Abstract
The Genome segment 7 (S7) of the 11 double stranded RNA genomes from Antheraea mylitta cypovirus (AmCPV) was converted to cDNA, cloned and sequenced. The nucleotide sequence showed that segment 7 consisted of 1789 nucleotides with an ORF of 530 amino acids and could encode a protein of approximately 61 kDa, termed P61. The 5′ terminal sequence, AGTAAT and the 3′ terminal sequence, AGAGC of the plus strand was found to be the same as genome segment 10 of AmCPV encoding polyhedrin. No sequence similarity was found by searching nucleic acid and protein sequence databases using BLAST. The secondary structure prediction showed the presence of 17 α-helices, 18 extended β-sheets along the entire length of P61. The ORF of segment 7 was expressed in E. coli as His-tagged fusion protein, purified through Ni-NTA chromatography, and polyclonal antibody was raised in rabbit indicating that P61 is immunogenic. Immunoblot analysis using this antibody on viral infected cells as well as purified polyhedra showed that P61 is a viral structural protein. Motif scan search showed some similarity of P61 with Inosine monophosphate dehydrogenase (IMPDH) cystathionine-β-synthase (CBS) domain at the C-terminus and it was hypothesized that by binding to single stranded viral RNA through its CBS domain P61 may help in virus replication or transcription.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cypoviruses (CPVs) belong to the genus Cypovirus in the family Reoviridae [1, 2]. These viruses infect midgut cells of a wide range of insects belonging to the order Diptera, Hymenoptera and Lepidoptera [3–7]. Viral infection is characterized by the production of large numbers of occlusion bodies called polyhedra in the cytoplasm of infected cells. The genomes of CPVs like those of other members of the Reoviridae are usually composed of 10 double stranded (ds) RNA segments (S1–S10) [1] although, in some cases, a small, eleventh segment (S11) has been reported [8]. Each dsRNA is composed of a plus-stranded mRNA and its complementary minus strand in an end to end base paired configuration except for a protruding 5′ cap on the plus strand. On the basis of electrophoretic migration patterns of the dsRNA segments in agarose or acrylamide gels, they have been classified into 14 different types [1, 5, 9]. Among the family Reoviridae, complete sequences of dsRNA genomes have been reported for members of the genera Orthoreovirus, Rotavirus, Orbivirus, Phytoreovirus, Coltivirus, Seadornavirus, Dinovernavirus, and putative members of Fijivirus and Cypovirus, [10–17].
From the Cypovirus, complete nucleotide sequence of type 14 Lymnatria dispar CPV (LdCPV-14) [18], type 1 Dendrolimus punctatus CPV (DpCPV-1) [19, 20], type 15 Trichoplusia ni CPV (TnCPV-15) [21], and type 1 Bombyx mori CPV (BmCPV-1) [22–27] have been reported and deposited in the Genbank. The elucidation of these cypovirus sequences has led to a better understanding of the possible functions that each dsRNA segment may play a role in viral pathogenesis. In case of BmCPV, segment 1, 3, 4, 6, 7, 10 encodes structural proteins VP1, VP2, VP3, VP4, VP5, polyhedrin, and segment 2, 5, 8, 9 encodes viral non-structural proteins RNA dependent RNA polymerase, p101, p44, NS5, respectively [22–27]. Segments 1, 3, 4, 6, 7, and 10 of DpCPV encode viral structural proteins while segments 2, 5, 8, 9 and 10 encode non-structural proteins [19, 20]. Besides these, polyhedrin genes of several other cypoviruses such as Euxoa scandens CPV (EsCPV) [28], Orgyia pseudotsugata CPV (OpCPV), Heliothis armigera CPV (HaCPV) [29], Choristoneura fumiferana CPV (CfCPV) [30], and Uranotaenia sapphirina CPV (UsCPV) [31] have been molecularly characterized but no sequence homology has been found among them.
The Indian non-mulberry saturniidae silkworm, Antheraea mylitta, produces an exotic variety of silk called tasar silk. Being wild in nature, a major population of these silkworms is getting destroyed by viral infection [32] and we have reported earlier that a type IV CPV called AmCPV containing 11 dsRNA segments is the causative agent [33]. AmCPV belongs to genus Cypovirus of the family Reoviridae [33]. Molecular cloning and characterization of AmCPV genome segment 9 and 10 of this CPV show that segment 9 codes for viral non-structural protein NSP38 having RNA binding property [34], and segment 10 codes for polyhedrin [35], respectively but no sequence similarity has been found with any other sequences in the public databases. To understand the role of each genome segment of AmCPV in virus replication and pathogenesis, complete characterization of all its genome segments is necessary but the structure and functions of no other genome segments except 9 and 10 have been reported. Here we report molecular cloning, sequencing and characterization of genome segment 7 of AmCPV, and show by immunoblot analysis that it encodes a 61 kDa viral structural protein.
Materials and methods
Silkworm and virus
CPV-infected Indian non-mulberry silkworm A. mylitta was collected from the tasar silk farms of West Bengal and Jharkhand states of India.
Purification of polyhedral bodies, isolation of total genomic RNA, and extraction of genome segment 7 RNA
Polyhedra were purified from the midguts of infected silkworm larvae by sucrose density gradient centrifugation according to the method of Hayashi and Bird [36] with some modification [33]. Genomic RNA was extracted from the purified polyhedra by the standard guanidinium isothiocyanate method [37] and fractionated in 1% agarose gel. The genome segment 7 was excised from ethidium bromide-stained gel and eluted using Qiaquick gel extraction kit (Qiagen).
Molecular cloning and sequencing of genome segment 7 RNA
Segment 7 genomic RNA of AmCPV was converted to cDNA as described by a sequence independent RT method [38] by using two primers (AG1 and AG2). The 3′-end of 5′-phosphorylated primer, AG1, (5′ PO4-CCCGGATCCGTCGACGAATTCTTT-NH2 3′) was blocked by NH2 to prevent its concatenation in subsequent dsRNA/DNA ligation reactions. Approximately 200 ng of purified segment 7 RNA was taken and primer AG1 was ligated to both 3′ends of dsRNA by using T4 RNA ligase for 1 h at 37°C. The tailed RNA was denatured by heating, annealed to primer AG2 (5′AAAGAATTCGTCGACGGATCCGGG 3′), which is complementary to AG1, and reverse transcribed at 55°C for 50 min by using Thermoscript reverse transcriptase (Invitrogen). The template RNA from RNA/cDNA hybrid was removed by digestion with RNaseH, and reannealing of the cDNA strands was done by incubating at 65°C for 2 h. The cDNA ends were repaired by incubation with Taq DNA polymerase (Bioline) at 72°C for 20 min and cloned into pCR2.1-TOPO vector (Invitrogen) to make plasmid pCR2.1 TOPO/AmCPV7. After transforming in E. coli TOP 10 cells, plasmids were isolated and characterized by EcoRI digestion. Recombinant plasmids containing proper size insert were then sequenced using Bigdye Terminator (ABI) in an ABI 3100 automated DNA sequencer with M13 forward and reverse primers as well as internal primers designed from deduced sequences.
Sequence analysis
Genome Sequence of AmCPV segment 7 was analyzed by Sequencher program and homology searches were done using BLAST [39]. Conserved motifs were identified using MotifScan program (http://myhits.isb-sib.ch/cgi-bin/motif_scan). The molecular weight of the deduced protein, isoelectric point, and composition were calculated using protein calculator program (http://www.scripps.edu/∼cdputnam/protcalc. html) [40]. Secondary structure was predicted using PHD program [41] and GOR4 [42].
Northern hybridization
In order to verify cloning of the segment 7 cDNA from the corresponding RNA of AmCPV, all of the genomic dsRNA segments were separated in a 10% polyacrylamide gel and observed by staining with ethidium bromide. The RNA segments in the gel were then denatured by brief treatment with 0.1 M NaOH, neutralized in TAE buffer and electroblotted onto nitrocellulose membrane. The membrane was then hybridized with 32P-labelled cloned segment 7 cDNA from AmCPV, washed and autoradiographed [34, 43].
Expression of AmCPV segment 7 in E. coli
The entire protein-coding region (530 amino acids) of AmCPV segment 7 cDNA, from nucleotide 18 to 1610, was amplified by PCR from plasmid pCR2.1 TOPO/AmCPV7 by using Accuzyme DNA polymerase (Bioline) and two synthetic primers, AGCPV54F (5′ AGTAATCGGGATCCGAAATG 3′; forward primer) and AGCPV62R (5′ TGTAGTCGTCGACCTTCATTA 3′; reverse primer), complementary to bases 1–20 and bases 1600–1620, respectively, and containing BamHI (in the forward primer) and SalI (in the reverse primer) restriction enzyme sites (underlined). The amplified PCR product was digested with BamHI and SalI, separated on a 1% agarose gel and purified from the gel by using a gel extraction kit (Qiagen). Purified DNA was ligated to BamHI/SalI-digested pQE-30 vector (Qiagen) in-frame with a sequence encoding six histidine residues at the N-terminus. The resulting recombinant plasmid, pQE-30/AmCPV7, was then transformed into E. coli M15 and colonies were screened following BamHI and SalI digestion.
For protein expression, recombinant bacteria was grown in 5 ml LB medium containing ampicillin (100 μg/ml) and kanamycin (25 μg/ml) for 3 h at 37°C and then induced with 1 mM IPTG for an additional 4 h at the same temperature. Bacteria were harvested by centrifugation, lysed by boiling with sample loading buffer (60 mM Tris–HCl, pH 6–8; 10% glycerol ; 2% SDS; 5% β-mercaptoethanol and 1 μg/ml bromophenol blue) for 3 min and then loaded onto a 5% stacking gel cast above a 10% resolving SDS-polyacrylamide gel. After electrophoresis, the protein bands in the gel were stained with Coomassie brilliant blue [37]. The molecular mass of the expressed recombinant protein was determined by comparison with standard protein molecular mass markers and by using Quantity One software in Gel-Doc 2000 (Bio-Rad).
Purification of His-tagged protein
Recombinant bacteria containing pQE-30/AmCPV7 were grown in 1 l LB medium and induced with IPTG as described above. The insoluble His-tagged fusion protein (P61) was first purified as inclusion bodies [44]. After solubilizing the inclusion bodies in 6 M guanidine hydrochloride, further purification of protein was carried out using a Ni-NTA agarose kit (Qiagen) according to the manufacturer’s protocol. The total amount of purified protein was quantitated by Bradford method [45] using BSA as the standard and purity was checked by SDS-10% PAGE [46].
Rabbit immunization and production of polyclonal antibodies
One rabbit was immunized with bacterially expressed, purified, recombinant His-tagged protein (P61) by standard methods [34, 47]. In brief, purified protein (550 mg) was mixed with Freund’s complete adjuvant, and injected subcutaneously at multiple sites. Three booster doses with Freund’s incomplete adjuvant and the same amount of protein were administered via the same route at 4-week intervals. Twelve days after the final booster, blood was collected, serum prepared and the antibody titer was determined by ELISA [47]. Specific antibody was purified by antigen (p61-Sepharose) affinity chromatography [47, 48].
Western blot analysis
For detecting the expression of P61 in infected cells, protein samples from dissected midgut of AmCPV infected and uninfected 5th instar larvae were prepared by homogenizing the tissue in PBS followed by centrifugation at 10,000g for 10 min [34]. Each protein sample in the supernatant was boiled in sample loading buffer and run on SDS-10% PAGE under reducing conditions. After electrophoresis, proteins from the gel were transferred onto a duralose membrane (Stratagene) using a transblot cell (Bio-Rad) according to manufacturer’s protocol. The membrane was then blocked in PBS containing 3% BSA for 1 h at room temperature followed by incubation with 500-fold diluted affinity purified anti-p61 polyclonal antibody for 1 h. After washing with PBS, the membrane was incubated with 200-fold diluted Protein A-conjugated horse raddish peroxidase for 1 h and then washed, and color development was done by using HPO color development kit (Bio-Rad).
Nucleotide sequence accession number
The nucleotide sequence data reported here for segment 7 of AmCPV have been deposited in Genbank database and have been assigned the Accession No: DQ512389.
Results
Cloning of AmCPV genome segment 7
AmCPV genome segment 7 was purified from the genomic RNA of AmCPV after separating by 1% agarose gel electrophoresis, converted to cDNA and cloned into pCR 2.1 TOPO vector to create plasmid pCR 2.1 TOPO/AmCPV7. Digestion of isolated plasmids from transformed E. coli with EcoRI and their analysis by agarose gel electrophoresis showed an insert of 1.8 kb (data not shown) indicating cloning of full length segment 7 cDNA.
Northern analysis
Among the 11 dsRNA segments present in AmCPV the cDNA of segment 7 specifically hybridized with segment 7 genomic RNA, confirming the cloning of segment 7 genomic RNA from AmCPV (Fig. 1).
Northern blot analysis of AmCPV genome RNA. AmCPV genome segments were resolved by 10% PAGE as discrete bands (Lane 1) (segment 11 was not visible as it ran out off the gel under these running conditions) and hybridized with 32P-labelled cloned segment 7 cDNA (Lane 2). Arrow indicates hybridization of segment 7 cDNA to segment 7 genomic RNA. The number and sizes of different RNA segments of AmCPV are shown in the left
Molecular analysis of genome segment 7
Sequencing of AmCPV segment 7 showed that it consisted of 1789 nucleotides with a long single ORF of 530 amino acids. The ORF started with an ATG codon at nucleotide 18 and terminated in a TAG codon at nucleotide 1608 (Fig. 2a). Seventeen nucleotides upstream of the start codon and 179 nucleotides downstream of stop codon were present as untranslated sequences. The molecular weight of the encoded protein was deduced as 61 kDa and we termed the protein as P61. The 5′ terminal sequences AGTAAT, and 3′ terminal sequence AGAGC were found to be same as the 5′ and 3′ terminal sequences of polyhedrin gene encoded by AmCPV genome segment 10. No significant homology was detected with any nucleotide or protein sequences available in the public databases using BLAST. The deduced amino acid composition resulted in an isoelectric point of 6.07 and showed that P61 is rich in isoleucine (8.5%), leucine (8.1%), aspartate (7.9%), lysine (7.7%), threonine (7.5%) and glutamate (6.7%) residues with an isoelectric point of 6.07. Four potential N-linked glycosylation sites (at positions 68–70, 124–126, 175–177 and 309–311) and several phosphorylation sites were found within the protein coding region.
(a) Complete nucleotide (nt) and deduced amino acid (aa) sequences of AmCPV genome segment 7. The CBS motif found between 440 and 497 amino acid residues is represented by  below the amino acid sequences. The initiation and termination codons are shown in bold. Four potential N-linked glycosylation sites are double underlined. (b) Secondary structure prediction was represented above the amino acid residues. The cylinders (
below the amino acid sequences. The initiation and termination codons are shown in bold. Four potential N-linked glycosylation sites are double underlined. (b) Secondary structure prediction was represented above the amino acid residues. The cylinders ( ) represent helix, arrows (
) represent helix, arrows ( ) represent the beta strands and the joining lines (―) represent random coil
) represent the beta strands and the joining lines (―) represent random coil
Secondary structure prediction using PHD and GOR4 programs showed that 45.8% of the residues are likely to form random coils, 34.2% would form α-helices and 20% would form extended sheets. A total of 17 α-helices and 18 extended β-sheets were found along the entire length of protein (Fig. 2b). MotifScan search showed significant similarity of P61 with Inosine monophosphate dehydrogenase (IMPDH) enzyme from 201 to 526 amino acid residues containing one Cystathionine Beta Synthase (CBS) domain at C-terminus (440–497 amino acid residues) with the characteristic of sheet/helix/sheet/sheet/helix topology (Fig. 3).
Alignment of CBS domains found in AmCPV segment 7 with that of enzymes, channel and few hypothetical proteins. The sequences are annotated with their SWISS-PROT identifier followed by the start point and end point of each domain. AmCPV_S7/440–497 denotes CBS domain found in AmCPV segment 7, from 440 to 497. IMDH_ARATH, IMDH_METKA, IMDH_DROME, IMD2_MESAU, IMH3_YEAST and IMH4_YEAST are inosine monophosphate dehydrogenases from Arabidopsis thaliana, Methanopyrus kandleri, Drosophila melanogaster, Mesocricetus aurateus and Saccharomyces cervisiceae, respectively. YR33_THEPE is a hypothetical protein from Thermophilum pendens. YTFL_HAEIN is a hypothetical protein from Haemophilus influenza.CLC3_HUMAN is a chloride channel protein from Homo sapiens. AAKG_RAT is a 5′ AMP activated protein kinase from Rattus norvegicus. KSF1_ECOLI is a polysialic acid capsule expression protein from E. coli. CBS_RAT is a cystathionine beta synthase H-450 protein from Rattus norvegicus. ACUB_BACSU is acetoin utilization protein B from Bascillus subtilis. The shaded boxes indicate the conserved residues in the CBS domains from different species
Expression and purification of genome segment 7 encoded protein
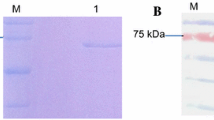
To express segment 7 encoded protein in E. coli, the ORF was cloned in pQE30 vector, expressed in E. coli and purified as 6X His-tag fusion protein through Ni-NTA affinity chromatography. Analysis of induced bacterial lysate and purified protein through SDS 10%-PAGE showed the production of ∼63 kDa protein (Fig. 4, lanes 2 and 3) but, no such protein band was found in uninduced bacterial lysate (Fig. 4, lane 1).
Detection of P61 expression in virions and viral infected cells
To demonstrate the expression of P61 in virions and virus infected cells, CPV infected A. mylitta midgut cells and purified polyhedral bodies were analyzed through SDS-10% PAGE and subjected to immunoblot analysis using affinity purified anti-P61 polyclonal antibodies. A major immunoreactive protein band of approximately 65 kDa was found in lane containing lysate of infected midgut cell (Fig. 5, lane 2) and polyhedral bodies (Fig. 5, lane 3), but not in lane containing lysate of uninfected cell (Fig. 5, lane 1)
SDS-PAGE and Western blot analysis of P61. Samples were separated by SDS-10% PAGE and stained with Coomassie brilliant blue (a) or transblotted onto nitrocellulose membrane and reacted with anti- P61 antibody (b) lane M, Molecular weight markers; lane 1, Uninfected and lane 2, CPV-infected midgut cell lysate; lane 3, Purified AmCPV polyhedra. Arrow indicates the position of immunoreactive protein
Discussion
AmCPV although infects and destroys an economically important insect species, Antheraea mylitta, which produces tasar silk, but except segments 9 and 10, none of its other genome segments have been fully characterized at the molecular level. But full molecular characterization of this CPV is very much needed to understand the viral life cycle, pathogenesis and to find target sites for the development of effective anti-viral compound. Here we describe molecular analysis of another of its genome segment, Segment 7, by cloning, sequencing and expressing in E. coli. The 1789 nucleotides long cloned cDNA consists of an ORF of 530 amino acids and its molecular mass is calculated to be approximately 61 kDa. Presence of conserved terminal sequences at 5′ and 3′ ends is a characteristic feature found in different genome segments of most of the virions of the Reoviridae family [49]. At the 5′ and 3′ end of AmCPV segment 7, AGTAAT and AGAGC sequences were found as observed at the 5′ and 3′ ends of AmCPV genome segment 10 encoding polyhedrin indicating the genome structure of this CPV may follow the same pattern as observed in other CPVs, but cloning and sequencing of other genome segments of AmCPV are required to confirm it. Absence of any significant sequence similarity of S7 either at nucleotide or amino acid level with the available database indicates that segment 7 of AmCPV encodes a novel protein and further confirms that a new type of CPV infects tasar silk worm, A. mylitta which has been reported earlier [33–35].
Cloning of S7 ORF in bacterial expression vector and analysis of bacterially expressed recombinant His-tagged fusion protein showed the production of a protein of 63 kDa, the molecular weight of which is ∼2 kDa higher than the calculated molecular weight of 61 kDa. This reduction in the mobility of the protein might be due to the presence of high proportions of hydrophobic and acidic amino acids residues such as isoleucine (8.5%), leucine (8.1%), aspartic acid (7.9%), and glutamic acid (6.7%) residues. Anomalous electrophoretic behavior of several proteins such as pig heart calpastatin have been found in SDS-PAGE, which are rich in hydrophilic and acidic amino acid residues [50].
Four potential N-linked glycosylation sites have been found in ORF of P61. When expression of this protein was analyzed in AmCPV infected midgut cells or in purified polyhedra by immunoblot analysis using anti-P61 polyclonal antibodies, an immunoreactive band of approximately 65 kDa was observed. This indicates that the protein has undergone glycosylation during expression in eukaryotic cells utilizing some of the potential glycosylation sites found in segment 7. Production of high titer antibodies (10−5) in rabbit as shown by ELISA (data not shown) and western blot indicate that P61 is highly antigenic to induce strong immune response. The antibodies generated in this study could aid in the development of a rapid and simple diagnostic test to detect AmCPV. Further work is required to demonstrate if this antibody possesses any neutralizing activity to this CPV. Detection of P61 protein expression in virus infected midgut cells and in polyhedra by Immunoblot analysis using anti-P61 antibody indicate that P61 codes for a viral structural protein.
Since a significant similarity of P61 was found with IMPDH enzyme containing one CBS domain by motif scan analysis it may be assumed that P61 may possess IMPDH like activity. It has been shown that IMPDH of human, T. foetus and E. coli can bind to single stranded nucleic acids with nanomolar affinity through its CBS domains (not by its catalytic domain) [51]. CBS domains are also found in chloride channels, ABC transporters and protein kinases where their function is unknown [52] but CBS domain mutations have been implicated in a wide array of inherited diseases including myotonia [53] and familial hypertropic cardiomyopathy [54]. Perhaps nucleic acid binding is a general property of CBS domains and P61 encoded by AmCPV segment 7, by binding to single stranded viral RNA through its CBS domain may help in viral replication or transcription.
References
C.C. Payne, P.P.C. Mertens, in Cytoplasmic Polyhedrosis Viruses. The Reoviridae, ed. by W.K. Joklik (Plenum, New York, 1983), pp. 425–504
M.H.V. van Regenmortel, C.M. Fauquet, D.H.L. Bishop, E.B. Carstens, M.K Estes, S.M. Lemon, J. Maniloff, M.A. Mayo, D.J. McGeoch, C.R. Pringle, R.B. Wickner (eds.), Virus Taxonomy, Seventh report of the International Committee on Taxonomy of Viruses (Academic Press, San Diego, 2000)
S. Belloncik in Cytoplasmic Polyhedrosis Viruses - Reoviridae. Advances in Virus Research, vol 36, ed. by K. Maramorsch, F.A. Murphy, A.J. Shatkin (Academic Press, New York, 1989), pp. 173–209.
S. Belloncik, K. Mori, in Cypoviruses, The insect viruses, ed. by L.K. Miller, L.A. Ball (Plenum, New York, 1998), pp. 337–369
M. Fouillaud, G. Morel, J. Invertebr. Pathol. 64, 89–95 (1994)
T. Hukuhara, J.R. Bonami, in Atlas of Invertebrates, ed. by J.R. Adams, J.R. Bonami (CRC Press, Boca Raton, Florida, 1991), pp. 394–430
C.C. Payne in: Pathogenesis of Invertebrate Microbial Diseases, E.W. Davidson, (Eds.). Cytoplasmic polyhedrosis viruses. Allanheld, Osmun, Totowa, New Jersey, (1981), pp. 61–100
M. Arella, C. Lavallee, S. Belloncik, Y. Furuichi, J. Virol. 62, 211–217 (1988)
S. Belloncik, J. Liu, D. Su, M. Arella, J. Invertebr. Pathol. 67, 41–47 (1996)
R. Duncan, Virology 260, 316–328 (1999)
M.K. Estes, J. Cohen, Microbiol. Rev. 53, 410–449 (1989)
P. Roy, J.J. Marshall, T.J. French, Curr. Top. Microbiol. Immunol. 162, 43–87 (1990)
N. Suzuki, Semin. Virol. 6, 89–95 (1995)
H. Attoui, F. Billoir, P. Biagini, J.F. Cantaloube, de R. Chesse, de P. Micco, de X. Lamballerie, Biochem. Biophys. Res. Commun. 273, 1121–1125 (2000)
H. Attoui, F. Billoir, P. Biagini, de P. Micco, de X. Lamballerie, J. Gen. Virol. 81, 1507–1515 (2000)
H. Attoui, F.M. Jaafar, M. Belhouchet, P. Biagini, J.-F.Cantaloube, de P. Micco, de X. Lamballerie, Virology 343, 212–223 (2005)
N. Nakashima, M. Koizumi, H. Watanabe, H. Noda, J. Gen. Virol. 77, 139–146 (1996)
S. Rao, M. Shapiro, D. Lynn, K. Hagiwara, R. Dean, G.R. Carner, GenBank Accession Nos. NC_003006 - NC_003015 (2001)
S.L. Zhao, C.Y. Liang, J.J. Hong, H.G. Xu, H.Y. Peng, Virus Res. 94, 17–23 (2003)
S.L. Zhao, C.Y. Liang, J.J. Hong, H.Y. Peng, Arch. Virol. 148, 1357–1368 (2003)
S. Rao, G. Carner, S. Scott, D. Heckel, GenBank Accession Nos. NC_002557-NC_002559, NC_002567, NC_002560-NC_002566 (2000)
K. Hagiwara, M. Tomita, K. Nakai, J. Kobayashi, S. Miyajima, T. Yoshimura, J. Virol. 72, 5762–5768 (1998)
K. Hagiwara, M. Tomita, J. Kobayashi, S. Miyajima, T. Yoshimura, Biochem. Biophys. Res. Commun. 247, 549–553 (1998)
K. Hagiwara, T. Matsumoto, J. Gen. Virol. 81, 1143–1147 (2000)
K. Hagiwara, J. Kobayashi, M. Tomito, T. Yoshimura, Arch. Virol. 146, 181–187 (2001)
K. Hagiwara, S. Rao, W.S. Scott, R.G. Carner, J. Gen. Virol. 83, 1477–1482 (2002)
K. Ikeda, H. Nakazawa, R. Alain, S. Belloncik, H. Mori, Arch. Virol. 143(2), 241–248 (1998)
F. Fossiez, S. Belloncik, M. Arella, Virology 169, 462–465 (1989)
M.S. Galinski, Y. Yu, B.R. Heminway, G.S. Beaudreau, J. Gen. Virol. 75, 1969–1974 (1994)
F. Echeverry, J. Bergeron, W. Kaupp, C. Guertin, M. Arella, Gene 198, 399–406 (1997)
A. Shapiro, T. Green, S. Rao, S. White, G. Carner, P.P. Mertens, J.J. Becnel, J. Virol. 79(15), 9430–9438 (2005)
M.S. Jolly, S.K. Sen, M.M. Ahsan, Tasar Culture. (Ambika publishers, Bombay, 1974)
K.R. Qanungo, S.C. Kundu, A.K. Ghosh, Acta Virol. 44, 349–357 (2000)
K.R. Qanungo, S.C. Kundu, J.I. Mullins, A.K. Ghosh, J. Gen. Virol. 83, 1483–1491 (2002)
Sinha-U. Datta, C.V.R. Murthy., A.K. Ghosh, Biochem. Biophys. Res. Commun. 332, 710–718 (2005)
Y. Hayashi, F.T. Bird, Can. J. Microbiol. 6, 695–701 (1970)
F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, K. Struhl (eds.), Current Protocols in Molecular Biology. (Wiley, New York, 1995)
P.R. Lambden, S.J. Cooke, E.O. Caul, I.N. Clarke, J. Virol. 66(3), 1817–1822 (1992)
S.F. Altschul, T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller, D.J. Lipman, Nucleic Acids Res. 25, 3389–3402 (1997)
P. Zhang, N. Briones, C. Liu, C.K. Brush, T. Powdrill, Y. Belosludtsev, M. Hogan, Nucleosides, Nucleotides Nucleic Acids 20(4–7), 1251–1254 (2001)
B. Rost, C. Sander, Proteins 19, 55–72 (1994)
J. Garnier, J.-F. Gibrat, B. Robson, Meth. Enzymol. 266, 540–553 (1996)
A.P. Feinberg, B. Vogelstein, Anal. Biochem. 132, 6–13 (1983)
J.E. Caligan, B.M. Dunn, H.L. Ploegh, D.W. Speicher, P.T. Wingfield, Current Protocols in Protein Science. (Wiley, New York, 1995)
M.M. Bradford, Anal. Biochem. 72, 248–254 (1976)
U.K. Laemmli, Nature 227, 680–685 (1970)
E. Harlow D. Lane, Using Antibodies: A Laboratory Manual. (Cold Spring Harbor Laboratory. Cold Spring Harbor, New York, 1988)
J. Sambrook E.F. Fritsch T. Maniatis, Molecular Cloning: A Laboratory Manual, 2nd edn. (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1989)
J.T. Patton, E. Spencer, Virology 277(2), 217–225 (2000)
M. Maki, E. Takano, T. Osawa, T. Ooi, T. Murachi, M. Hatanaka, J. Biol. Chem. 263(21), 10254–10261 (1988)
J.E. McLean, N. Hamaguchi, P. Belenky, S.E. Mortimer, M. Stanton, L. Hedstrom, Biochem. J. 379, 243–251 (2004)
A. Bateman, Trends Biochem. Sci. 22, 12–13 (1997)
C. Ponting, J. Mol. Med. 75, 160–163 (1997)
E. Blair, C. Redwood, H. Ashrafian, M. Oliveira, J. Broxholme, B. Kerr, A. Salmon, I. Ostman-Smith, H. Watkins, Hum. Mol. Genet. 10, 1215–1220 (2001)
Acknowledgments
The authors thank the Director(s) of Central Tasar Research and Tranining Institute, Ranchi and Jhargram for providing the virus infected A. mylitta larvae. Venkata Ramana Murthy Chavali is the recipient of fellowship from Indian Institute of Technology, Kharagpur and Council of Scientific and Industrial Research, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chavali, V.R.M., Ghosh, A.K. Molecular cloning, sequence analysis and expression of genome segment 7 (S7) of Antheraea mylitta cypovirus (AmCPV) that encodes a viral structural protein. Virus Genes 35, 433–441 (2007). https://doi.org/10.1007/s11262-006-0070-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-006-0070-z