Abstract
We evaluated the predicted biochemical properties of Gag proteins from a diverse group of feline immunodeficiency viruses (FIV) to determine how different evolutionary histories of virus and host have changed or constrained these important structural proteins. Our data are based on FIV sequences derived from domestic cat (FIVfca), cougar (FIVpco), and lions (FIVple). Analyses consisted of determining the selective forces acting at each position in the protein and the comparing predictions for secondary structure, charge, hydrophobicity and flexibility for matrix, capsid and nucleocapsid, and the C-terminal peptide, which comprise the Gag proteins. We demonstrate that differences among the FIV Gag proteins have largely arisen by neutral evolution, although many neutrally evolving regions have maintained biochemical features. Regions with predicted differences in biochemical features appear to involve intramolecular interactions and structural elements that undergo conformational changes during particle maturation. In contrast, the majority of sites involved in intermolecular contacts on the protein surface are constrained by purifying selection. There is also conservation of sites that interact with host proteins associated with cellular trafficking and particle budding. NC is the only protein with evidence of positive selection, two of which occur in the N-terminal region responsible for RNA binding and interaction with host proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retroviruses genomes vary in length and genome organization but all minimally contain three open reading frames. The structural proteins required to form the viral particle are encoded by gag at the 5′ end of the genome, enzymes required for virus replication are encoded by pol in the central portion of the genome, and the viral envelope proteins, which determine cell tropism, are encoded by env at the genome 3′ end. Members of the Retroviridae differ in the enzymes encoded by pol and there is extensive nucleotide sequence diversity and, consequently, receptor specificity associated with env. However, based on three-dimensional structures of Gag proteins [1–4], the morphological characteristics of the viral particle formed from gag gene products are similar across this diverse viral family [5, reviewed in 6]. Because Gag orchestrates many steps in the virus life cycle and encoded proteins elicit an adaptive immune response, the viral gag gene products are of interest from both a structural and therapeutic perspective.
The Gag polyprotein is produced from the gag gene on unspliced full-length viral transcripts. The intact polyprotein functions during the late stages of viral replication to direct virus assembly and budding. There are three protein domains in Gag that function as components of the intact polyprotein during particle assembly and as independent proteins during maturation of the viral particle. The N-terminal portion contains the matrix protein (MA), which localizes the polypeptide to the membrane via a myristoylated residue and a patch of basic residues [7–9] and is responsible for incorporation of the envelope glycoprotein during particle assembly [10–12]. Capsid (CA) is the central protein in Gag and is composed of a C-terminal domain, which initiates Gag dimerization, and an N-terminal domain, which forms a lattice of hexamers that yields the morphology characteristic of each retrovirus core [13]. The nucleocapsid (NC) is the C-terminal protein of Gag. The ability of this basic protein to bind RNA confers several important functions on Gag including packaging of the genome [14], initiating Gag dimerization [15, 16] and compaction of the viral genome [reviewed in 17]. Cellular proteins that can confer antiviral activity interact with the basic domains of NC [18, 19]. The C-terminal peptide of Gag is highly variable within and between retrovirus groups. Based on several conserved motifs, the principal function of this region appears to be its interaction with cellular factors. In the primate lentiviruses, this region yields a peptide, p6, that facilitates budding and release of the virion via the cellular endocytic sorting pathway [20, 21].
Maturation of the newly formed virion requires proteolytic cleavage of the Gag polyprotein by the virus encoded protease and is an ordered process [22]. The viral nucleocapsid is cleaved first to allow coating and compaction of the genomic RNA followed by proteolysis of the MA/CA junction. The released MA forms trimers that line the inner lipid membrane and CA assembles into hexamers via the N-terminal domains, which form the viral core [4]. Finally, peptides are removed from the C-termini of CA and NC [22, 23]. The sequential cleavage of proteins allows for distinct protein interactions that can facilitate both particle assembly and particle dissociation at temporally defined steps of the virus life cycle.
In this study, we evaluate how predicted biochemical properties and structure of Gag proteins and peptides have evolved in FIV derived from different host species. Feline immunodeficiency viruses infect many members in the family Felidae [24, 25] and like primate immunodeficiency viruses, the co-evolutionary histories and clinical consequences of infection vary in each species [26–28]. During evolution of the virus in each species of host feline, proteins may change by drift or they may derive new characteristics that confer greater fitness in the host environment due to positive selection. In contrast, sequences in which any change decreases viral fitness will be constrained by purifying or negative selection. Evaluating the biochemical properties of proteins in an evolutionary context provides insights on structural constraints on viral proteins and highlights regions of proteins in which different structural or biochemical, and hence potential functional, properties have evolved in each virus family.
Materials and methods
Samples and nucleotide sequences
Cougar (Puma concolor) PBMC and LN samples were prepared as described by Biek et al. [26, 29]. The 5′ half of FIVpco genomes were amplified by nested PCR from genomic DNA using high-fidelity ExTaq polymerase (Takara), cloned, and sequenced as described [30]. First-round primers were 5oLTF (TAACCGTAAACCGCAAGTG) and 5oPCLR (TAGCCATAAGTTCCCATAAGG) and second-round primers were 5iXhF (ACTCGAGTGCTTGCATGCAAGAAATGACG) and 5iNotR (ATGCGGCCGCTGTTGATTCCACTC). The underlined regions indicate restriction sites for XhoI and NotI, respectively, which were incorporated to facilitate cloning of longer PCR fragments. Conditions for the first round amplification were 94°C for 3 min, then 35 cycles of 94°C for 30 s, 52°C for 30 s, and extension at 70°C for 5 min. Second-round PCR conditions were identical except that annealing occurred at 54°C. Second-round PCR products were digested with XhoI and NotI cloned into pBluescript (Stratagene, USA), which was cut with the same enzymes. PBMC and serum from infected African lions (Panthera leo) were kindly provided by Dr. C. Packer (University of Minnesota). Primers used to amplify FIVPleB614 from genomic DNA were 5iPleF (CTTCACGGATCTTCAAGCCAGG ) and sPle2560R (CTCTCCTGGTCCTTGATTATTGA). Genomic DNA was amplified using the following conditions: 94°C for 3 min, then 35 cycles of 94°C for 30 s, 54°C for 30 s, and extension at 70°C for 2 min. PCR products were cloned into the pDrive vector (Qiagen, USA) using standard protocols and sequenced. TA cloning strategies were used for FIVple fragments because there was insufficient sequence information to design specific sites for cloning into the primers. Sequence from lion PleC914 was obtained from plasma. Viral RNA was isolated using the QIAamp viral RNA kit (Qiagen, USA), cDNA was prepared using Ple46760R (TGGTCCATTATCWGWTTGTA) [31] and PCR amplification used this primer with 384F (CCGAACAGGGACTTGAA) for first round amplification. Conditions were: 94°C for 3 min, then 40 cycles of 94°C for 30s, 51°C for 30 s, and extension at 70°C for 4 min. Second-round PCR was conducted with 5iPleF and PleC3350R (AAAGCATCTCCCTAAGTTCATT) using identical conditions except that extension time was reduced to 2 min. Other sequences used for analyses were acquired from Genbank and limited to only those with full-length genomic sequences. FIVfca sequences from domestic cat (Felis catus) were representative of clade A: FIVCG (M25381), FIV-Z1 (X57002), FIV-PPR (M36968); clade B: FIV-GVEPX (E03581), FIV-USIL2489 (U11820), and clade C: FIV-C (AF474246), FIV-C36 (AY600517). Additional non-domestic cat FIV sequences came from a Florida Panther (Puma concolor coryi) PLV-14 (U03982) [32], a cougar from Vancouver Island BC, PLV (DQ192583) [30] and a Pallas’ cat (Otocolobus manul) OMA (AY713445) [33].
Protein structural analysis
Computational analyses were used to predict chemical and physical properties of translated gag sequences using a number of public servers. Molecular weight and pI were calculated using the EXPASY Compute pI/MW tool [34]. The EXPASY PROTSCALE [34] server was used to determine hydrophobicity [35] and flexibility [36] indices via a sliding window analysis. Sliding window analysis for charge was performed using the CHARGE program in EMBOSS (v2.8.0) [37]. Sliding window of average identity of amino acids within a window size of ten amino acids was performed using values calculated by the SWAAP program [38]. All other sliding window analyses used a window size of nine residues. Raw data output from these analyses was compiled and data points were correlated to residues in the multiple sequence alignment. Secondary structure predictions were performed using the PSIPRED server [39] because this algorithm was the most consistent of those evaluated (JPRED, PHD, PREDATOR, Z-PRED) for predicting the secondary structural elements identified in solved crystal structures. Prediction of disordered regions within proteins using the DISOPRED server [40]. Pattern recognition of conserved and repeated sequences was performed using the EMBL PRATT server [41].
Phylogenetic and selection analyses
To determine the phylogenetic relationships of the approximately 1,500 bp gag gene, a maximum likelihood (ML) tree was created in PAUP* (4.10b; Sinauer Associates, Inc.) [42], using a GTR + G model as determined in Modeltest [43]. Support for topology was determined using 100 bootstrap replicates with a heuristic search algorithm. All main nodes had bootstrap values of 90–100.
To determine sites under positive selection, five models of codon-based substitution available in the CODEML module of PAML [44, 45] were considered that allowed different values for the parameter ω, the ratio of the rates of non-synonymous and synonymous substitutions. A likelihood ratio test with the Bonferroni correction for multiple tests was used to determine the model that best fit the data. Model M0 (single value of ω) was contrasted with M1 (ω < 1, ω = 1), M1 was compared to M2, which allows ω > 1). M7 and M8 have values of ω comparable to those in M1 and M2 and include a beta distribution to model the frequency of sites with ω ≤ 1. The Bayes empirical Bayes (BEB) calculation of posterior probabilities for site classes was used to calculate the probabilities of sites under positive selection [45]. This analysis was performed for the entire dataset and for the FIVpco and FIVfca sets individually.
The sequence alignments and trees were also submitted to the Hyphy package available from DataMonkey [46, 47]. Both SLAC (single-likelihood ancestor counting) and FEL (fixed effects likelihood) analyses were conducted. Data on purifying selection is based on FEL analyses using a value of P < 0.05 to establish significance.
Structural models
Homology models of proteins were based on Gc34 Gag proteins and were produced using the SCWRL server [48]. The Gc34 Gag sequence was used because it is the closest sequence to the FIVpco consensus and is therefore a good representative of this group. Potential structural homologues were determined using the FUGUE sequence-structure homology prediction server [49]. For use in homology models, optimal structural templates with highest Z-score from FUGUE analysis were selected based on sequence similarity to FIV proteins, size of the protein sequence, and structure resolution. Structures for Equine infectious anemia virus (EIAV) capsid (2EIA) [2] and Human immunodeficiency virus type 1 (HIV-1) nucleocapsid (1A1T) [50] proteins were used to produce homology models. As sequence identity of Gc34 MA was low compared to sequences from solved structures, the PRALINE profile alignment program [51] was used to align and map to analogous positions on protein EIAV matrix structure (1HEK) [52]. Model visualization, annotation, and generation of figures was performed using the PyMOL open source software [53].
Results
Phylogenetic relationship of the FIV gag gene
The gag gene of FIV varies in length from an average of 1350 bp in FIVfca and 1386 bp in FIVpco to as long as 1506 bp in FIVple. Phylogenetic reconstruction of the gag sequences demonstrates that there are three principal groups, which cluster according to host species (Fig. 1). The topology of taxa within each species-specific cluster is consistent with the clade structure previously identified for each of FIV [26, 29, 54, 55] (Fig. 1). There was phylogenetic support for inclusion of the Pallas’ cat sequence (OMA) in the lion lentivirus clade, as was previously shown for phylogenies based on pol gene sequences [25]. As a result, the OMA gag sequence was included in the FIVple group for analysis of protein structural properties.
Maximum likelihood tree of FIV gag nucleotide sequences. Sequences used in this study were bootstrapped with 100 trials and significant (90–100%) branch support values are shown. (a) Inset describes the phylogenetic nucleotide divergence for intra- and inter-group comparisons of FIVs (cougar (FIVpco), domestic cat (FIVfca), and African lion (FIVple)
The overall nucleotide identity within the FIVfca gag sequences is 93.6% (range 90.3–99.8%). There is greater diversity within FIVpco sequences than in FIVfca; nucleotide identity between FIVpco gag ranges from 61.8% to 99.8% and has an average of 89.7% (Fig. 1a). The sequences from FIVpleB614 and FIVpleC914 share only 71% nucleotide identity. OMA gag is 80% identical to that of FIVpleB614. Between groups, FIVpco gag shares 56–58% nucleotide identity with both FIVfca and FIVple while FIVfca and FIVple are 66.8% identical. The predicted intra-group Gag protein identity is 97%, 94%, and 81% for FIVfca, FIVpco, and FIVple, respectively. FIVpco Gag proteins are 70.3% and 73.3% identical to FIVple and FIVfca Gag proteins, respectively and FIVple and FIVfca share 75.8% identity. Thus, FIV Gag proteins have diversified by 25–30% as these viruses have evolved in each host species.
Predicted biochemical properties of matrix (MA)
The FIV Gag polyprotein is proteolytically cleaved to produce three structural proteins, matrix (MA), capsid (CA), and nucleocapsid (NC). Position of proteolytic cleavage sites in each sequence was based on sequence homology to those determined for FIVCG [56] (Fig. 2). The Tyr-Pro residues that mark the junction of MA and CA in FIVCG are conserved in all FIVfca and FIVple sequences except FIVpleC914, which has an Asn-Pro at the putative cleavage site. All FIVpco have Ala-Pro except for PLV-14, which has the Tyr-Pro sequence typical of the FIVfca clade. The predicted molecular masses of MA from FIVfca and FIVpco are similar at 14.5–14.6 kDa (Table 1). The MA from FIVple viruses possess an insertion of between 11 and 20 amino acids at the C-terminus of MA relative to the other FIV. Thus, the predicted molecular mass of FIVple MA is 15.6–17.5 kDa (Table 1).
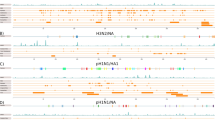
Schematic diagrams of FIV Gag proteins from: cougar (FIVpco), domestic cat (FIVfca), and African lion (FIVple). (a) Bar diagram indicates position of codons identified to be under positive selection (full bars) for all FIVs (full black bars; codon 404: PAML: P < 0.05; FEL: P < 0.007; codon 471: FEL: P < 0.001) and for FIVpco sequences (full gray bars; codon 399 PAML: P < 0.03), and half black bars represent sites that are invariant. (b) Percent identity sliding window plot for all FIV sequences. (c) Each FIV group displays: schematic diagram of the relative lengths of the Gag polyproteins and their subsequent products with breaks in blocks representing gaps for optimal sequence alignment (myristoylation of N-terminal Met is indicated by “Myr”) and majority consensus predicted secondary structures of each subunit protein are shown, where, lines represent unstructured regions, cylinders represent α-helices, and arrows represent β-sheets
Four of the five α-helices (H1 and H3–5) represented in the HIV-1 MA structure [52] are predicted to occur in FIV MA. Secondary structure predictions support the presence of α-helix 2 (H2) in all FIVpco MA, but this region is predicted to be a β-sheet (S1) in both FIVfca and FIVple MA proteins (Fig. 2). The secondary structural prediction confidence of the β-sheet for FIVfca and FIVple MA is low, but this region of the protein is not predicted to be disordered in any sequence (data not shown). It is notable that the hydrophobicity index for H2 from FIVpco MA is higher than for the other FIV (Fig. 3a) because H2 contains important determinants for membrane association [57]. Six of the seven N-terminal amino acids of H2/S1 are under purifying selection in all FIVs but the C-terminal region is neutrally evolving (Fig. 4a). Thus, there is no evidence that any functional difference associated with different intrinsic hydrophobicity of H2 in FIV MA is currently under selective pressure.
Secondary structure and graphical representation of sliding window analysis of (a) MA and (b) CA proteins, and (c) NC proteins. Secondary structures represent majority consensus structures for each FIV group (cougar (FIVpco), domestic cat (FIVfca), and African lion (FIVple) where cylinders represent α-helices and arrows represent β-sheets. Panels show sliding window analysis of: charge, hydrophobicity, and flexibility. Gaps were introduced for alignment of peaks according to multiple sequence alignment. Dashed boxes indicate region of inset and insets show values with standard error bars
Homology models of the (a) matrix (MA), (b) capsid (CA), and (c) nucleocapsid (NC) proteins. On the blue homology model structure, yellow residues indicate negatively selected sites and red residues indicate invariant sites for the FIVs (cougar (FIVpco), domestic cat (FIVfca), and African lion (FIVple). (a) Selected sites for FIVs relative to the EIAV MA protein crystal structure (1HEK). Shown are model images rotated approximately 180° about the Y-axis. (b). Homology model of Gc34 using the EIAV CA crystal structure (2EIA) shown in two orientations 180° about the Y-axis. C-terminal and N-terminal domains (NTD & CTD, respectively) are delineated by dashed line. (c) Gc34 homology model for stemloop-bound conformation of NC using the NMR structure from HIV-1 NC (1A1T) two coordinated zinc ions are represented by white spheres and the cyan residues denoted by asterisks (*) indicate sites determined to be under positive selection for all FIV sequences by FEL and PAML analyses. Note: one residue under positive selection by FEL analysis in the C-terminal region of NC is not indicated as model template does not extend to this residue. For all models, structural elements on the models highlighted by italicized and red outlined text denote regions discussed in text and boxed regions in Fig. 2
The pI of MA (Table 1) ranged from 8.3 to 9.5 in FIVpco and FIVfca, and 9.0–9.4 in FIVple indicating that areas of localized charge differences exist within and between FIV groups. Because charge is important for intra- and inter-molecular interactions, FIV MA proteins were evaluated for charge distribution using a sliding window analysis. Overall, the MA proteins of all FIV groups consist of positively charged N- and C- termini and a negatively charged central H3. The N-terminal region is basic in all FIV. There are four conserved acidic residues in H3, which contribute to the net negative charge of this helix. The charged residues at either end of H3 are evolving neutrally and the two in the central portion of the helix are under purifying selection in all FIV. FIVpco sequences have three additional acidic residues in H3 that are all under purifying selection. Thus, FIVpco MA sequences are more negatively charged in H3 than other FIV (Fig. 3a). FIVfca and FIVple sequences have acidic H5 and FIVpco sequences vary in this region. However, the long C-terminal extension characteristic of FIVple MA sequences carries a net positive charge, which contributes to the more basic pI of MA from this group. There was no evidence for positive selection on any sites associated with charge differences in FIV MA.
Myristoylation of the N-terminus of retrovirus MA proteins is important for membrane localization [58] but some lentiviruses, such as EIAV, Visna/Maedi virus, and Caprine arthritis-encephalitis virus (CAEV) do not require myristoylated MA for assembly [59, 60]. FIVCG MA has been previously reported to be myristoylated [56] and all FIV MA proteins evaluated in this study contain similar recognition sites for myristoylation (Fig. 2).
Predicted biochemical properties of capsid (CA)
The capsid proteins of these three groups of FIV have similar length and predicted secondary structural elements. The C-terminal cleavage site of capsid (Val-Gln) is present in OMA and all FIVfca and FIVpco sequences but is absent from the two FIVple. The approximate C-terminus of FIVple capsid was estimated from the alignment, which is conserved except for the last five amino acids surrounding the putative cleavage site. Molecular masses of FIVpco, FIVfca, and FIVple CA proteins are approximately 25.2, 24.7, and 25.3 kDa, respectively (Table 1).
There are nine α-helices and one short β-sheet (Figs. 2, 3b) predicted for each FIV CA protein. Based on the crystal structure of CA from HIV-1, these secondary structural elements are arranged in two distinct domains that are delineated by the predicted β-sheet [61–63]. The N-terminal domain (NTD) forms the surface of the core particle and encompasses predicted helices 1–5 and a proline-rich loop structure situated between H4 and H5. We note that the two small helices in the HIV-1 structure, H5 and H6, were not predicted for any of the FIV CA. Thus H5 in FIV CA is equivalent to H7 in the HIV-1 CA structure. The first three helices (H1-H3) share common properties for charge, hydrophobicity, and flexibility; 70% of these residues are under purifying selection. The central portion of H5 from FIVpco CA is predicted to be less hydrophobic and more flexible than that of FIVfca or FIVple H5 (Fig. 3b).
The C-terminal domain (CTD) is responsible for Gag multimerization and is comprised of helices 6–9 (corresponding to H8–11 in HIV-1). The region spanning the C-terminus of H8 through H9 from FIVpco CA proteins is predicted to have lower hydrophobicity than the other FIVs. FIVple and FIVpco H7 have greater flexibility than FIVfca H7 (Fig. 3b).
The majority of sites in CA from the three groups of FIV are under purifying selection, which is consistent with the observed conservation of sequence and biochemical properties. Although selection against changes in amino acid sequence is anticipated for key structural elements, it is of interest that the loop region between H4 and H5 is also under purifying selection (Fig. 4b). This region is highly flexible and disordered and shows absolute conservation of a PRPLPY motif.
Predicted biochemical properties of nucleocapsid (NC)
The molecular mass of FIV NC ranges from 7 kDa to 9 kDa (Table 1). Variation in mass is due principally to length differences in the N- and C- terminal domains. The FIVple and FIVpco N-terminal domains are basic and range in length from 15 to 34 amino acids. There are two sites in this region predicted to be under positive selection in FIVpco sequences (Figs. 2, 4c). The N-terminal domain of FIVfca is only nine amino acids long, is not basic, and has no sites under positive selection. The longest N-terminal domain is in the PLV-14 NC protein. This sequence has an insertion of approximately 20 amino acids consisting of Arg-Gly repeat [32] that is unique among FIV NC.
The central region of FIV NC consists of two CCHC zinc-finger (ZF; C–X2–C–X4–H–X4–C) motifs (Figs. 2, 4c) that are both under purifying selection. The five amino acid linker, which separates the two zinc-finger motifs, has lower sequence homology but maintains a conserved motif consisting of a hydrophobic residue flanked by charged residues. In FIVple and FIVpco, all charged residues are basic and there is a conserved Pro central in the sequence whereas FIVfca lacks the Pro and there is an acidic residue N-terminal of the central hydrophobic residue (Fig. 3c).
The C-terminus of NC following the second zinc-finger motif has the lowest inter-feline group sequence identity of any region of Gag (Fig. 2). However, within each FIV group, the identity is high. In FIVfca NC, the region from the C-terminal end of ZF2 to the end of NC is basic whereas both FIVple and FIVpco NC are acidic to neutral in this region. The Val-Gln-Gln cleavage site at the end of NC is conserved in all sequences examined except for FIVCG, which has Met-Gln-Gln.
Predicted biochemical properties of the C-terminal peptide (p2C)
The C-terminal peptide produced from proteolytic cleavage of Gag is approximately 19 amino acids, or 2.2 kDa (p2C). Although this peptide is smaller than the corresponding p6 peptide from primate lentiviruses, the FIV p2C peptides share two motifs that are found in the primate p6 and that are under purifying selection. The PS/TAPP motif, which is involved in endocytic pathway sorting, is present in all FIV sequences. The C-terminal LXXL motif is associated with clathrin binding [64] and is conserved in FIVpco and FIVfca. FIVple motifs at this site are variable and consist of LXXI/M. The PLV-14 p2C sequence is truncated before the LXXL motif. However, the N-terminus of the peptide is approximately eight residues longer than that of the other FIVpco sequences and contains a YPXL motif analogous to that described for the EIAV p9 peptide [65]. The OMA and FIVpleB614 p2C peptides also are longer in the N-terminus of p2C but these extended domains do not contain any characterized motifs.
Discussion
The feline immunodeficiency viruses form a monophyletic group within the lentiviruses. Our phylogenetic analysis demonstrates that there is greater nucleotide diversity within the cougar (FIVpco) and lion (FIVple) lineages than within FIVfca from domestic cats. In addition, 200 of the 515 sites are under purifying selection across the FIVpco gag gene but there are only 78 sites under purifying selection in FIVfca gag. The greater diversity and number of sites under purifying selection in FIVpco are consistent with a longer evolutionary history of FIV with wild felid hosts [29, 54].
We evaluated predicted secondary structure and charge, hydrophobicity, and flexibility profiles for each of the Gag proteins from the different FIV. The variation for each of these predictions was low within each virus group and, in general, there was also substantial conservation of biochemical features between virus groups. However, we identified several regions that differed among FIV groups due to charge, hydrophobicity or flexibility on a segment, or that conferred a unique property on a protein due to length variation.
Charge differences were detected in the third and fifth helices and C-terminal extension of matrix. H3 is responsible for precise presentation of the trimerization domain of matrix and affect particle formation [4, 66, 67]. In HIV-1 MA, the loop between H3 and H4 is a hinge that facilitates displacement of the intervening region upon trimerization [68]. The differences in charge associated with H3 and loop among the FIVs may affect the nature of intermolecular interactions and subsequently impact either particle assembly or disassembly. In the immature Gag polyprotein, H5 and C-terminal sequences of MA form a random coil [69] that acts as a flexible linker that tethers MA to CA. This C-terminal extension of MA can accommodate mutations [70] and insertions such as those in FIVple without significantly impacting infectivity or assembly [71, 72].
The matrix protein is a globular protein stabilized by extensive hydrophobic interactions. The H2 of MA forms one edge of the protein and makes hydrophobic contacts with other helices [7]. There are also basic residues on the outer surface of this helix that may facilitate interaction with the host membrane [73]. Whereas the charge profile in H2 is similar among the FIV and three of the four basic residues are under purifying selection, the H2 of FIVpco is predicted to be more hydrophobic. Further, the secondary structure prediction for H2 differs among the FIV MA. Thus, if there is a functional consequence of H2 structure or hydrophobicity, it is likely to be on intramolecular contacts within FIVpco MA.
The charge profiles of all FIV capsid proteins were similar. Differences between FIV CA involved a decrease in hydrophobicity and an increase in flexibility of H5 (H7 in the HIV-1 crystal), flexibility differences in H7 (H9 in the HIV-1 crystal) and decreased hydrophobicity in the C-terminal helix. These differences are of interest because capsid proteins undergo conformational changes following proteolytic cleavage in the newly released virus particle giving the virus core its characteristic morphology. The rearrangements that occur after proteolytic processing of HIV-1 CA do not involve movement of either H7 or adjacent H6 [69, 74]. However, H7 and H2 form antiparallel four-helix bundles at the dimer interface in EIAV capsid crystals [2], suggesting that movement of H7 in the NTD may be associated with maturation in some lentiviruses. Similarly, greater flexibility is predicted for FIVpco in the CTD helices, which are involved in capsid dimerization in HIV-1 [62]. Thus, differences in both MA and CA among the FIV can potentially influence restructuring of the molecules that occur during maturation in the virus particle due to changes in charge or hydrophobic interactions or flexibility.
There were notable charge differences in both N- and C-terminal domains of nucleocapsid from the FIV. Both FIVpco and FIVple sequences contain long N-terminal regions with four to six basic residues reminiscent of HIV-1 NC. In contrast the N-terminal region of FIVfca is shorter on average by 10 residues and has only one basic residue. However, unlike the other FIV, FIVfca NC carries a net positive charge at the C-terminal end of the second Zn finger. The basic residues in NC are important for its multiple roles as an RNA binding protein. To package the viral genome, the N-terminal basic residues in HIV-1 form electrostatic and hydrogen bonds with the phosphodiester backbone in the major groove of the stem-loop of the Ψ-packaging sequence and the zinc-fingers bind to the exposed based of the RNA tetraloop [50]. Removal of basic residues significantly reduces RNA encapsidation [75–77], whereas increasing the positive charge in this region decreases selectivity of NC binding to viral RNA [78]. PLV-14, which is the most divergent of the FIVpco, possesses an N-terminal insertion in NC consisting of a Gly-Arg repeat sequence [32]. Although this is unlike any sequence found in other lentiviruses, a similar domain exists in the nucleic acid binding domain of NC for spumaretroviruses [79]. In addition to the role in RNA binding, the N-terminal region of NC adjacent to the first zinc-finger motif binds several cellular components, including a cytidine deaminase (APOBEC3G) [19]. It is notable that two of the sites predicted to be under positive selection in FIVpco are in this multi-functional N-terminal region. Thus, differences in the N-terminus of NC may dictate different strategies for encapsidation of the viral genome or interaction with cellular proteins among the FIV.
The diversity within gag sequences in these FIV groups also provides an opportunity to assess the structural constraints imposed on Gag proteins that have different evolutionary histories in each host species. For example, the loop between H2 and H3 in MA forms part of the trimer interface [4, 80]. Residues in this region are absolutely conserved among the FIV. Our data suggests there are limited opportunities to change sequence of CA because not only are predicted biochemical properties largely conserved, over half of the codons in FIVpco are under purifying selection. In particular, the surface of hexameric rings formed by α-helices 1 and 2 and the CypA binding loop is essentially identical among the FIV. These data indicate that the structure of the capsid is likely to be similar among the FIV but, as discussed above, the transition upon maturation and intramolecular packing may be slightly different.
In addition to conservation of structural faces of capsid, there is also notable conservation of key residues that have been identified in studies of other retroviruses. For example, upon cleavage from MA, CA refolds to present an N-terminal β-hairpin structure of the NTD that forms CA-CA contacts in the viral core. This interface is stabilized by a salt bridge between Pro 1 and Asp 51 in HIV-1 [61, 63]. There is a Pro and Asp at the corresponding sites in all FIV CA, which are under purifying selection, suggesting that these residues are also critical to stabilize the maturing capsid structures in FIV.
Although nucleocapsid is the most divergent of the three FIV Gag proteins, it retains the two zinc-finger motif found in other lentivirus NC. The key Cys and His residues, which coordinate the zinc ions within the zinc-fingers, are conserved and under purifying selection. Mutational studies have shown that the zinc-fingers are required for nucleic acid binding but can be functionally replaced by a cluster of basic residues [81]. It is of interest that some sites that appear to have a function dictated by a general biochemical property, such as charge, are absolutely constrained to specific amino acids in this group of lentiviruses because properties of a protein can also be retained by neutrally evolving sites.
Gag proteins also interface with host cellular proteins. Several motifs that are responsible for these interactions are present and highly conserved in FIV Gag proteins. The cellular peptidylprolyl isomerase A, cyclophilin A (CypA), is incorporated into HIV-1 particles [82] via interaction with a proline residue in the loop between H5 and H6 of HIV-1 CA [61, 82–84]. Although all FIVs possess conserved Pro residues within this proline-rich loop that are under purifying selection, interaction of non-primate lentivirus CA with CypA has not yet been investigated. However, cyclosporin is a potent antiviral agent for FIV [85], suggesting that CypA plays a role in the FIV life cycle. This may relate to a newly described function of CypA in primate lentiviruses, which is to inhibit binding of post-entry primate host restriction factors, such as TRIM-5α, Ref-1, and Lv1 [86–88].
Despite extensive variability in the C-terminal peptide (p2C) of Gag, there are two motifs that are conserved among the FIV. All FIV have a PT/SAPP domain, which is present in all lentiviruses except EIAV. The cellular protein, Tsg101, binds to this motif [21, 89], is responsible for recognition and sorting via the cellular endosomal [20, 90] and is required for virus release or “pinching off”. A second conserved motif located at the C-terminus of the peptide is the LXXL motif, which also exists in most lentiviruses. LXXL motifs are required for viral budding via interaction with clathrin adaptor proteins [reviewed in 91]. However, PLV-14 has a YXXL motif, which is equivalent to an L domain used by EIAV [92] that is upstream of the PT/SAPP domain. L domains of diverse retroviruses have been shown to be functionally interchangeable and positionally independent [93, 94].
References
C.P. Hill, D. Worthylake, D.P. Bancroft, A.M. Christensen, W.I. Sundquist, Proc. Natl. Acad. Sci. USA 93, 3099 (1996)
Z. Jin, L. Jin, D.L. Peterson, C.L. Lawson, J. Mol. Biol. 286, 83 (1999)
S. Khorasanizadeh, R. Campos-Olivas, M.F. Summers, J. Mol. Biol. 291, 491 (1999)
Z. Rao, A.S. Belyaev, E. Fry, P. Roy, I.M. Jones, D.I. Stuart, Nature 378, 743 (1995)
N. Riffel, K. Harlos, O. Iourin, Z. Rao, A. Kingsman, D. Stuart, E. Fry, Structure 10, 1627 (2002)
B.G. Turner, M.F. Summers, J. Mol. Biol. 285, 1 (1999)
M.R. Conte, M. Klikova, E. Hunter, T. Ruml, S. Matthews, Embo. J. 16, 5819 (1997)
S.S., Rhee, E. Hunter, Cell 63, 77 (1990)
M.F. Verderame, T.D. Nelle, J.W. Wills, J. Virol. 70, 2664 (1996)
S.A. Gonzalez, J.L. Affranchino, H.R. Gelderblom, A. Burny, Virology 194, 548 (1993)
E.O. Freed, J.M. Orenstein, A.J. Buckler-White, M.A. Martin, J. Virol. 68, 5311 (1994)
T. Dorfman, F. Mammano, W.A. Haseltine, H.G. Gottlinger, J. Virol. 68, 1689 (1994)
M.V. Nermut, D.J. Hockley, J.B. Jowett, I.M. Jones, M. Garreau, D. Thomas, Virology 198, 288 (1994)
D.T. Poon, G. Li, A. Aldovini, J. Virol. 72, 1983 (1998)
S. Baba, K. Takahashi, Y. Nomura, S. Noguchi, Y. Koyanagi, N. Yamamoto, H. Takaku, G. Kawai, Nucleic Acids Res Suppl, 155 (2001)
M.J. Rist, J.P. Marino, Biochemistry 41, 14762 (2002)
R. Berkowitz, J. Fisher, S.P. Goff, Curr. Top. Microbiol. Immunol. 214, 177 (1996)
K. Luo, B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, X.F. Yu, J. Virol. 78, 11841 (2004)
T.M. Alce, W. Popik, J. Biol. Chem. 279, 34083 (2004)
J.E. Garrus, U.K. von Schwedler, O.W. Pornillos, S.G. Morham, K.H. Zavitz, H.E. Wang, D.A. Wettstein, K.M. Stray, M. Cote, R.L. Rich, D.G. Myszka, W.I. Sundquist, Cell 107, 55 (2001)
L. VerPlank, F. Bouamr, T.J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, C.A. Carter, Proc. Natl. Acad. Sci. USA 98, 7724 (2001)
S.C. Pettit, J.N. Lindquist, A.H. Kaplan, R. Swanstrom, Retrovirology 2, 66 (2005)
K. Wiegers, G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, H.G. Krausslich, J. Virol. 72, 2846 (1998)
M.A. Carpenter, E.W. Brown, M. Culver, W.E. Johnson, J. Pecon-Slattery, D. Brousset, S.J. O’Brien, J. Virol. 70, 6682 (1996)
J.L. Troyer, J. Pecon-Slattery, M.E. Roelke, W. Johnson, S. VandeWoude, N. Vazquez-Salat, M. Brown, L. Frank, R. Woodroffe, C. Winterbach, H. Winterbach, G. Hemson, M. Bush, K.A. Alexander, E. Revilla, S.J. O’Brien, J. Virol. 79, 8282 (2005)
R. Biek, T.K. Ruth, K.M. Murphy, C.R. Anderson Jr., M. Poss, Can. J. Zool. 84, 365 (2006)
M.J. Burkhard, G.A. Dean, Curr. HIV Res. 1, 15 (2003)
C. Packer, S. Alitzer, M. Appel, E. Brown, J. Martenson, S.J. O’Brien, M. Roelke-Parker, R. Hofmann-Lehmann, H. Lutz, J. Anim. Ecol. 68, 1161 (1999)
R. Biek, A.G. Rodrigo, D. Holley, A. Drummond, C.R. Anderson Jr., H.A. Ross, M. Poss, J. Virol. 77, 9578 (2003)
M. Poss, H.A. Ross, S.L. Painter, D.C. Holley, J.A. Terwee, S. Vandewoude, A. Rodrigo, J. Virol. 80, 2728 (2006)
J.L. Troyer, J. Pecon-Slattery, M.E. Roelke, L. Black, C. Packer, S.J. O’Brien, J. Virol. 78, 3777 (2004)
R.J. Langley, V.M. Hirsch, S.J. O’Brien, D. Adger-Johnson, R.M. Goeken, R.A. Olmsted, Virology 202, 853 (1994)
M.C. Barr, L. Zou, F. Long, W.A. Hoose, R.J. Avery, Virology 228, 84 (1997)
E. Gasteiger, C. Hoogland, A. Gattiker, S. Duvaud, M.R. Wilkins, R.D. Appel, A. Bairoch, in The Proteomics Protocols Handbook, ed. by J.M. Walker (Human Press, Totowa, NJ, USA, 2005), 571 pp
S.D. Black, D.R. Mould, Anal. Biochem. 193, 72 (1991)
R. Bhaskaran, P.K. Ponnuswamy, Int. J. Peptide Protein Res. 32, 242 (1988)
P. Rice, I. Longden, A. Bleasby, Trends Genet. 16, 276 (2000)
D.T. Pride, M.J. Blaser, Genome Lett. 1, 2 (2002)
L.J. McGuffin, K. Bryson, D.T. Jones, Bioinformatics 16, 404 (2000)
J.J. Ward, J.S. Sodhi, L.J. McGuffin, B.F. Buxton, D.T. Jones, J. Mol. Biol. 337, 635 (2004)
I. Jonassen, Comput. Appl. Biosci. 13, 509 (1997)
D.L. Swofford. PAUP* 4.0b10: phylogenetic analysis using parsimony (and other models) (Sinauer Associates, Sunderland, Mass., 2002)
D. Posada, K.A. Crandall, Bioinformatics 14, 817 (1998)
Z. Yang, Comp. Appl. Biosci. 13, 555 (1997)
Z. Yang, W.S. Wong, R. Nielsen, Mol. Biol. Evol. 22, 1107 (2005)
S.L. Pond, S.D. Frost, Bioinformatics 21, 2531 (2005)
S. Pond, S. Frost, Mol. Biol. Evol. 22, 1208 (2005)
A.A. Canutescu, A.A. Shelenkov, R.L. Dunbrack Jr., Protein Sci. 12, 2001 (2003)
J. Shi, T.L. Blundell, K. Mizuguchi, J. Mol. Biol. 310, 243 (2001)
R.N. De Guzman, Z.R. Wu, C.C. Stalling, L. Pappalardo, P.N. Borer, M.F. Summers, Science 279, 384 (1998)
V.A. Simossis, J. Heringa, Nucleic Acids Res. 33, W289 (2005)
H. Hatanaka, O. Iourin, Z. Rao, E. Fry, A. Kingsman, D.I. Stuart, J. Virol. 76, 1876 (2002)
W.L. DeLano, The PyMol Molecular Graphics System (DeLano Scientific, San Carlos, CA, USA, 2002)
M.A. Carpenter, E.W. Brown, D.W. MacDonald, S.J. O’Brien, Virology 251, 234 (1998)
E.W. Brown, N. Yuhki, C. Packer, S.J. O’Brien, J. Virol. 68, 5953 (1994)
J.H. Elder, M. Schnolzer, C.S. Hasselkus-Light, M. Henson, D.A. Lerner, T.R. Phillips, P.C. Wagaman, S.B. Kent, J. Virol. 67, 1869 (1993)
P. Spearman, R. Horton, L. Ratner, I. Kuli-Zade, J. Virol. 71, 6582 (1997)
M. Bryant, L. Ratner, Proc. Natl. Acad. Sci. USA 87, 523 (1990)
L.E. Henderson, R.C. Sowder, G.W. Smythers, S. Oroszlan, J. Virol. 61, 1116 (1987)
A.M. Schultz, L.E. Henderson, S. Oroszlan, Annu. Rev. Cell Biol. 4, 611 (1988)
T.R. Gamble, F.F. Vajdos, S. Yoo, D.K. Worthylake, M. Houseweart, W.I. Sundquist, C.P. Hill, Cell 87, 1285 (1996)
T.R. Gamble, S. Yoo, F.F. Vajdos, U.K. von Schwedler, D.K. Worthylake, H. Wang, J.P. McCutcheon, W.I. Sundquist, C.P. Hill, Science 278, 849 (1997)
R.K. Gitti, B.M. Lee, J. Walker, M.F. Summers, S. Yoo, W.I. Sundquist, Science 273, 231 (1996)
A.R. Ramjaun, P.S. McPherson, J. Neurochem. 70, 2369 (1998)
B.A. Puffer, S.C. Watkins, R.C. Montelaro, J. Virol. 72, 10218 (1998)
M.L. Manrique, C.C. Celma, S.A. Gonzalez, J.L. Affranchino, Virus Res. 76, 103 (2001)
Y. Morikawa, T. Kishi, W.H. Zhang, M.V. Nermut, D.J. Hockley, I.M. Jones, J. Virol. 69, 4519 (1995)
M.A. Massiah, D. Worthylake, A.M. Christensen, W.I. Sundquist, C.P. Hill, M.F. Summers, Protein Sci. 5, 2391 (1996)
C. Tang, Y. Ndassa, M.F. Summers, Nat. Struct. Biol. 9, 537 (2002)
P.M. Cannon, S. Matthews, N. Clark, E.D. Byles, O. Iourin, D.J. Hockley, S.M. Kingsman, A.J. Kingsman, J. Virol. 71, 3474 (1997)
B. Muller, J. Daecke, O.T. Fackler, M.T. Dittmar, H. Zentgraf, H.G. Krausslich, J. Virol. 78, 10803 (2004)
M.R. Auerbach, C. Shu, A. Kaplan, I.R. Singh, Proc. Natl. Acad. Sci. USA 100, 11678 (2003)
P.S. Murray, Z. Li, J. Wang, C.L. Tang, B. Honig, D. Murray, Structure 13, 1521 (2005)
U.K. von Schwedler, T.L. Stemmler, V.Y. Klishko, S. Li, K.H. Albertine, D.R. Davis, W.I. Sundquist, Embo. J. 17, 1555 (1998)
T.L. South, M.F. Summers, Protein Sci. 2, 3 (1993)
L. Berthoux, C. Pechoux, M. Ottmann, G. Morel, J.L. Darlix, J. Virol. 71, 6973 (1997)
J. Dannull, A. Surovoy, G. Jung, K. Moelling, Embo. J. 13, 1525 (1994)
A. Cimarelli, S. Sandin, S. Hoglund, J. Luban, J. Virol. 74, 3046 (2000)
A.W. Schliephake, A. Rethwilm, J. Virol. 68, 4946 (1994)
S. Matthews, M. Mikhailov, A. Burny, P. Roy, Embo. J. 15, 3267 (1996)
J.B. Bowzard, R.P. Bennett, N.K. Krishna, S.M. Ernst, A. Rein, J.W. Wills, J. Virol. 72, 9034 (1998)
E.K. Franke, H.E. Yuan, J. Luban, Nature 372, 359 (1994)
D.A. Bosco, D. Kern, Biochemistry 43, 6110 (2004)
J. Luban, K.L. Bossolt, E.K. Franke, G.V. Kalpana, S.P. Goff, Cell 73, 1067 (1993)
E. Mortola, Y. Endo, K. Ohno, T. Watari, H. Tsujimoto, A. Hasegawa, Vet. Res. Commun. 22, 553 (1998)
L. Berthoux, S. Sebastian, E. Sokolskaja, J. Luban, J. Virol. 78, 11739 (2004)
G.J. Towers, T. Hatziioannou, S. Cowan, S.P. Goff, J. Luban, P.D. Bieniasz, Nat. Med. 9, 1138 (2003)
L. Berthoux, S. Sebastian, E. Sokolskaja, J. Luban, Proc. Natl. Acad. Sci. USA 102, 14849 (2005)
O. Pornillos, S.L. Alam, D.R. Davis, W.I. Sundquist, Nat. Struct. Biol. 9, 812 (2002)
A. Goff, L.S. Ehrlich, S.N. Cohen, C.A. Carter, J. Virol. 77, 9173 (2003)
O. Pornillos, J.E. Garrus, W.I. Sundquist, Trends Cell Biol. 12, 569 (2002)
B.A. Puffer, L.J. Parent, J.W. Wills, R.C. Montelaro, J. Virol. 71, 6541 (1997)
G. Medina, Y. Zhang, Y. Tang, E. Gottwein, M.L. Vana, F. Bouamr, J. Leis, C.A. Carter, Traffic 6, 880 (2005)
L.J. Parent, R.P. Bennett, R.C. Craven, T.D. Nelle, N.K. Krishna, J.B. Bowzard, C.B. Wilson, B.A. Puffer, R.C. Montelaro, J.W. Wills, J. Virol. 69, 5455 (1995)
Acknowledgements
This study was funded by NIH AI054303. EB was supported by NIH P20 RR015583-06. We gratefully acknowledge contributions by Lindsey Person for data acquisition and analysis and by Kari Samuel and Sally Painter for assistance with sequence acquisition.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this paper have been submitted to the GenBank nucleotide sequence database and have been assigned the accession number EF106722–EF106737.
Rights and permissions
About this article
Cite this article
Burkala, E., Poss, M. Evolution of feline immunodeficiency virus Gag proteins. Virus Genes 35, 251–264 (2007). https://doi.org/10.1007/s11262-006-0058-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-006-0058-8