inhibited gene expression under certain circumstances. Some of the elements have also been shown to interact with various cellular factors. For example, the inhibitory elements in the HIV-1 gag have been shown to interact with PABP [21], PSF [22], hnRNP A1 [23], and hnRNP H [24], and it has been suggested that similar inhibitory RNA elements in HIV-1 pol binds hnRNP A1 [25]. In addition, RNA elements in the HTLV long terminal repeats also inhibit gene expression [26, 27]. Also in this case, did the RNAs interact with hnRNP A1 [27]. Although the exact function of these elements have not yet been determined, it is reasonable to assume that they play important roles in the viral gene expression programmes, perhaps regulating RNA processing steps. Other RNA elements on HIV-1 mRNAs located in the close vicinity of splice sites have been shown to act as splicing silencer of suboptimal HIV-1 splice sites [28]. In some cases, these silencers interacted with hnRNP A1. In one model, cooperative binding of hnRNP A1 to multiple binding sites on the HIV-1 mRNA extended over splice sites, thereby inhibiting splicing [29]. One may speculate that the inhibitory RNA elements described above are involved in viral gene expression by regulating RNA splicing or nuclear RNA export. HIV-1 and HTLV-1 have evolved to contain the viral Rev and Rex proteins that bind to the partially spliced mRNAs and induce nuclear export in order to overcome nuclear retention [30]. Indeed, it has been shown that over-expression of hnRNP A1 enhances nuclear retention of HIV-1 mRNAs [31]. One report suggested that HIV-1 Rev and hnRNP A1 acted in concert to induce export of HIV-1 mRNAs [23], suggesting that HIV-1 Rev could overcome nuclear retention of hnRNP A1 binding mRNAs. In the experiments described here, we mapped an inhibitory RNA element to a 36-nucleotide sequence located between positions 94–129 in the HPV-16 L1 gene. This RNA is one of several splicing silencers that are located in HPV-16 L1 that bind to hnRNP A1. In contrast, a splicing enhancer that we recently mapped to nucleotides 1–17 in L1 [8] did not display inhibitory activity when fused to the CAT gene (Fig. 6b), nor did it bind to hnRNP A1 (Fig. 5e). Interestingly, Taniguchi et al. recently reported that splicing enhancers may retain mRNAs in the nucleus through a nuclear retention factor [32]. Splicing silencer and enhancer elements may both prevent further processing of immature mRNAs, perhaps in a position-dependent manner.
HPV-16 has been shown to contain regulatory RNA elements in the L1 and L2 coding regions [3–5, 33, 34]. These RNA elements inhibit expression of L1 and L2 from mammalian expression plasmids by reducing RNA and/protein production [3–5, 34]. These cis-acting elements also displayed a sequence specific inhibitory activity when placed downstream of a reporter gene, demonstrating that they acted in a position independent manner [4]. For HPV-16 L1, two ways to overcome inhibition have been demonstrated. Either, one could provide HIV-1 Rev in trans and the Rev responsive element RRE in cis [3] or one could mutationally destroy the RNA element by alteration of the wobbling base in the L1 coding region [5]. In either case was the production of HPV-16 L1 protein achieved [3, 5]. We later showed that these inhibitory elements in HPV-16 L1 were multiple splicing silencers that interacted with hnRNP A1 and acted on the upstream, late 3′ splice on HPV-16 [7, 8]. Here we show that the inhibitory activity of these splicing silencers coincides with their interactions with hnRNP A1. The HPV-16 L2 coding region also contains inhibitory RNA elements [4, 5, 34]. These elements could be destroyed by the alteration of the woobling bases [34] as we described for HPV-16 L1 [5]. In the case of HPV-16 L2, we found that the inhibitory elements coincided with a downstream polyadenylation element that was required for full polyadenylation at the early polyadenylation signal of HPV-16 [34]. The function of this element correlated with the binding of hnRNP H [34] Interestingly, hnRNP H has also been implicated in negative regulation of mRNA splicing [35, 36], suggesting that cellular factors that are involved in splicing inhibition, inhibit gene expression when they interact with mRNAs that are unable to splice.
A few investigators have “codon optimised” HIV-1 [37], BPV-1 [38] and HPV-16 [39–41] genes. Translation codons in the viral genes have been replaced by codons that are used more frequently in the human genome. As these experiments in many cases have resulted in increased levels of gene expression, it has in most cases been attributed to an increase in mRNA translation efficiency, unfortunately many times in the absence of RNA analysis [39], not to mention the complete lack of consideration of the effect of the mutations on RNA protein interactions. We predict that an analysis of the proteins binding to a wild type RNA compared to a corresponding “codon optimised” RNA will invariably show a qualitative or quantitative difference in the RNA-protein interactions. We propose that the increase in gene expression in mammalian cells observed for most if not all “codon optimised” mRNAs is a result of the destruction of protein binding sites on the mRNAs, in many cases splicing silencer factors, and to a minor extent, if any, is the result of enhanced translation of more common codons.
References
P.M. Howley in Fields Virology, eds. by B.N. Fields, D.M. Knipe, P.M. Howley (Lippincott-Raven Publishers, Philadelphia, 1996), pp. 2045–2076
M.S. Longworth, L.A. Laimins, Microbiol. Mol. Biol. Rev. 68, 362–372 (2004)
W. Tan, B.K. Felber, A.S. Zolotukhin, G.N. Pavlakis, S. Schwartz, J. Virol. 69, 5607–5620 (1995)
M. Sokolowski, W. Tan, M. Jellne, S. Schwartz, J. Virol. 72, 1504–1515 (1998)
B. Collier, D. Öberg, X. Zhao, S. Schwartz, J. Virol. 76, 2739–2752 (2002)
E. Rollman, L. Arnheim, B. Collier, D. Öberg, H. Hall, J. Klingström, J. Dillner, D.V. Pastrana, C.B. Buck, J. Hinkula, B. Wahren, S. Schwartz, Virology 322, 182–189 (2004)
X. Zhao, M. Rush, S. Schwartz, J. Virol. 78, 10888–10905 (2004)
X. Zhao, J. Fay, H. Lambkin, S. Schwartz, Virology 369, 351–363 (2007)
S.R. Billakanti, C.E. Calef, A.D. Farmer, A.L. Halpern, G.L. Myers, Human Papillomaviruses: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. (Theoretical biology and biophysics, Los Alamos National Laboratory, Los Alamos, 1996)
X. Zhao, D. Öberg, M. Rush, J. Fay, H. Lambkin, S. Schwartz, J. Virol. 79, 4270–4288 (2005)
I.M. Kennedy, J.K. Haddow, J.B. Clements, J. Virol. 65, 2093–2097 (1991)
K. Spångberg, L. Wiklund, S. Schwartz, Virology 274, 378–390 (2000)
J.D. Dignam, R.M. Lebovitz, R.G. Roeder, Nucleic Acids Res. 11, 1475–1489 (1983)
D.D. Chang, P.A. Sharp, Cell 59, 789–795 (1989)
W. Tan, M. Schalling, C. Zhao, M. Luukkonen, M. Nilsson, E.M. Fenyo, G.N. Pavlakis, S. Schwartz, J. Virol. 70, 3645–3658 (1996)
W. Zhou, R.F. Cook, S.J. Cook, S.A. Hammond, K. Rushlow, N.N. Ghabrial, S.L. Beger, R.C. Montelaro, C.J. Issel, Veterinary Micorbiol. 88, 127–151 (2002)
S. Schwartz, M. Campbell, G. Nasioulas, J. Harrison, B.K. Felber, G.N. Pavlakis, J. Virol. 66, 7176–7182 (1992)
G. Nasioulas, A.S. Zolotukhin, C. Tabernero, L. Solomin, C.P. Cunningham, G.N. Pavlakis, B.K. Felber, J. Virol. 65, 2986–2993 (1994)
A.W. Cochrane, K.S. Jones, S. Beidas, P.J. Dillon, A.M. Skalka, C.A. Rosen, J. Virol. 65, 5305–5313 (1991)
F. Maldarelli, M.A. Martin, K. Strebel, J. Virol. 65, 5732–5743 (1991)
E.N.M. Afonina, G.N. Pavlakis, J. Biol. Chem. 272, 2307–2311 (1997)
A.S. Zolotukhin, D. Michalowski, J. Bear, S.V. Smulevitch, A.M. Traish, R. Peng, J. Patton, I.N. Shatsky, B.K. Felber, Mol. Cell. Biol. 23, 6618–6630 (2003)
I. Najera, M. Krieg, J. Karn, J. Mol. Biol. 285, 1951–1964 (1999)
M. Caputi, A.M. Zahler, J. Biol. Chem. 276, 43850–43859 (2001)
A.C. Black, J. Luo, S. Chun, A. Bakker, J.K. Fraser, J.D. Rosenblatt, Virus Genes 12, 275–285 (1996)
M. Seiki, A. Hikikoshi, M. Yoshida, Virology 176, 81–86 (1990)
A.C. Black, J. Luo, C. Watanabe, S. Chun, A. Bakker, J.K. Fraser, J.P. Morgan, J.D. Rosenblatt, J. Virol. 69, 6852–6858 (1995)
T.O. Tange, C.K. Damgaard, S. Guth, J. Valcarel, J. Kjems, EMBO J 20, 5748–5758 (2001)
C.K. Damgaard, T.O. Tange, J. Kjems, RNA 8, 1401–1415 (2002)
V.W. Pollard, M.H. Malim, Annu. Rev. Microbiol. 52, 491–532 (1998)
K. Asai, C. Platt, A. Cochrane, Virology 314, 229–242 (2003)
I. Taniguchi, K. Masuyama, M. Ohno, Proc. Natl. Acad. Sci. USA 104, 13684–13689 (2007)
S. Schwartz, X. Zhao, D. Öberg, M. Rush, Recent Res. Develop. Virol. 6, 29–45 (2004)
D. Oberg, J. Fay, H. Lambkin, S. Schwartz, J. Virol. 79, 9254–9269 (2005)
S. Jacquenet, A. Méreau, P.S. Bilodeau, L. Damier, C.M. Stoltzfus, C. Branlant, J. Biol. Chem. 276, 40464–40475 (2001)
C.D. Chen, R. Kobayashi, D.M. Helfman, Genes Dev 13, 593–606 (1999)
L. Vinner, H.V. Nielsen, K. Bryder, S. Corbet, C. Nielsen, A. Fomsgaard, Vaccine 17, 2166–2175 (1999)
J. Zhou, W.J. Liu, S.W. Peng, X.Y. Sun, I. Frazer, J. Virol. 73, 4972–4982 (1999)
C. Leder, J.A. Kleinschmidt, C. Wiethe, M. Müller, J. Virol. 75, 9201–9209 (2001)
W.J. Liu, F. Gao, K.N. Zhao, W. Zhao, G.J. Fernandp, R. Thomas, I.H. Frazer, Virology 301, 43–52 (2002)
G.L. Disbrow, I. Sunitha, C.C. Baker, J. Hanover, R. Schlegel, Virology 311, 105–114 (2003)
Acknowledgements
We thank Margaret Rush, Daniel Öberg and members of Göran Magnussons, Catharina Svenssons and Göran Akusjärvis labs for discussions. We are also grateful to Anna Grynfeld for contributing to some experiments. Research supported by the Swedish Cancer Society, by the Swedish Research Council-Medicine and by Linneus Support from the Swedish Research Council to Uppsala RNA Research Center.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, X., Schwartz, S. Inhibition of HPV-16 L1 expression from L1 cDNAs correlates with the presence of hnRNP A1 binding sites in the L1 coding region. Virus Genes 36, 45–53 (2008). https://doi.org/10.1007/s11262-007-0174-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-007-0174-0